Abstract
Topoisomerase II removes supercoils and catenanes generated during DNA metabolic processes such as transcription and replication. Vertebrate cells express two genetically distinct isoforms (α and β) with similar structures and biochemical activities but different biological roles. Topoisomerase IIα is essential for cell proliferation, whereas topoisomerase IIβ is required only for aspects of nerve growth and brain development. To identify the structural features responsible for these differences, we exchanged the divergent C-terminal regions (CTRs) of the two human isoforms (α 1173-1531 and β 1186-1621) and tested the resulting hybrids for complementation of a conditional topoisomerase IIα knockout in human cells. Proliferation was fully supported by all enzymes bearing the α CTR. The α CTR also promoted chromosome binding of both enzyme cores, and was by itself chromosome-bound, suggesting a role in enzyme targeting during mitosis. In contrast, enzymes bearing the β CTR supported proliferation only rarely and when expressed at unusually high levels. A similar analysis of the divergent N-terminal regions (α 1-27 and β 1-43) revealed no role in isoform-specific functions. Our results show that it is the CTRs of human topoisomerase II that determine their isoform-specific functions in proliferating cells. They also indicate persistence of some functional redundancy between the two isoforms.
INTRODUCTION
The enzyme topoisomerase II is responsible for resolving catenanes and supercoils in chromosomal DNA that are generated during DNA metabolic processes. It plays an essential role in condensation and segregation of chromosomes at mitosis (1–3). Topoisomerase II is of considerable interest to human medicine, because it is an important target for cancer therapy (4). It is also suspected that topoisomerase II can be converted into a potent DNA toxin by minor (and as such repairable) DNA lesions frequently induced by environmental factors (e.g. UV radiation, oxidative stress) (5,6). Moreover, certain nutritional constituents (e.g. flavonoids) are known to disturb the enzyme's normal catalytic cycle (7,8), which is thought to contribute to translocations within the MLL-locus that trigger infant leukemia (9). These adverse properties of topoisomerase II could be the ultimate reason why vertebrates maintain two genetically distinct isoforms (denoted α and β) (10–14), while lower eukaryotes have only one.
The divergence of vertebrate topoisomerase II into α and β isoforms remains enigmatic, because the two enzymes are very similar in structure and function. They share a high degree of overall sequence homology with 68% identity and 86% similarity (15,16). So far, the only major in vitro difference between the two isoforms is a preferential relaxation of positive supercoils by the IIα isoform (17), whereas other basic catalytic aspects are very similar (18–21). Moreover, they have the same capacity for complementing essential topoisomerase II functions in temperature-sensitive Δtop2 yeast mutants (22,23). Despite these similarities, the two isozymes apparently play different biological roles in vertebrate cells (24). Human cell lines lacking the α isoform encounter serious problems at mitosis because chromosome segregation is deficient (25,26). For similar reasons, mouse embryos lacking the TOP2α gene, fail to develop beyond the 4–8-cell stage (27). In contrast, mammalian cell lines lacking topoisomerase IIβ pass normally through mitosis, and their most prominent phenotype is a decreased sensitivity towards topoisomerase II poisons (28–30).
These findings indicate that all essential topoisomerase II functions in cell-cycle-related events, such as DNA replication and sister chromatid segregation, can be performed by the α isozyme, while the β isozyme does not play an essential role in proliferating cells. And yet, TOP2β −/− mice are not viable. They suffocate shortly after birth due to developmental defects of motor and sensory neurons (31) and the brain (32). These defects most likely reflect a requirement of topoisomerase IIβ activity in regulating the expression of genes important at later stages of neuronal differentiation (33). This view has convincingly been confirmed by the recent finding that the β isoform plays an important role in the regulation of gene transcription, in as much as it introduces double-strand breaks at promoter regions of several genes, which are required for the proper signal-dependent activation of these genes (34). In this respect, it is of interest that topoisomerase IIβ is constitutively expressed in all cells of the mammalian organism (35), probably because expression is driven by a promoter with features characteristic of housekeeping genes (36), whereas expression of topoisomerase IIα is repressed as soon as cells stop proliferating (37,38). Therefore, the β isoenzyme is the only type II topoisomerase available in quiescent cells. In synopsis, the data available clearly suggest that the two isoforms have different biological functions in vertebrate organisms.
Here, we address the question of precisely which features render topoisomerase IIα essential for cell proliferation, and conversely, lack of which features prevents topoisomerase IIβ from adopting these functions. We have approached this problem by identifying those parts of topoisomerase IIα and IIβ that are responsible for isoform-specific functioning inside the living mammalian cell. We started from the assumption that those portions of the enzymes that are most divergent between α and β forms would be most likely to mediate isoform-specific functions. Sequence comparisons, limited proteolysis experiments and crystallographic studies suggest that topoisomerase II is composed of three major structural and functional domains (15,16,39–42). With the exception of brief stretches at the N-terminal ends (first 27 or 43 amino acids of α and β, respectively), the ATPase and the central breakage/reunion domains are similar between the two isoforms, whereas the C-terminal regions differ both in size and sequence (16). Thus, divergent and homologous portions of human topoisomerase IIα and IIβ were combined in a varied manner to form α/β-chimeric enzymes that were tested for unique, isoform-specific functions in human cells, most notably for localization of the enzymes at mitosis (43,44) and for complementation of a conditional topoisomerase IIα knockout in human cells (26).
MATERIALS AND METHODS
Alignment, plasmid construction, cell culture and live cell imaging
We aligned amino acid sequences of topoisomerase IIα and IIβ using the default settings for ClustalW v.1.83 (WWW Service at the European Bioinformatics Institute, http://www.ebi.ac.uk/clustalw) (45). Exchange of defined regions between cDNAs of human topoisomerase IIα and IIβ at the positions indicated in Figure 1B was accomplished by overlap-extension PCR (46). Fragments encompassing the divergent C-terminal regions alone were generated by PCR. Chimeric and truncated cDNAs were inserted into a bicistronic expression vector (47) used previously for stable expression of biofluorescent human topoisomerase IIα and IIβ in human cells (44). Here, the vector was modified to provide C-terminal fusion with enhanced yellow fluorescent protein (YFP). To facilitate simultaneous visualization of topoisomerase IIα and IIβ, a tricistronic expression plasmid was generated, in which topoisomerase IIα fused to CFP was placed in the first, and topoisomerase IIβ fused to YFP in the second cistron (47). A vector expressing YFP alone served as a control. All new constructs were checked by DNA sequencing. Human embryonal kidney 293 cells (# DSMZ ACC 305, German Collection of Microorganisms and Cell Culture, Braunschweig, Germany) were transfected with these constructs. Stable transgenic cell clones were selected and maintained in medium containing 0.4 µg ml−1 puromycin (details see:48). Epifluorescence microscopy was done with a Zeiss Axiovert 100 inverted light microscope equipped with an on-stage heating chamber (ΔTC3 from Bioptechs, Butler, PA, USA), a heated 63×/1.4 NA oil immersion objective system, a mercury lamp and appropriate filter sets. Confocal imaging was done with a Zeiss LSM 510 META inverted confocal laser-scanning microscope equipped with a 63×/1.4 NA oil immersion objective. To maintain a constant temperature of 37°C for live cell imaging, the confocal microscope was built in a ZEISS Incubator XL. Cells were cultured under the microscope in CO2-independent medium (Invitrogen, Karlsruhe, Germany). To analyze complementation of topoisomerase IIα function, we used human HT-1080 cells, in which both alleles of the TOP2A gene are disrupted. The cells are rescued by transgenic expression of human topoisomerase IIα from a tetracycline repressible construct stably integrated into the genome (26). Upon transfection of these cells (designated HTETOP) with the various chimeric constructs, complementation of topoisomerase IIα function was determined by comparing the number of stable cell clones obtained by selection with tetracycline (1 µg ml−1) versus puromycin (0.4 µg ml−1) (details see:26). For fluorescence activated cell sorting, cells were grown to ∼80% confluence, washed in PBS, trypsinized and resuspended in ice cold PBS at 106 cells ml−1. For each measurement, 20 000 cells were analyzed in a FACScan flow cytometer (BD bioscience) at a high flow rate setting with an Argon ion laser tuned to 488 nm.
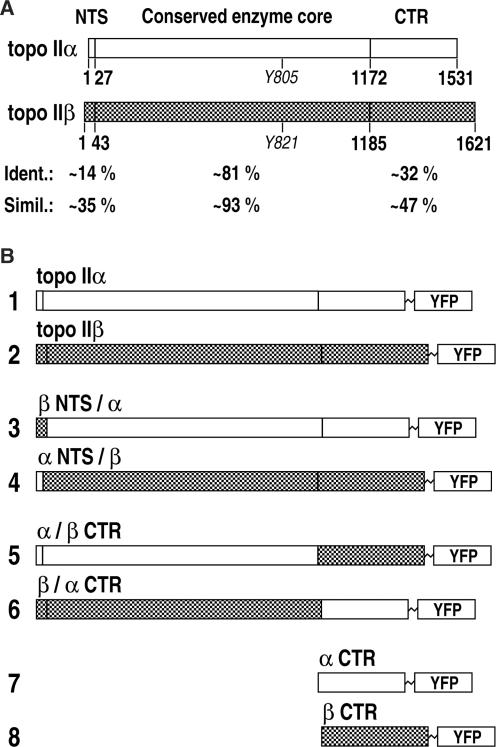
Figure 1.
(A) Comparison of human topoisomerase IIα and IIβ. Amino acid sequence alignment of the two isoforms indicates two regions with low and one with high sequence homology. Short stretches of the N-terminal ends (denoted ‘NTS’) and the CTR have highly divergent sequences, whereas most of the ATPase domains and the catalytic cores (denoted ‘Conserved enzyme core’) are similar. Y805 and Y821 are the active site tyrosines. Numbers indicate last residues of the respective regions selected for fission/fusion and truncation of enzyme variants. Amino acid identities and similarities for each region are indicated below. (B) Schematic synopsis of the constructs studied. All constructs were fused to YFP at their C-terminal ends; topo IIα, topo IIβ: full-length human topoisomerases IIα and IIβ; β NTS/α: chimeric human topoisomerase IIβ (1–43) –> IIα (28–1531); α NTS/β: chimeric human topoisomerase IIα (1–27) −> IIβ (44–1621); α/β CTR: chimeric human topoisomerase IIα (1–1172) −> IIβ (1186–1621); β/α CTR: chimeric human topoisomerase IIβ (1–1185) −> IIα (1173–1531); α CTR: truncated human topoisomerase IIα (Δ1–1172); β CTR: truncated human topoisomerase IIβ (Δ1–1185).
Immunoblotting and band depletion assay
Isolation of cell nuclei and extraction of nuclear proteins (400 mM NaCl) followed published procedures (48). Whole cell lysate was prepared from 3 × 106 cells suspended in D-PBS (Invitrogen, Karlsruhe, Germany) by addition of an equal volume of 2-fold lysis buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 4% SDS, 20 mM DTT, 500 mM urea, 5 mM AEBSF, 0.04% bromophenol blue) followed by ultrasound treatment (15 s, 14 W, 20 kHz, tip diameter 2 mm). For assessment of in vivo activity by immunoband depletion, cells were cultured with 100 µM VM26 (teniposide, Bristol-Myers Squibb, Munich, Germany) for 30 min prior to harvesting. Cell lysate (5 × 104 cells/lane) was subjected to SDS PAGE and transferred to PVDF membranes (Immobilon P, Millipore, Bedford, Maryland, USA). Blots were probed with the following antibodies: (i) mouse monoclonal antibodies against GFP (clone JL8, Clontech, Heidelberg, Germany), which cross react with YFP and subsequently are referred to as ‘YFP antibodies’; (ii) rabbit peptide antibodies raised against a peptide of amino acid residues 1514–1531 of human topoisomerase IIα (CIC, Genosys Cambridge, England); (iii) rabbit polyclonal antibodies raised against a peptide of amino acid residues 1586–1621 of human topoisomerase IIβ (designated 670) (49); (iv) various other antibodies against C-terminal epitopes of human topoisomerase IIβ serving as a control for results obtained with 670. These include rabbit polyclonal antibodies against amino acid residues 1341–1621 (designated H-286, Santa Cruz, Heidelberg, Germany) and 1611–1621 (designated 779) (49), and mouse monoclonal antibodies against amino acid residues 1583–1601 (clone 3H10) (50).
Immunoprecipitation and kDNA decatenation
Magnetic beads (Dynabeads M-280, Dynal/Invitrogen, Oslo, Norway) coupled to sheep anti-mouse IgG were loaded with YFP antibodies (Anti-GFP, mixture of two mouse monoclonal antibodies, Roche, Basel, Switzerland) according to the manufacturer's instructions (60 µg of antibodies per 108 beads). Loaded beads (4 × 107) were incubated (2 h at 4°C) with nuclear extract (200 µg total protein) in a final volume of 400 µl binding buffer (5.5 mM Na2HPO4, 1.2 mM NaH2PO4, pH 7.4, 265 mM NaCl, 5% FCS, 13.75% glycerol, 2.25 mM EDTA, 0.35 mM DTT, 10 μg ml−1 aprotinin, 1 mM AEBSF). Subsequently, beads were washed once with three volumes of binding buffer (20 min, 4°C), followed by four washes (10 min, 4°C) with three volumes of washing buffer (5.5 mM Na2HPO4, 1.2 mM NaH2PO4, pH 7.4, 890 mM NaCl, 13.75% Glycerol, 2.25 mM EDTA, 0.35 mM DTT, 10 μg ml−1 aprotinin, 1 mM AEBSF). These washing steps were found crucial for disrupting non-covalent interactions of YFP-fused topoisomerase II with endogenous topoisomerase species. Immunoprecipitates were finally eluted from the beads by boiling for 10 min in sample buffer (31.25 mM Tris-HCl, pH 6.8, 5% glycerol, 3% SDS, 2 mM DTT, 2 mM EDTA, 10 μg ml−1 aprotinin, 1 mM AEBSF). Eluates were subjected to SDS PAGE and silver staining (4 × 106 beads per lane) or western blotting (2 × 106 beads per lane). Alternatively, immunoprecipitates were washed twice with decatenation buffer (50 mM Tris-HCl, pH 7.6, 100 mM KCl, 10 mM MgCl2, 1 mM ATP, 0.5 mM DTT, 0.5 mM EDTA, 30 μg ml−1 BSA) and incubated (2 h, 37°C) with 300 ng catenated kinetoplast DNA from Crithidia fasciculata (kDNA, TopoGen Inc., Columbus, USA), with or without 1 mM ICRF-187 (Zinecard, Pharmacia & Upjohn, Kalamazoo, MI, USA), in a final volume of 27 µl decatenation buffer. The reaction was stopped by adding 1% SDS and 0.1 mg ml−1 proteinase K, and DNA reaction products were analyzed by agarose gel electrophoresis.
RESULTS
Construction of topoisomerase IIα/β chimeras
Topoisomerase IIα and IIβ are composed of three functional domains, which are bordered by protease-sensitive sites. Limited proteolysis experiments revealed that the C-terminal domains begin at residues 1263 (IIα) and 1296 (IIβ), respectively (39,41). However, sequence alignment shows that the region of high diversity between the two isoforms extends beyond this domain border and reaches up to amino acid positions 1171–1179 (IIα) or 1185–1191 (IIβ) (16,51). Interestingly, type II enzymes from chlorella viruses lack this divergent C-terminal region (52–54), and truncation of human topoisomerase IIα at amino acid 1175 produced a catalytically active variant in vitro (53). An extended truncation at position 1121, however, destroyed enzyme activity (55) probably due to deletion of the primary dimerization region (56). In summary, these findings suggested to us residues 1173–1531 and 1186–1621 of human topoisomerase IIα and IIβ, respectively, as candidate regions for the isoform-specific regulation. These regions encompass a maximum of heterogeneity (bearing only ∼32% identical amino acid residues) and can be exchanged between the two isoforms without interfering with the basic enzymatic functions. In fact, it has been reported that exchanging of the C-terminal regions of murine topoisomerase II isoforms at positions corresponding to amino acids 1173 and 1186 of human topoisomerase IIα and IIβ, respectively, gives rise to chimeric enzymes that are fully active upon heterologous expression in yeast (57). Therefore, we chose to exchange the same C-terminal regions of the human enzymes. Because these regions extend beyond the C-terminal domains defined by limited proteolysis (39,41), they are herein referred to as C-terminal regions (CTRs). We also selected short divergent stretches at the N-terminal ends of human topoisomerase IIα and IIβ (first 27 or 43 amino acids, respectively) to be exchanged between isoforms. These regions are only ∼14% identical, whereas the rest of the N-terminal domains and the core domains (amino acids 28–1172 and 44–1185 of IIα and IIβ, respectively) are very similar with ∼81% identical amino acid residues. A synopsis of sequence homologies and sites chosen for exchanging regions between topoisomerase IIα and IIβ is shown in Figure 1A.
Constitutive expression of active topoisomerase IIα/β-chimeras in HEK 293 cells
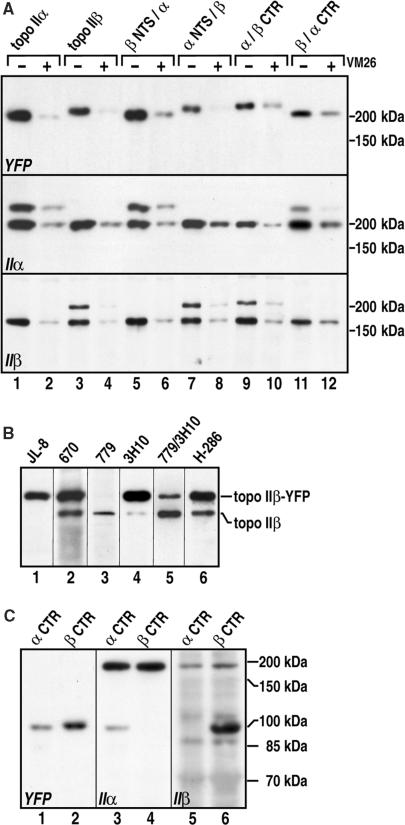
When transfected into HEK 293 cells, each of the constructs depicted in Figure 1B gave rise to viable cell lines supporting stable expression of the YFP-fused proteins. Cell clones with intermediate expression levels were selected and expanded for further analysis. All clones exhibited growth rates and gross morphologies similar to cells not transfected or expressing YFP alone. To assess the integrity of the fusion proteins and to compare their relative expression levels, we subjected the cells to western blotting and probed the blots with YFP antibodies. YFP-fused full-length, non-chimeric topoisomerase IIα and IIβ and the various topoisomerase IIα/β chimeras were detected as single protein bands of expected size. There were no smaller bands detected in addition by YFP antibodies (Figure 2A, top, odd numbered lanes). Thus, we could exclude rearrangements of the transgenes and safely assume that yellow fluorescence of the cells was entirely due to the desired YFP-fused protein. All constructs supported similar expression levels allowing a comparison of data between these cells. To compare YFP-fused and endogenous enzymes, blots were probed with isoform-specific antibodies against C-terminal epitopes of topoisomerase IIα or IIβ (Figure 2A, middle and bottom, respectively). The YFP-fused proteins could clearly be discriminated from the corresponding endogenous enzymes as additional bands of slower migration. From comparison of lanes it became evident that endogenous levels of topoisomerase IIα and IIβ were similar in all transfected cell clones (Figure 2A, middle and bottom, odd numbered lanes) and similar to those in untransfected cells (not shown), indicating that none of the YFP-fused enzymes interfered with endogenous topoisomerase II expression. It should also be noted that the desired exchanges of CTRs outlined in Figure 1B were confirmed by the presence of unique C-terminal epitopes of topoisomerase IIα and IIβ in the products of the various constructs shown in middle and bottom panel of Figure 2A. To compare expression levels of endogenous and YFP-fused proteins within each clone, we intended to compare signal intensity within the lanes of western blots stained with isoform-specific topoisomerase II antibodies (Figure 2A, middle and bottom, odd numbered lanes). However, upon testing several antibodies directed at various unique epitopes of human topoisomerase IIβ, we found that all antibodies tested did not exhibit the same preference for the endogenous form and heterologously expressed YFP-fused variants of the enzyme. On the contrary, the antibodies preferentially recognized one or the other form of the enzyme (Figure 2B). Thus, conclusions about expression levels drawn from such analyses are in our eyes unreliable. However, it should be noted that all cell clones expressing YFP-fused enzyme constructs exhibited growth rates and morphologies indistinguishable from untransfected cells, suggesting that the enzymes were at least expressed at physiologically tolerable levels. Similar analyses were carried out on cell clones expressing YFP fusion proteins of the CTRs of topoisomerase IIα and IIβ alone (Figure 2C). These constructs also gave rise to single protein bands. However, the apparent molecular weight (∼100 kDa in both cases) was larger than expected from the amino acid sequence (70 kDa for α CTR-YFP and 80 kDa for β CTR-YFP). This could be due to phosphorylation, since these regions harbor the majority of phosphorylation sites (38).
Figure 2.
Immunoblot analysis of YFP-fused topoisomerase II constructs expressed in HEK 293 cells. Positions of marker proteins are indicated on the right margin. See Figure 1B for nomenclature of the constructs. (A) Topoisomerase II immunoband depletion assay. Lysates of whole cells expressing full-length human topoisomerase IIα (lanes 1 and 2), or IIβ (lanes 3 and 4), or enzyme chimeras (lanes 5–12) were subjected to SDS PAGE (6% gels) and western blotting. Blots were stained with YFP antibodies (top), or antibodies against human topoisomerase IIα (middle), or IIβ (mixture of antibodies 779 and 3H10, bottom). Cells in even lanes were first cultured with 100 µM VM26 for 30 min. (B) Specificity of topoisomerase IIβ antibodies in immunostaining of western blots. Whole cell lysates of cells expressing full-length human topoisomerase IIβ, were subjected to SDS PAGE (6% gels) and western blotting. Blot membranes were probed with YFP antibodies (lane 1, JL-8) or antibodies against topoisomerase IIβ, as follows. Lane 2 (670), lane 3 (779), lane 4 (3H10), lane 5 (a mixture of antibodies 779 and 3H10) and lane 6 (H286). (C) Lysates of whole cells expressing human topoisomerase IIα Δ1–1172 (α CTR, odd numbered lanes) or IIβ Δ1–1185 (β CTR, even numbered lanes) were subjected to SDS PAGE (10% gels) and western blotting. Blots were probed with YFP-antibodies (left), or antibodies against human topoisomerase IIα (middle), or IIβ (right).
To determine whether the exogenous enzymes were as active in the cells as the endogenous enzymes, we employed immunoband depletion. The assay is based on the stabilization of covalent complexes of topoisomerase II and genomic DNA by specific poisons (4). As a consequence, signals specific for active topoisomerase II molecules are depleted from immunoblots due to retention of topoisomerase II•DNA complexes in the gel slots. As demonstrated in Figure 2A, treatment with the topoisomerase II poison VM26 depleted YFP-fused non-chimeric full-length enzymes and topoisomerase IIα/β chimeras from immunoblots to a similar extent (Figure 2A, top, compare lanes 1–4 with lanes 5–12). The extent of depletion was also comparable to that of endogenous topoisomerase IIα (Figure 2A, middle) and IIβ (Figure 2A, bottom). It should be noted that in this analysis antibody-derived biases (compare Figure 2B) did not play a role since they apply in the same manner to the bands to be compared in quantitative terms. These results confirm that all the topoisomerase IIα/β chimeras were active in the cells. Moreover, it could be deduced that activity levels were comparable to those of non-chimeric enzymes (exogenous or endogenous) because bands were depleted to similar extent. Corresponding analyses of cells expressing the YFP-tagged α CTR or β CTR confirmed that these proteins were catalytically inactive, as predicted (not shown).
Impact of non-conserved regions on localization of topoisomerase II in living cells
We have previously shown by immunohistochemistry (43) as well as in vivo localization of biofluorescent topoisomerase IIα and IIβ (44) that the most obvious difference between the isoforms is their association with metaphase chromosomes. This observation is demonstrated in the first two rows of Figure 3B. YFP-fused topoisomerase IIα (row 1) and IIβ (row 2) exhibit a similar distribution in the interphase nucleus (left). However, at metaphase (right) the α isoenzyme accumulates on the condensed chromosomes, whereas the β isoenzyme diffuses mostly into the cytosol and shows only a marginal association with chromosomes. The phenomenon is even more clearly seen in Figure 3A showing cells co-expressing topoisomerase IIα and IIβ fused to CFP and YFP, respectively. At interphase (left panel), the two isoenzymes colocalize, whereas at mitosis (right panel) topoisomerase IIα (shown in red) is chromosome bound, and topoisomerase IIβ (shown in green) predominantly resides in the cytosol. Thus, binding to metaphase chromosomes can be used as experimental readout of isoform-specific functioning of topoisomerase IIα and IIβ in proliferating mammalian cells, and we have used it to characterize the various topoisomerase IIα/β chimeras (Figure 3B, rows 3–6) and the two CTRs alone (Figure 3B, rows 7 and 8). It becomes readily apparent, that the ability to bind to metaphase chromosomes is fully retained in all enzyme varieties bearing the α CTR (Figure 3B, rows 1, 3 and 6) and to some extent also in this enzyme portion alone (Figure 3B, row 7). Moreover, enrichment in mitotic chromosomes is lost from topoisomerase IIα, when its CTR is replaced with that of the β isoform (Figure 3B, row 5). Conversely, this property is fully gained by topoisomerase IIβ, when its CTR is replaced with that of the α isoform (Figure 3B, row 6). Similar effects are not apparent upon exchanging the non-conserved N-terminal stretches of the isoenzymes (Figure 3B, compare rows 3 and 4). It should be noted that the β CTR alone had a diffuse distribution in the cell at metaphase, but it was not entirely excluded from chromosomes, as was YFP alone (Figure 3B, row 9). In summary, these observations suggest that (i) the non-conserved CTR of topoisomerase IIα promotes binding of the enzyme to metaphase chromosomes, (ii) the corresponding CTR of topoisomerase IIβ is much less capable of performing this function, and (iii) the non-conserved N-terminal stretches of the two isozymes do not play a role in targeting the enzymes to metaphase chromosomes.
Figure 3.
In vivo localization of topoisomerase II constructs in interphase nuclei and during metaphase. (A) Representative examples of HEK 293 cells stably co-expressing topoisomerase IIα fused to CFP (pseudo-colored in red) and topoisomerase IIβ fused to YFP (green) visualized by confocal imaging at interphase (left column) and mitosis (right column). (B) Each row of images shows representative images of living HEK 293 cells stably expressing the YFP-fused topoisomerase II construct indicated on the left margin (see Figure 1B for nomenclature) or YFP alone (row 9). Each pair of images visualizes the same cell by transmitted light (left) and confocal imaging of YFP-fluorescence in mid plane (right). Left and right columns of image pairs show representative examples of cells at interphase and metaphase, respectively.
The dimerization state of topoisomerase IIα/β chimeras
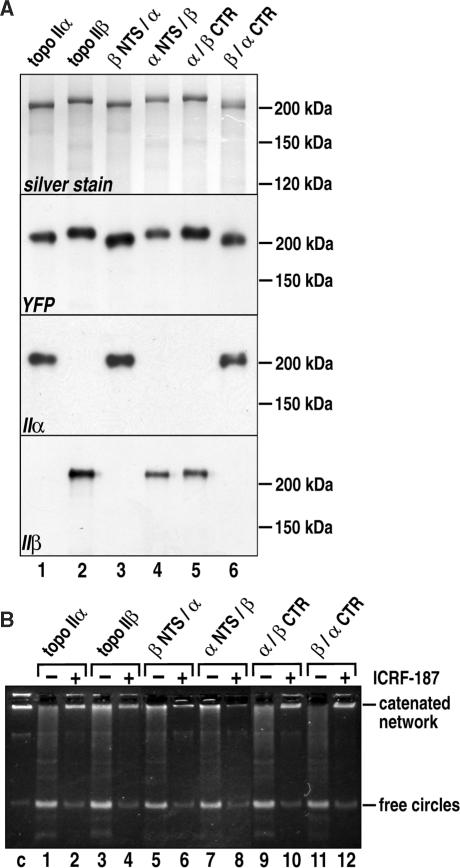
Given that topoisomerase II functions as a dimer, unambiguous deductions from the data in Figure 3 can only be made, when dimerization of topoisomerase IIα/β chimeras with endogenous enzyme varieties can be excluded, because this would render the behavior of the YFP-linked moiety attributable to an unpredictable mixture of homo- and heterodimers. In fact, we have previously assumed dimerization between GFP-fused, wild-type topoisomerase II and corresponding endogenous enzyme molecules based on GFP-directed immunoprecipitation (IP) protocols (44). We therefore wanted to clearly define the dimerization state of the topoisomerase IIα/β chimeras by YFP-directed IP followed by SDS PAGE, protein staining and immunoblotting. When we applied previously described low-salt IP conditions (44) to extracts from cell lines investigated here, the results were highly inconsistent, and non-reproducible; often both endogenous topo II isoforms were detected in the IPs by immunoblotting (data not shown). To address these problems we applied harsher buffer conditions with higher salt concentrations. Under these conditions, no prominent protein bands, other than the expected YFP-chimeras, were detected in silver-stained gels of any of the IPs (Figure 4A, top), at least not in the size range where endogenous topoisomerase IIα or IIβ would migrate (i.e. 170–180 kDa). Similarly, endogenous topoisomerase IIα or IIβ were undetectable in immunoblots of the IPs (Figure 4A, middle-bottom and bottom), the only protein species detected being the ones directly targeted by the precipitating YFP antibody. It is unlikely that this difference to our previous results was brought about by experimental dissociation of topoisomerase II dimers because they are known to be extremely salt-stable (58). We find it more likely that the presence of endogenous topoisomerase II isoforms in IPs of GFP-tagged topoisomerase II, as reported previously by us (44), was due to the multimerization of topoisomerase II homodimers (59), which is known to decrease with increasing ionic strength (59). To further demonstrate that topoisomerase II dimers were not disrupted during high salt IP, we tested whether the enzymes retained their catalytic activity (indicative of functional dimeric enzymes) in the final IPs. As demonstrated in Figure 4B, IPs of all the YFP-tagged proteins exhibited strong DNA decatenation activity, thus attesting to the integrity of the enzyme dimers throughout the IP procedure. Thus, the most plausible conclusion is that all the exogenous topoisomerase II species heterologously expressed in HEK 293 cells undergo homodimer formation. Because these observations were based on IPs of YFP-tagged proteins, they do not rule out the formation of endogenous topoisomerase II heterodimers (60). They do, however, allow for an unambiguous interpretation of the data obtained here with the various topoisomerase IIα/β chimeras. Thus, in vivo localization of these proteins (Figure 3B) clearly indicates a decisive role of the non-conserved α CTR in targeting the core portion of topoisomerase II to metaphase chromosomes.
Figure 4.
Immunoprecipitation of YFP-fused topoisomerase II constructs followed by determination of enzyme activity in vitro. (A) Topoisomerase constructs indicated at the top (see Figure 1B for nomenclature) were stably expressed in HEK 293 cells and subjected to YFP-directed immunoprecipitation. Precipitates were analyzed by SDS PAGE (6% gels) and protein silver staining (top), or subjected to western blotting and probed with YFP antibodies (middle top), or antibodies against topoisomerase IIα (middle bottom), or IIβ (bottom). Migration distances of molecular weight marker proteins are indicated on the right margin. (B) Alternatively, precipitates were reacted with 300 ng kDNA in the absence (odd numbered lanes) or presence (even numbered lanes) of 1 mM ICRF-187. DNA-reaction products were separated by agarose gel electrophoresis and visualized with ethidium bromide. Positions of catenated DNA network and free DNA circles are indicated on the right margin. The first lane on the left (c) shows the kDNA substrate alone.
The CTR of topoisomerase IIα is required for the efficient support of cell proliferation
Our observation that targeting to metaphase chromosomes is promoted by the α CTR (Figure 3) suggests that this region may also enable topoisomerase IIα to perform its essential functions in proliferating cells. We tested this hypothesis by complementation studies making use of human HT-1080 cells, in which both alleles of the TOP2α gene are disrupted and cell proliferation is supported by expression of topoisomerase IIα from a tetracycline repressible vector stably integrated into the genome (26). These HTETOP cells, which die when topoisomerase IIα is depleted by the addition of tetracycline, were stably transfected with the YFP-fused topoisomerase II constructs investigated here (Figure 1B) and the frequency with which colonies could form in the presence of tetracycline was measured. The results are summarized in Table 1. In the absence of any transfected constructs, no viable cell clones emerged in the presence of tetracycline (row 7), but the YFP-fused version of topoisomerase IIα was able to support proliferation of the cells in the presence of tetracycline, as expected (row 1). The YFP-fused version of topoisomerase IIβ, however, consistently gave rise to a much lower frequency of clones (row 2). Most interestingly, topoisomerase IIα lost most of its ability to support cell proliferation upon replacement of its CTR with that of the β isoform (row 5), whereas topoisomerase IIβ furnished with the α CTR gained this ability (row 6). A similar effect was not observed upon exchanging the non-conserved N-terminal stretches of the isozymes (rows 3 and 4). In summary, all versions of human topoisomerase II bearing the α CTR were capable of supporting cell proliferation (rows 1, 3 and 6), whereas all those furnished with the β CTR (rows 2, 4 and 5) were highly inefficient in this respect. It is, however, important to note that the latter constructs were not completely incapable of supporting cell proliferation.
Table 1.
HTETOP complementation with topoisomerase IIα, IIβ and IIα/β-chimerasa
| Constructb | TETc (%) | PUROc (%) | CId |
|---|---|---|---|
| Topo IIα | 100 | 100 | 0.91 ± 0.20 |
| Topo IIβ | 11.3 ± 5.6 | 69.8 ± 18.1 | 0.14 ± 0.10 |
| β NTS/α | 128.6 ± 18.2 | 73.0 ± 11.6 | 1.64 ± 0.61 |
| α NTS/β | 5.1 ± 3.3 | 85.6 ± 16.8 | 0.11 ± 0.05 |
| α/β CTR | 3.9 ± 4.7 | 120.9 ± 42.1 | 0.14 ± 0.11 |
| β/α CTR | 69.5 ± 32.2 | 71.1 ± 13.3 | 1.08 ± 0.35 |
| Mock transfection | 0 | 0 | – |
aHTETOP: human HT-1080 cells in which both alleles of the TOP2α gene are disrupted and which are salvaged by transgenic expression of human topoisomerase IIα from a tetracycline repressible construct stably integrated into the genome (26).
bConstructs tested for complementation of tetracycline-induced shutdown of heterologous topoisomerase IIα expression. All constructs also confer puromycin resistance. For nomenclature of constructs refer to Figure 1B.
cNumber of stable cell clones obtained after transfection of 3 × 106 HTETOP cells and selection with puromycin (PURO, 0.4 µg · ml−1,48 h) or tetracycline (TET, 1 µg · ml−1, 48 h). Numbers are normalized to those obtained with topoisomerase IIα (row 1). Mean values ± SEM of three similar experiments are given.
dComplementation index: ratio of stable cell clones obtained upon selection with tetracycline versus puromycin. Values are not normalized. Mean values ± SEM of at least three similar experiments are given.
Support of cell proliferation by topoisomerase IIβ requires highly elevated enzyme levels
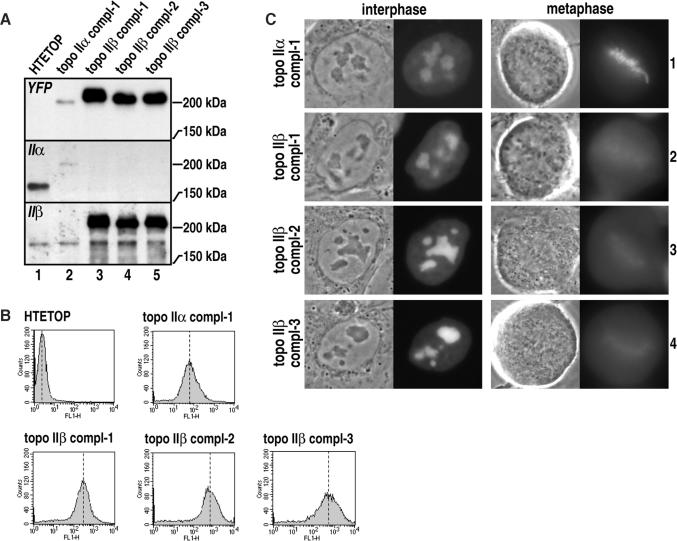
The average complementation index for all constructs bearing the α CTR was 1.21 (derived from Table 1, rows 1, 3 and 6). In contrast, the average complementation index for all constructs bearing the β CTR was 0.13 (derived from Table 1, rows 2, 4 and 5). Because no viable cell clones emerged from mock transfection (Table 1, row 7), these observations suggest that topoisomerase IIβ (or enzyme chimeras bearing the β CTR) can also support cell proliferation to some extent. Two explanations for this result can be considered: (i) topoisomerase IIβ could be able to substitute for the α isoform, when present at much higher levels; (ii) binding sites normally occupied by topoisomerase IIα (e.g. sites on mitotic chromosomes) could become freely accessible to the β isoform when the α isoform is absent. These sites could then be occupied by topoisomerase IIβ irrespective of its concentration in the cell. To investigate these hypotheses, we compared expression levels and localization of YFP-fused topoisomerase IIα or IIβ in clones supporting cell growth in the presence of tetracycline (Figure 5). From comparison of representative cell clones subjected to western blotting and probing with YFP antibodies (Figure 5A, top) it became readily apparent that complementation by YFP-fused topoisomerase IIβ (lanes 3, 4 and 5) requires much higher expression levels than complementation by YFP-fused topoisomerase IIα (lane 2). Flow cytometry confirmed this finding, showing that YFP-fluorescence was ∼10-fold brighter in various cell clones complemented by YFP-fused topoisomerase IIβ than in a reference cell clone complemented by YFP-fused topoisomerase IIα (Figure 5B). Notwithstanding our hesitations about quantitative comparisons between YFP-fused and endogenous topoisomerase II species based on topoisomerase II-directed immunoblotting (see Figure 2B), it should be noted that the cellular complement of YFP-fused topoisomerase IIα enabling full cell proliferation was hardly detectable by topoisomerase-directed immunoblotting (Figure 5A, middle, compare lane 1 with lane 2), whereas YFP-fused topoisomerase IIβ complementing topoisomerase IIα function gave a much more intense signal than corresponding bands of endogenous topoisomerase IIβ (Figure 5A, compare bands within lanes 3, 4 and 5). Thus, in our model system, the β isoform seems only able to substitute for topoisomerase IIα when it is highly (at least 10-fold) overexpressed. Figure 5C further suggests that, upon repression of topoisomerase IIα expression, binding sites on mitotic chromosomes normally occupied by this enzyme do not become freely accessible to the β isoform and are not readily occupied by it. If that were the case, topoisomerase IIβ should accumulate on metaphase chromosomes upon repression of topoisomerase IIα. However, this was clearly not the case in any of the cell clones complemented by YFP-fused topoisomerase IIβ (Figure 5C, rows 2–4). In all these cell clones, YFP-fused topoisomerase IIβ was mostly localized in the cytoplasm during mitosis. We failed to detect an accumulation of the enzyme on metaphase chromosomes, as seen in the same cell model with YFP-fused topoisomerase IIα (Figure 5C, row 1). It should also be noted that localization of YFP-fused topoisomerase IIβ during interphase (Figure 5C, left) and metaphase (Figure 5C, right) was identical to that observed in HEK 293 cells where endogenous expression of topoisomerase IIβ was not silenced (Figure 3A and B). In summary, these morphological data suggest that the binding equilibrium of topoisomerase IIβ at metaphase chromosomes is (i) independent of the absence or presence of topoisomerase IIα, and (ii) not significantly influenced by the total cellular level of topoisomerase IIα and IIβ. Thus, the weak but notable ability of the β isoform to complement the function of the α isoform cannot be due to an increase in its propensity to interact with mitotic chromosomes upon removal of topoisomerase IIα.
Figure 5.
Characterization of HTETOP clones rescued by topoisomerase IIα or IIβ. (A) Immunoblot analysis of YFP-fused topoisomerase II constructs expressed in HTETOP cells. Whole cell lysates of untransfected HTETOP (lane 1) or tetracycline resistant HTETOP clones expressing topoisomerase IIα (lane 2) or topoisomerase IIβ (lanes 3, 4 and 5) were subjected to SDS PAGE (6% gels) and western blotting. Blots were stained with YFP antibodies (top), or antibodies against human topoisomerase IIα (middle), or IIβ (bottom). Positions of marker proteins are indicated on the right margin. (B) Yellow fluorescence level of untransfected HTETOP (HTETOP) or tetracycline resistant HTETOP clones expressing topoisomerase IIα (topo IIα compl-1) or topoisomerase IIβ (topo IIβ compl-1, topo IIβ compl-2 and topo IIβ compl-3) was measured by flow cytometry and shown as histograms. (C) In vivo localization of YFP-fused topoisomerase II constructs in interphase nuclei and during metaphase. Tetracycline resistant HTETOP clones stably expressing topoisomerase IIα (row 1) or IIβ (rows 1, 2 and 3) were visualized by fluorescence microscopy. Corresponding phase contrast images (left) and YFP images (right) are shown. Left and right of image pairs show representative examples of cells at interphase and metaphase respectively.
DISCUSSION
It is well established that the C-terminal domains of both yeast and human topoisomerase II are dispensable for the enzyme's basic catalytic activity (55,61). On the other hand, a large body of evidence suggests that the C-terminal domain plays a role in regulating the cellular functioning of topoisomerase II. Most notably, it contains crucial nuclear localization signals (55,62,63) and sites phosphorylated in a cell-cycle-related manner (38). Since the C-terminal domains are the most divergent portions of the two mammalian isoforms of topoisomerase II (16), it has been proposed that they determine specific functions differing between these two isoforms in vivo (39). Here, we provide direct evidence in support of this view. We demonstrate that the divergent CTRs of topoisomerase IIα and IIβ govern two features in which the two isoforms characteristically differ, namely binding to mitotic chromosomes and support of cell proliferation. We show that YFP-fused topoisomerase IIα is preferentially chromosome-bound during mitosis and fully supports proliferation of cells lacking endogenous topoisomerase IIα. In contrast, the majority of YFP-fused topoisomerase IIβ is not chromosome-bound at mitosis, and clones emerged from complementation experiments at greatly reduced frequencies. The specific features of the α isoform were stringently linked to the presence of the α CTR. Replacement of the CTR in topoisomerase IIβ with the α CTR produced an enzyme chimera that behaved like topoisomerase IIα, whereas the converse experiment produced an enzyme chimera behaving like topoisomerase IIβ.
The requirement of topoisomerase IIα for proper cell division has been suggested by indirect evidence showing an essential role in chromosome segregation, which is not readily adopted by the β-isoform (e.g. 25). More recently, depletion of topoisomerase IIα by various experimental strategies resulted in each case in an impaired separation of chromosomes in anaphase. (26,27,64). Here we describe a striking coincidence between the ability of all versions of topoisomerase II furnished with the α CTR to complement such a lack of endogenous topoisomerase IIα and the propensity of the complementing construct to bind to mitotic chromosomes. We even observe that the α CTR alone preferentially binds to metaphase chromosomes. It remains unclear whether chromosome binding is due to direct DNA-interactions, as suggested for various prokaryotic type II topoisomerases (65,66), or to interactions with other proteins, e.g. condensins (67) or HSP90 (68). However, the strict correlation between the binding to metaphase chromosomes and the support of cell proliferation suggests a mechanistic connection between the two. It can be hypothesized (i) that efficient separation of sister chromatids and proper cell division depend on a high, local concentration of active topoisomerase II at the mitotic chromosome, (ii) that under physiological conditions only the α isoform accumulates in sufficient concentrations at the mitotic chromosome, and (iii) that this feature is promoted by an intrinsic ability of the α CTR to bind to metaphase chromosomes. These hypotheses would assign to the α CTR the function of an adaptor that shifts the binding equilibrium of the entire enzyme molecule towards the bound state and thus provides the chromosome at mitosis with sufficient topoisomerase II activity to perform extensive DNA-decatenation in the course of sister chromatid segregation. Our finding that topoisomerase IIβ can also support cell proliferation when expressed at extremely high levels supports such a hypothesis: sufficient local concentration of active topoisomerase II at the mitotic chromosome cannot only be acquired by expressing normal levels of an enzyme having a high affinity (due to the α CTR), but also by expressing highly increased levels of an enzyme having a much lower one (due to the β CTR).
The above interpretation is more difficult to fit with complementation studies carried out in yeast that show that both mammalian isoforms are equally capable of rescuing temperature-sensitive Δtop2 yeast mutants (22,23). One explanation could be that yeast might be unable to discriminate between the two mammalian isoforms. However, this explanation is unlikely because mouse topoisomerase IIα and IIβ can be discriminated by yeast, in as much as they are distributed in a distinguishable manner in yeast cell nuclei (57). Another explanation could be that yeast is more tolerant to changes in topoisomerase II expression levels than human cells, which are readily killed by overexpression of these enzymes (69). Since high copy number vectors were used in the yeast studies for expression of the complementing enzymes, expression levels of the β isoform could have been high enough to enable efficient complementation of topoisomerase II functions in the same manner as seen here in a human cell line.
Another possible interpretation of our data is that the topoisomerase IIα plays an essential role during replication. It has been shown in yeast that topoisomerase II is required for DNA replication, when topoisomerase I is lacking (70), because movement of DNA replication complexes through the DNA double helix induces positive supercoils ahead of this machinery. Recent work in yeast demonstrates that topoisomerase II relaxes chromatin even more efficiently than topoisomerase I (71). In mammals, topoisomerase IIα, but not IIβ, appears to be a key player in removal of this type of torsional stress during replication (17), and it was postulated that this isoform-specificity is determined by the divergent C-terminal regions (72). The residues that were suggested to play this role in replication are all within the α CTRs analyzed here. In addition, a study in chicken fibroblast showed that topoisomerase IIα, but not IIβ, co-localizes with sites of replication, and this targeting was also mediated by the α CTR (73). Unfortunately, it is difficult to determine whether topoisomerase IIα plays a truly essential role during replication that cannot be complemented by other proteins (e.g. topoisomerase I or topoisomerase IIβ). Although silencing of topoisomerase IIα in human cells (26) and mice (27) causes a defect in chromosome segregation, suggesting that its essential role is during mitosis rather than S-phase, this phenotype could conceivably be caused by loss of an essential topoisomerase IIα function during the late phases of replication. Lack of such a function could still allow for a progression into metaphase followed by mitotic catastrophes due to unresolved DNA catenanes. Thus, an essential role of topoisomerase IIα in relaxation of positive supercoils generated at late stages of replication cannot be excluded, and our observation that the α CTR is required for efficient support of cell proliferation by topoisomerase II may just as well reflect a specific involvement of the α isoform in DNA replication (72).
Regardless of the exact role of the α CTR, we demonstrate in this article that it confers a unique, proliferation-associated functionality to the topoisomerase II core enzyme (either version of it), whereas the β CTR is much less efficient in this respect. It is therefore plausible that the two versions of the CTR cooperate in differential targeting of topoisomerase IIα and IIβ, thus providing unique functionality of the two isoforms in proliferating cells.
ACKNOWLEDGEMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (grants BO 910/3-2; SFB 503, GRK 1033) and the Cancer Research UK (grant C9700/A5962). Funding to pay the Open Access publication charge was provided by the above mentioned grants
Conflict of interest statement. None declared.
REFERENCES
- 1.Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 2.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 3.Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 4.Liu LF. DNA topoisomerase poisons as antitumor drugs. Annu. Rev. Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 5.Kingma PS, Osheroff N. The response of eukaryotic topoisomerases to DNA damage. Biochim. Biophys. Acta. 1998;1400:223–232. doi: 10.1016/s0167-4781(98)00138-9. [DOI] [PubMed] [Google Scholar]
- 6.Mielke C, Christensen MO, Barthelmes HU, Boege F. Enhanced processing of UVA-irradiated DNA by human topoisomerase II in living cells. J. Biol. Chem. 2004;279:20559–20562. doi: 10.1074/jbc.C400032200. [DOI] [PubMed] [Google Scholar]
- 7.Constantinou A, Mehta R, Runyan C, Rao K, Vaughan A, Moon R. Flavonoids as DNA topoisomerase antagonists and poisons: structure-activity relationships. J. Nat. Prod. 1995;58:217–225. doi: 10.1021/np50116a009. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita Y, Kawada S, Nakano H. Induction of mammalian topoisomerase II dependent DNA cleavage by nonintercalative flavonoids, genistein and orobol. Biochem. Pharmacol. 1990;39:737–744. doi: 10.1016/0006-2952(90)90153-c. [DOI] [PubMed] [Google Scholar]
- 9.Strick R, Strissel PL, Borgers S, Smith SL, Rowley JD. Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc. Natl. Acad. Sci. USA. 2000;97:4790–4795. doi: 10.1073/pnas.070061297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drake FH, Zimmerman JP, McCabe FL, Bartus HF, Per SR, Sullivan DM, Ross WE, Mattern MR, Johnson RK, et al. Purification of topoisomerase II from amsacrine-resistant P388 leukemia cells. Evidence for two forms of the enzyme. J. Biol. Chem. 1987;262:16739–16747. [PubMed] [Google Scholar]
- 11.Tsai-Pflugfelder M, Liu LF, Liu AA, Tewey KM, Whang-Peng J, Knutsen T, Huebner K, Croce CM, Wang JC. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21-22. Proc. Natl. Acad. Sci. USA. 1988;85:7177–7181. doi: 10.1073/pnas.85.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung TD, Drake FH, Tan KB, Per SR, Crooke ST, Mirabelli CK. Characterization and immunological identification of cDNA clones encoding two human DNA topoisomerase II isozymes. Proc. Natl. Acad. Sci. USA. 1989;86:9431–9435. doi: 10.1073/pnas.86.23.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Austin CA, Fisher LM. Isolation and characterization of a human cDNA clone encoding a novel DNA topoisomerase II homologue from HeLa cells. FEBS Lett. 1990;266:115–117. doi: 10.1016/0014-5793(90)81520-x. [DOI] [PubMed] [Google Scholar]
- 14.Tan KB, Dorman TE, Falls KM, Chung TD, Mirabelli CK, Crooke ST, Mao J. Topoisomerase II alpha and topoisomerase II beta genes: characterization and mapping to human chromosomes 17 and 3, respectively. Cancer Res. 1992;52:231–234. [PubMed] [Google Scholar]
- 15.Austin CA, Sng JH, Patel S, Fisher LM. Novel HeLa topoisomerase II is the II beta isoform: complete coding sequence and homology with other type II topoisomerases. Biochim. Biophys. Acta. 1993;1172:283–291. doi: 10.1016/0167-4781(93)90215-y. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins JR, Ayton P, Jones T, Davies SL, Simmons DL, Harris AL, Sheer D, Hickson ID. Isolation of cDNA clones encoding the beta isozyme of human DNA topoisomerase II and localisation of the gene to chromosome 3p24. Nucleic Acids Res. 1992;20:5587–5592. doi: 10.1093/nar/20.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 18.Drake FH, Hofmann GA, Bartus HF, Mattern MR, Crooke ST, Mirabelli CK. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 19.Cornarotti M, Tinelli S, Willmore E, Zunino F, Fisher LM, Austin CA, Capranico G. Drug sensitivity and sequence specificity of human recombinant DNA topoisomerases IIalpha (p170) and IIbeta (p180) Mol. Pharmacol. 1996;50:1463–1471. [PubMed] [Google Scholar]
- 20.Leontiou C, Lightowlers R, Lakey JH, Austin CA. Kinetic analysis of human topoisomerase IIalpha and beta DNA binding by surface plasmon resonance. FEBS Lett. 2003;554:206–210. doi: 10.1016/s0014-5793(03)01172-4. [DOI] [PubMed] [Google Scholar]
- 21.Marsh KL, Willmore E, Tinelli S, Cornarotti M, Meczes EL, Capranico G, Fisher LM, Austin CA. Amsacrine-promoted DNA cleavage site determinants for the two human DNA topoisomerase II isoforms alpha and beta. Biochem. Pharmacol. 1996;52:1675–1685. doi: 10.1016/s0006-2952(96)00516-3. [DOI] [PubMed] [Google Scholar]
- 22.Meczes EL, Marsh KL, Fisher LM, Rogers MP, Austin CA. Complementation of temperature-sensitive topoisomerase II mutations in Saccharomyces cerevisiae by a human TOP2 beta construct allows the study of topoisomerase II beta inhibitors in yeast. Cancer Chemother. Pharmacol. 1997;39:367–375. doi: 10.1007/s002800050585. [DOI] [PubMed] [Google Scholar]
- 23.Jensen S, Redwood CS, Jenkins JR, Andersen AH, Hickson ID. Human DNA topoisomerases IIα and IIβ can functionally substitute for yeast TOP2 in chromosome segregation and recombination. Mol. Gen. Genet. 1996;252:79–86. [PubMed] [Google Scholar]
- 24.Wang JC. Cellular roles of dna topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 25.Grue P, Grasser A, Sehested M, Jensen PB, Uhse A, Straub T, Ness W, Boege F. Essential mitotic functions of DNA topoisomerase IIα are not adopted by topoisomerase IIβ in human H69 cells. J. Biol. Chem. 1998;273:33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 26.Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol. Biol. Cell. 2004;15:5700–5711. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akimitsu N, Adachi N, Hirai H, Hossain MS, Hamamoto H, Kobayashi M, Aratani Y, Koyama H, Sekimizu K. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 28.Errington F, Willmore E, Tilby MJ, Li L, Li G, Li W, Baguley BC, Austin CA. Murine transgenic cells lacking DNA topoisomerase IIbeta are resistant to acridines and mitoxantrone: analysis of cytotoxicity and cleavable complex formation. Mol. Pharmacol. 1999;56:1309–1316. doi: 10.1124/mol.56.6.1309. [DOI] [PubMed] [Google Scholar]
- 29.Khelifa T, Casabianca-Pignede MR, Rene B, Jacquemin-Sablon A. Expression of topoisomerases II alpha and beta in Chinese hamster lung cells resistant to topoisomerase II inhibitors. Mol. Pharmacol. 1994;46:323–328. [PubMed] [Google Scholar]
- 30.Dereuddre S, Delaporte C, Jacquemin-Sablon A. Role of topoisomerase II beta in the resistance of 9-OH-ellipticine-resistant Chinese hamster fibroblasts to topoisomerase II inhibitors. Cancer Res. 1997;57:4301–4308. [PubMed] [Google Scholar]
- 31.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIβ and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 32.Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIbeta. Proc. Natl. Acad. Sci. USA. 2003;100:7123–7128. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyu YL, Lin CP, Azarova AM, Cai L, Wang JC, Liu LF. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol. Cell. Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 35.Turley H, Comley M, Houlbrook S, Nozaki N, Kikuchi A, Hickson ID, Gatter K, Harris AL. The distribution and expression of the two isoforms of DNA topoisomerase II in normal and neoplastic human tissues. Br. J. Cancer. 1997;75:1340–1346. doi: 10.1038/bjc.1997.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng SW, Liu Y, Schnipper LE. Cloning and characterization of the 5′-flanking sequence for the human DNA topoisomerase IIβ gene. Gene. 1997;203:113–119. doi: 10.1016/s0378-1119(97)00500-3. [DOI] [PubMed] [Google Scholar]
- 37.Isaacs RJ, Harris AL, Hickson ID. Regulation of the human topoisomerase IIα gene promoter in confluence-arrested cells. J. Biol. Chem. 1996;271:16741–16747. doi: 10.1074/jbc.271.28.16741. [DOI] [PubMed] [Google Scholar]
- 38.Isaacs RJ, Davies SL, Sandri MI, Redwood C, Wells NJ, Hickson ID. Physiological regulation of eukaryotic topoisomerase II. Biochim. Biophys. Acta. 1998;1400:121–137. doi: 10.1016/s0167-4781(98)00131-6. [DOI] [PubMed] [Google Scholar]
- 39.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase II beta. Bioessays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 40.Lindsley JE, Wang JC. Proteolysis patterns of epitopically labeled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc. Natl. Acad. Sci. USA. 1991;88:10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Austin CA, Marsh KL, Wasserman RA, Willmore E, Sayer PJ, Wang JC, Fisher LM. Expression, domain structure, and enzymatic properties of an active recombinant human DNA topoisomerase II beta. J. Biol. Chem. 1995;270:15739–15746. doi: 10.1074/jbc.270.26.15739. [DOI] [PubMed] [Google Scholar]
- 42.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 43.Meyer KN, Kjeldsen E, Straub T, Knudsen BR, Hickson ID, Kikuchi A, Kreipe H, Boege F. Cell cycle-coupled relocation of types I and II topoisomerases and modulation of catalytic enzyme activities. J. Cell Biol. 1997;136:775–788. doi: 10.1083/jcb.136.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergaard O, Boege F, et al. Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J. Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 47.Mielke C, Tummler M, Schubeler D, von Hoegen I, Hauser H. Stabilized, long-term expression of heterodimeric proteins from tricistronic mRNA. Gene. 2000;254:1–8. doi: 10.1016/s0378-1119(00)00294-8. [DOI] [PubMed] [Google Scholar]
- 48.Christensen MO, Barthelmes HU, Boege F, Mielke C. Residues 190-210 of human topoisomerase I are required for enzyme activity in vivo but not in vitro. Nucleic Acids Res. 2003;31:7255–7263. doi: 10.1093/nar/gkg927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boege F, Andersen A, Jensen S, Zeidler R, Kreipe H. Proliferation-associated nuclear antigen Ki-S1 is identical with topoisomerase IIα. Delineation of a carboxyterminal epitope with peptide antibodies. Am. J. Pathol. 1995;146:1302–1308. [PMC free article] [PubMed] [Google Scholar]
- 50.Kimura K, Nozaki N, Enomoto T, Tanaka M, Kikuchi A. Analysis of M phase-specific phosphorylation of DNA topoisomerase II. J. Biol. Chem. 1996;271:21439–21445. doi: 10.1074/jbc.271.35.21439. [DOI] [PubMed] [Google Scholar]
- 51.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 52.Dickey JS, Choi TJ, Van Etten JL, Osheroff N. Chlorella virus Marburg topoisomerase II: high DNA cleavage activity as a characteristic of Chlorella virus type II enzymes. Biochemistry. 2005;44:3899–3908. doi: 10.1021/bi047777f. [DOI] [PubMed] [Google Scholar]
- 53.Dickey JS, Osheroff N. Impact of the C-terminal domain of topoisomerase IIalpha on the DNA cleavage activity of the human enzyme. Biochemistry. 2005;44:11546–11554. doi: 10.1021/bi050811l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lavrukhin OV, Fortune JM, Wood TG, Burbank DE, Van Etten JL, Osheroff N, Lloyd RS. Topoisomerase II from Chlorella virus PBCV-1. Characterization of the smallest known type II topoisomerase. J. Biol. Chem. 2000;275:6915–6921. doi: 10.1074/jbc.275.10.6915. [DOI] [PubMed] [Google Scholar]
- 55.Jensen S, Andersen A, Kjeldsen E, Biersack H, Olsen E, Andersen T, Westergaard O, Jakobsen B. Analysis of functional domain organization in DNA topoisomerase II from humans and saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:3866–3877. doi: 10.1128/mcb.16.7.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bjergbaek L, Jensen S, Westergaard O, Andersen AH. Using a biochemical approach to identify the primary dimerization regions in human DNA topoisomerase IIalpha. J. Biol. Chem. 1999;274:26529–26536. doi: 10.1074/jbc.274.37.26529. [DOI] [PubMed] [Google Scholar]
- 57.Adachi N, Miyaike M, Kato S, Kanamaru R, Koyama H, Kikuchi A. Cellular distribution of mammalian DNA topoisomerase II is determined by its catalytically dispensable C-terminal domain. Nucleic Acids Res. 1997;25:3135–3142. doi: 10.1093/nar/25.15.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tennyson RB, Lindsley JE. Type II DNA topoisomerase from Saccharomyces cerevisiae is a stable dimer. Biochemistry. 1997;36:6107–6114. doi: 10.1021/bi970152f. [DOI] [PubMed] [Google Scholar]
- 59.Vassetzky YS, Dang Q, Benedetti P, Gasser SM. Topoisomerase II forms multimers in vitro: effects of metals, beta-glycerophosphate, and phosphorylation of its C-terminal domain. Mol. Cell. Biol. 1994;14:6962–6974. doi: 10.1128/mcb.14.10.6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biersack H, Jensen S, Gromova I, Nielsen IS, Westergaard O, Andersen AH. Active heterodimers are formed from human DNA topoisomerase II alpha and II beta isoforms. Proc. Natl. Acad. Sci. USA. 1996;93:8288–8293. doi: 10.1073/pnas.93.16.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shiozaki K, Yanagida M. A functional 125-kDa core polypeptide of fission yeast DNA topoisomerase II. Mol. Cell. Biol. 1991;11:6093–6102. doi: 10.1128/mcb.11.12.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caron PR, Watt P, Wang JC. The C-terminal domain of Saccharomyces cerevisiae DNA topoisomerase II. Mol. Cell. Biol. 1994;14:3197–3207. doi: 10.1128/mcb.14.5.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mirski SE, Gerlach JH, Cole SP. Sequence determinants of nuclear localization in the alpha and beta isoforms of human topoisomerase II. Exp. Cell Res. 1999;251:329–339. doi: 10.1006/excr.1999.4587. [DOI] [PubMed] [Google Scholar]
- 64.Sakaguchi A, Kikuchi A. Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J. Cell Sci. 2004;117:1047–1054. doi: 10.1242/jcs.00977. [DOI] [PubMed] [Google Scholar]
- 65.Hsieh TJ, Farh L, Huang WM, Chan NL. Structure of the topoisomerase IV C-terminal domain: a broken beta-propeller implies a role as geometry facilitator in catalysis. J. Biol. Chem. 2004;279:55587–55593. doi: 10.1074/jbc.M408934200. [DOI] [PubMed] [Google Scholar]
- 66.Qi Y, Pei J, Grishin NV. C-terminal domain of gyrase A is predicted to have a beta-propeller structure. Proteins. 2002;47:258–264. doi: 10.1002/prot.10090. [DOI] [PubMed] [Google Scholar]
- 67.Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J. Cell Biol. 2003;160:645–655. doi: 10.1083/jcb.200209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barker CR, McNamara AV, Rackstraw SA, Nelson DE, White MR, Watson AJ, Jenkins JR. Inhibition of Hsp90 acts synergistically with topoisomerase II poisons to increase the apoptotic killing of cells due to an increase in topoisomerase II mediated DNA damage. Nucleic Acids Res. 2006;34:1148–1157. doi: 10.1093/nar/gkj516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McPherson JP, Goldenberg GJ. Induction of apoptosis by deregulated expression of DNA topoisomerase IIα. Cancer Res. 1998;58:4519–4524. [PubMed] [Google Scholar]
- 70.Kim RA, Wang JC. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J. Mol. Biol. 1989;208:257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- 71.Salceda J, Fernandez X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McClendon AK, Dickey JS, Osheroff N. Ability of viral topoisomerase II to discern the handedness of supercoiled DNA: bimodal recognition of DNA geometry by type II enzymes. Biochemistry. 2006;45:11674–11680. doi: 10.1021/bi0520838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niimi A, Suka N, Harata M, Kikuchi A, Mizuno S. Co-localization of chicken DNA topoisomerase IIalpha, but not beta, with sites of DNA replication and possible involvement of a C-terminal region of alpha through its binding to PCNA. Chromosoma. 2001;110:102–114. doi: 10.1007/s004120100140. [DOI] [PubMed] [Google Scholar]