Abstract
Agrobacterium tumefaciens encodes a single NAD+-dependent DNA ligase and six putative ATP-dependent ligases. Two of the ligases are homologs of LigD, a bacterial enzyme that catalyzes end-healing and end-sealing steps during nonhomologous end joining (NHEJ). Agrobacterium LigD1 and AtuLigD2 are composed of a central ligase domain fused to a C-terminal polymerase-like (POL) domain and an N-terminal 3′-phosphoesterase (PE) module. Both LigD proteins seal DNA nicks, albeit inefficiently. The LigD2 POL domain adds ribonucleotides or deoxyribonucleotides to a DNA primer-template, with rNTPs being the preferred substrates. The LigD1 POL domain has no detectable polymerase activity. The PE domains catalyze metal-dependent phosphodiesterase and phosphomonoesterase reactions at a primer-template with a 3′-terminal diribonucleotide to yield a primer-template with a monoribonucleotide 3′-OH end. The PE domains also have a 3′-phosphatase activity on an all-DNA primer-template that yields a 3′-OH DNA end. Agrobacterium ligases C2 and C3 are composed of a minimal ligase core domain, analogous to Mycobacterium LigC (another NHEJ ligase), and they display feeble nick-sealing activity. Ligation at DNA double-strand breaks in vitro by LigD2, LigC2 and LigC3 is stimulated by bacterial Ku, consistent with their proposed function in NHEJ.
INTRODUCTION
DNA ligases are ubiquitous enzymes that seal DNA nicks via three sequential nucleotidyl transfer reactions (1). In the first step, nucleophilic attack on the α phosphorus of ATP or NAD+ by ligase results in release of PPi or NMN and formation of a covalent ligase-adenylate intermediate in which AMP is linked via a phosphoamide (P–N) bond to Nζ of a lysine. In the second step, the AMP is transferred to the 5′ end of the 5′ phosphate-terminated DNA strand to form a DNA-adenylate intermediate. In the third step, ligase catalyzes attack by the 3′-OH of the nick on DNA-adenylate to join the two polynucleotides and liberate AMP.
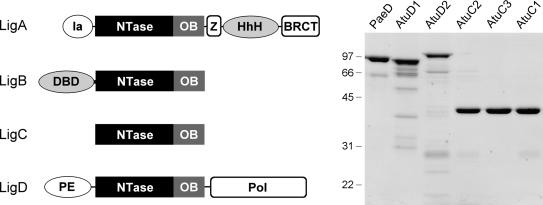
DNA ligases are grouped into two families, ATP-dependent ligases and NAD+-dependent ligases, according to the cofactor required for ligase-adenylate formation (1). An NAD+-dependent DNA ligase (LigA) is found in every bacterial species (2). The structures of several LigA enzymes have been determined by X-ray crystallography (3–6). They contain the ligase catalytic core, composed of a nucleotidyltransferase (NTase) domain and an OB-fold domain, flanked by an N-terminal domain (called domain Ia) and three C-terminal domains: a tetracysteine Zn-finger, a helix-hairpin-helix (HhH) domain and a BRCT domain (Figure 1). Domain Ia is unique to NAD+-dependent ligases and required for the reaction of LigA with NAD+ to form the ligase-adenylate intermediate (7). LigA is essential in all bacteria tested to date (8–13).
Figure 1.
Multiple DNA ligases of A. tumefaciens. The Atu LigA, LigB, LigC and LigD polypeptides are depicted (left panel) in cartoon form with the N-termini on the left and the C-termini on the right. The core ligase catalytic domains, composed of nucleotidyltransferase (NTase) and OB modules, are shown as rectangles. Flanking domains of known structure or imputed function (variously drawn as ellipses or capsules) are discussed in the text. Aliquots (5 µg) of the nickel-agarose preparations of PaeLigD, AtuLigD1, AtuLigD2, AtuLigC2, AtuLigC3 and AtuLigC1 were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown in the right panel. The positions and sizes (in kDa) of marker polypeptides are indicated.
The first identification and characterization of a bacterial ATP-dependent ligase was reported in 1997 for Haemophilus influenzae (14). In the ensuing years, putative ATP-dependent ligases have been annotated in scores of bacterial species that have been subjected to genome sequencing, though only a few of them have been studied biochemically (15–19). Mycobacterium tuberculosis and Mycobacterium smegmatis have an exceptionally rich assortment of ATP-dependent ligases (named LigB, LigC and LigD) in addition to NAD+-dependent LigA. Whereas mycobacterial LigA and LigB display vigorous ligase activity, LigD and LigC are relatively feeble at sealing nicked DNAs in vitro (16). Genetic and biochemical analyses implicate LigD and LigC in a bacterial pathway of nonhomologous end joining (NHEJ) that is strictly dependent on a bacterial homolog of the NHEJ factor Ku (16–20). The fact that Ku and LigD are jointly encoded by dozens of diverse bacterial genera implies that NHEJ is broadly relevant to bacterial physiology, notwithstanding that neither Ku nor LigD are essential in Mycobacterium, Bacillus or Pseudomonas under laboratory growth conditions (16–18,21). However, Ku and LigD are important in protecting Bacillus spores against DNA damage incurred under conditions of environmental stress (22).
Agrobacterium tumefaciens is a soil bacterium that parasitizes plants and causes crown gall disease. The Agrobacterium genome (23,24) consists of a 2.8-Mb circular chromosome, a 2.1-Mb linear chromosome, a 543-kb plasmid (pAT) and a 214-kb virulence plasmid (pTi). Transfer of the Ti plasmid to the host and integration of T DNA into the plant genome are the critical events underlying crown gall disease. T DNA integration is believed to occur via NHEJ, yet ablation of the Ku and Lig4 proteins that comprise the NHEJ machinery of the plant host has no effect on T DNA integration (25,26). One of the striking features of the A. tumefaciens proteome is the presence of six putative ATP-dependent DNA ligases in addition to the canonical NAD+-dependent LigA (Figure 1). The predicted product of ORF 0840 on the circular chromosome (which we now designate AtuLigB) is a 541-aa polypeptide homologous to M. tuberculosis LigB. AtuLigB (like MtuLigB) consists of a core ATP-dependent ligase domain fused to an N-terminal segment that resembles the DNA-binding domain (DBD) of human DNA ligase 1 (27) (Figure 1). AtuLigB and MtuLigB are members of a distinct ligase clade that includes mammalian DNA ligase III, archaeal ligases, poxvirus ligases and LigB homologs from diverse bacteria (e.g. Caulobacter crescentus, Pseudomonas putida, Xanthomonas campestris, Myxococcus xanthus and many others).
In the present study, we focus on two Agrobacterium homologs of Pseudomonas aeruginosa LigD, which we designated AtuLigD1 and AtuLigD2, respectively. Like PaeLigD, AtuLigD1 (771-aa) and AtuLigD2 (840-aa) consist of a central ATP-dependent ligase domain fused to an N-terminal phosphoesterase module and a C-terminal polymerase-like domain (28–33) (Figure 1). Mycobacterium LigD is composed of the same three domains, albeit arranged in a different order (16–19,34,35). AtuLigD1 is encoded by ORF 4632 on the linear chromosome, which is immediately adjacent to ORF 4631 that encodes a homolog of Ku (hereafter named AtuKu1). This operon arrangement suggests an NHEJ function for AtuLigD1 and AtuKu1. AtuLigD2 is encoded by ORF 5055 on the AT plasmid. The AtuLigD2 gene is located next to an oppositely transcribed gene cluster (a putative operon) that encodes two other Ku homologs (ORFs 5049 and 5050; designated AtuKu2 and AtuKu3) and a homolog of mycobacterial LigC (ORF 5051, designated LigC2). There are two additional LigC paralogs in A. tumefaciens: one encoded on the AT plasmid (ORF 5097; LigC1) and another on the Ti plasmid (ORF 6090; LigC3). The AtuLigC paralogs are minimal ligases (345–353-aa) consisting only of the nucleotidyltransferase and OB modules with no auxiliary flanking domains (Figure 1). Genetic evidence implicates mycobacterial LigC (which is nonessential for bacterial growth) in a minor pathway of Ku-dependent NHEJ that is evident when the major LigD-dependent NHEJ pathway is ablated (17). We can surmise that neither AtuLigD2, AtuKu2, AtuKu3, AtuLigC2, AtuLigC1 nor AtuLigC3 are essential for viability of A. tumefaciens, insofar as strains cured of pAT or pTi, or both plasmids, were reported to grow as well as a wild-type strain containing the pAT and pTi plasmids (36).
As an initial step in understanding how ligases D and C might contribute to a putative Agrobacterium NHEJ pathway, we purified and characterized Atu ligases D1, D2, C1, C2 and C3. We find that four of the five proteins (LigC1 being the exception) are able to seal a nicked duplex DNA substrate in vitro, albeit with relatively low turnover. We focus in detail on the additional enzymatic functions associated with AtuLigD1 and AtuLigD2, especially their catalysis of 3′ end-healing reactions relevant to DNA repair, as described previously for Pseudomonas LigD (28–33). Whereas AtuLigD2 has an associated polymerase activity, AtuLigD1 does not. We report that bacterial Ku stimulates the repair of DNA double-strand breaks in vitro by Agrobacterium ligases D2, C2 and C3.
MATERIALS AND METHODS
Agrobacterium DNA ligases
A DNA encoding AtuLigD1 was PCR-amplified from A. tumefaciens strain C58 genomic DNA (a gift of Dr Andrew Binns) using primers designed to introduce NheI sites at the start codon and 3′ of the stop codon. The PCR product was digested with NheI and inserted into pET28b (Novagen). DNA encoding AtuLigD2 was amplified using primers designed to introduce NdeI sites at the start site and 3′ of the stop codon. The PCR product was digested with NdeI and inserted into pET16b. DNAs encoding AtuLigC1, AtuLigC2 and AtuLigC3 were amplified by PCR with primers designed to introduce an NdeI site at the start site and a BamHI site 3′ of the stop codon. The PCR products were digested with NdeI and BamHI and inserted into pET16b. The inserts were sequenced completely to exclude the acquisition of unwanted changes during amplification and cloning.
The pET-AtuLig plasmids were transformed into Escherichia coli BL21(DE3). Cultures (1 l) of E. coli BL21(DE3)/pET-AtuLig were grown at 37°C in Luria–Bertani medium containing either 0.05 mg/ml kanamycin (for AtuLigD1) or 0.1 mg/ml ampicillin (for AtuLigD2) until the A600 reached 0.6. The cultures were adjusted to 0.5 mM isopropyl-β-d-thiogalactopyranoside and then incubated at 17°C for 15 h. Cells were harvested by centrifugation and the pellets were stored at −80°C. All subsequent steps were performed at 4°C. Thawed bacteria were resuspended in 50 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 1.5 M NaCl, 10% glycerol, 15 mM imidazole). Lysozyme and Triton X-100 were added to final concentrations of 50 µg/ml and 0.1%, respectively. The lysates were sonicated to reduce viscosity and insoluble material was removed by centrifugation. The supernatants were applied to 2-ml columns of Ni2+-nitrilotriacetic acid-agarose (Qiagen, Chatsworth, CA, USA) that had been equilibrated with lysis buffer. The columns were washed with 50 ml of lysis buffer and then eluted stepwise with 4-ml aliquots of buffer A (50 mM Tris-HCl, pH 7.5, 0.4 M NaCl, 10% glycerol) containing 50, 100, 200 and 500 mM imidazole. The polypeptide compositions of the fractions were monitored by SDS-PAGE. The LigC and LigD proteins were recovered predominantly in the 200 and 500 mM imidazole eluate fractions, respectively. Protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard. The recombinant protein preparations were stored at −80°C. The yields were as follows: AtuLigC1 (32 mg), AtuLigC2 (1.3 mg), AtuLigC3 (2 mg), AtuLigD1 (14 mg) and AtuLigD2 (1 mg).
Phosphoesterase domains of LigD1 and LigD2
Gene fragments encoding LigD1-(1–186) and LigD2-(1–193) (the PE domains) were amplified by PCR with sense strand primers that introduced an NdeI site at the start codon and antisense primers that introduced a new stop codon and a flanking BamHI site. The PCR products were digested with NdeI and BamHI and inserted into pET16b. Alanine mutations were introduced into the pET-AtuPE plasmids by overlap extension PCR. The pET-AtuPE plasmids were transformed into E. coli BL21(DE3). Induction of protein expression, preparation of soluble bacterial lysates, and purification of the recombinant PE proteins by Ni-agarose affinity chromatography were performed as described above for full-length Atu ligases. The PE proteins were recovered predominantly in the 200 and 500 mM imidazole eluates. The yields from 200-ml cultures were as follows: AtuLigD1 PE (14 mg) and AtuLigD2 PE (6 mg).
Polymerase domains of LigD1 and LigD2
Gene fragments encoding LigD1-(536–771) and LigD2-(569–884) (the POL domains) were amplified by PCR from pET-AtuLigD plasmids with sense strand primers that introduced an NheI site at the new start codon for AtuLigD1 POL and an NdeI site at the new start codon for AtuLigD2 POL. The PCR products were restricted and inserted into pET28b (for AtuLigD1 POL) or pET16b (for AtuLigD2 POL). Protein expression, preparation of soluble bacterial lysates, and purification of the recombinant PE proteins by Ni-agarose affinity chromatography were performed as described above. The POL proteins were recovered predominantly in the 200 mM imidazole eluates. The yields from 1-l cultures were as follows: AtuLigD1 POL (6 mg) and AtuLigD2 POL (15 mg).
Purification of Ku
The P. aeruginosa gene encoding the 293-aa Ku protein (PA2150) was amplified by PCR from genomic DNA with primers designed to introduce an NdeI site at the start site and a BamHI site 3′ of the stop codon. The PCR product was digested with NdeI and BamHI and inserted into pET16b. Induction of Ku expression, preparation of a soluble bacterial lysate from a 1-l culture, and purification of the recombinant Ku by Ni-agarose affinity chromatography were performed as described above, except that the Ni-agarose column was eluted stepwise with buffers containing 50, 100, 200, 500 and 1300 mM imidazole. Ku was recovered predominantly in the 500 and 1300 mM imidazole eluates, which contained 2.3 and 1.2 mg of protein, respectively.
Glycerol gradient sedimentation
Aliquots (40 µg) of the Ni-agarose preparations of AtuLigC, AtuLigD or AtuLigD PE or POL proteins, or PaeKu were mixed with catalase (30 µg), bovine serum albumin (30 µg) and cytochrome c (30 µg). The mixtures were applied to 4.8-ml 15–30% glycerol gradients containing 50 mM Tris-HCl (pH 8.0), 0.2 M NaCl, 1 mM EDTA, 2.5 mM DTT, 0.1% Triton X-100. The gradients were centrifuged at 50 000 r.p.m. in a Beckman SW50 rotor for 16 h at 4°C. Fractions (∼0.2 ml) were collected from the bottoms of the tubes. The polypeptide compositions of the gradient fractions were analyzed by SDS-PAGE. Aliquots of the fractions were assayed for ligase, POL or PE activity as specified in the figure legends.
Nick-sealing assay
A 24-bp DNA duplex containing a centrally placed 3′-OH/5′-PO4 nick was formed by annealing a 5′ 32P-labeled 12-mer DNA strand and an unlabeled 12-mer 3′-OH strand to a complementary 24-mer DNA strand as described previously (28). Ligation reaction mixtures (20 µl) containing 50 mM Tris buffer as specified, 5 mM DTT, 5 mM MnCl2, 1 pmol of 32P-labeled nicked DNA substrate, 250 µM ATP where specified, and ligases as specified were incubated for 20 min at 37°C. The reactions were quenched by adjusting the mixtures to 10 mM EDTA and 48% formamide. The products were resolved by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 M urea in TBE (90 mM Tris-borate, 2.5 mM EDTA). The products were visualized by autoradiography and quantified by scanning the gel with a Fujifilm BAS-2500 imaging apparatus.
3′-Ribonuclease and 3′-phosphomonoesterase assays
The 5′ 32P-labeled D10R2 and D11p primer-templates were prepared as described previously (31,32). Reaction mixtures (10 µl) containing 50 mM Tris-acetate (pH 6.0), 5 mM DTT, 0.5 mM MnCl2, 0.5 pmol 32P-labeled D10R2 or D11p primer-templates, and AtuLig PE domain as specified were incubated at 37°C for 20 min. The reactions were quenched by adjusting the mixtures to 7 mM EDTA and 31% formamide. The products were resolved by electrophoresis through a 40-cm 18% polyacrylamide gel containing 7 M urea in TBE.
Polymerase assay
Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 100 µM each of ATP, GTP, CTP and UTP (rNTPs) or 100 µM each of dATP, dGTP, dCTP and dTTP (dNTPs), 1 pmol 5′ 32P-labeled 12-mer/24-mer primer-template, and AtuPOL proteins were incubated at 37°C for 20 min. The reactions were quenched by adjusting the mixtures to 10 mM EDTA and 48% formamide. The products were resolved by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 M urea in TBE.
RESULTS
Agrobacterium LigC paralogs
Ligases C1, C2 and C3 are 350-aa, 345-aa and 353-aa polypeptides composed of N-terminal nucleotidyltransferase and C-terminal OB domains (Figure 1). Within the nucleotidyltransferase module is an adenylate-binding pocket composed of six motifs (I, Ia, III, IIIa, IV and V; highlighted in Supplemental Figure S1) that define the polynucleotide ligase/mRNA capping enzyme superfamily of covalent nucleotidyltransferases. Motif I (KWDGYR) includes the lysine nucleophile to which AMP becomes covalently linked in the first step of the ligase reaction. Motifs Ia, III, IIIa, IV and V contain conserved amino acids that contact AMP and play essential roles in one or more steps of the ligation pathway. Located near the C-terminus of the OB domain is motif VI, which is important for formation of the ligase-adenylate intermediate. Motif VI of the AtuLigC paralogs (RExQ) deviates from the canonical motif VI (RxDK) found in the majority of ATP-dependent ligases.
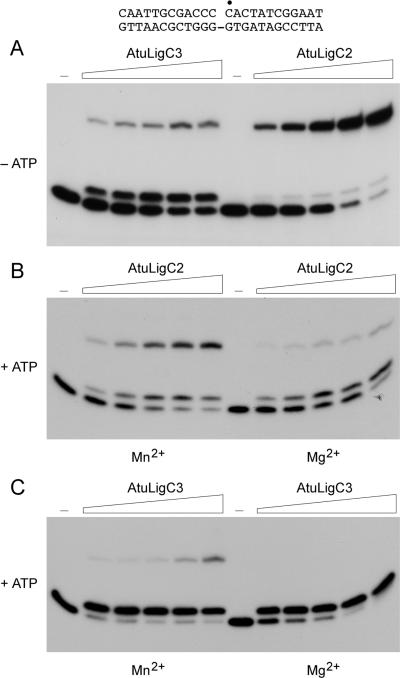
To evaluate the biochemical properties of the AtuLigC proteins, we produced them in E. coli as His10-AtuLigC fusions and purified them from soluble extracts by adsorption to nickel-agarose resin and elution with buffer containing imidazole. SDS-PAGE revealed that each preparation contained a predominant 40 kDa polypeptide corresponding to His10-AtuLigC (Figure 1). A synthetic duplex DNA substrate containing a single nick (Figure 2) was used to gauge the DNA-sealing activity of the AtuLigC proteins, which was evinced by the conversion of the 5′ 32P-labeled 12-mer strand to a 24-mer product. We observed no nick-sealing activity for two independent preparations of recombinant AtuLigC1. Therefore, all further studies of AtuLigC were conducted with the LigC2 and LigC3 isozymes. AtuLigC2 catalyzed nick sealing in the absence of added ATP (Figure 2A); this activity was ascribed to preformed LigC-AMP in the enzyme preparations. The extent of ATP-independent sealing was proportional to input AtuLigC2 and there was little accumulation of the AppDNA intermediate (which migrates ∼1.5 nt steps slower than the 12-mer pDNA strand). From the slope of the titration curve, we estimated that ligase-AMP comprised 16% of the AtuLigC2 preparation. ATP-independent nick sealing by AtuLigC2 required a divalent cation cofactor, which could be manganese, magnesium or cobalt (not shown). Calcium, copper and zinc did not support nick sealing (not shown). Manganese-dependent sealing was optimal over a range of pH values from 6.0 to 8.0 in Tris-acetate or Tris-HCl buffers (not shown). Lowering the pH to 4.5 in Tris-acetate resulted in complete inhibition of sealing and the accumulation of DNA-adenylate (AppDNA) as the sole product (not shown).
Figure 2.
Nick-sealing activity of AtuLigC2 and AtuLigC3. (A) Reaction mixtures (20 µl) containing either 50 mM Tris-HCl (pH 7.5) (AtuLigC2) or 50 mM Tris-acetate (pH 6.0) (AtuLigC3), 5 mM DTT, 5 mM MnCl2, 1 pmol 32P-labeled nicked 24-mer DNA substrate (depicted at the top with the labeled 5′ nucleotide at the nick indicated by filled circle), and increasing amounts of AtuLigC2 (38, 75, 150, 300 and 600 ng) or AtuLigC3 (50, 100, 200, 400 and 800 ng) were incubated for 20 min at 37°C. Enzyme was omitted from a control reaction mixture (lane −). The products were resolved by PAGE and visualized by autoradiography. (B) Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2 (left panel) or 5 mM MgCl2 (right panel), 250 µM ATP, 1 pmol 32P-labeled nicked 24-mer DNA substrate, and increasing amounts of AtuLigC2 (38, 75, 150, 300 and 600 ng) were incubated at 37°C for 20 min. Enzyme was omitted from a control reaction mixture (lane −). (C) Reaction mixtures (20 µl) containing 50 mM Tris-acetate (pH 6.0), 5 mM DTT, 5 mM MnCl2 (left panel) or MgCl2 (right panel), 250 µM ATP, 1 pmol 32P-labeled nicked 24-mer DNA substrate, and increasing amounts of AtuLigC3 (50, 100, 200, 400 and 800 ng) were incubated at 37°C for 20 min. Enzyme was omitted from a control reaction mixture (lane −).
ATP failed to stimulate AtuLigC2 nick-sealing activity under otherwise optimal conditions because it elicited the accumulation of high levels of the DNA-adenylate intermediate (Figure 2B). This effect was more pronounced when magnesium was the divalent cation cofactor. At the highest level of input AtuLigC2 tested in this experiment, in which nearly all the pDNA strand had reacted, AppDNA comprised 56% of total labeled DNA (Figure 2B, Mg2+). This ATP-dependent trapping phenomenon, which was described previously for eukaryotic viral DNA ligases (37,38) and mycobacterial DNA ligases C and D (16), results from dissociation of the enzyme from the DNA-adenylate and immediate reaction of the free ligase with ATP to yield ligase-AMP, which cannot rebind to AppDNA and thus cannot catalyze the phosphodiester formation step (there being only one adenylate-binding pocket on the enzyme).
AtuLigC3 reacted with the nicked substrate in the absence of ATP, but the major product was the AppDNA intermediate rather than the ligated 24-mer (Figure 2A). At the highest level of LigC3 tested, AppDNA comprised 41% of the labeled material while only 16% of the label was sealed 24-mer. From the slope of the titration curve, we estimated that 12% of the AtuLigC3 preparation was preformed ligase-AMP. Thus, AtuLigC3 appeared to dissociate readily from the AppDNA intermediate and was loathe to rebind and seal the adenylylated strand even in the absence of an ATP trap. ATP-independent sealing by AtuLigC3 was optimal at pH 6.0–6.5 with manganese as the cofactor (not shown). Cobalt supported nick sealing by AtuLigC3, but magnesium was less active and calcium, copper and zinc were ineffective (not shown). Inclusion of ATP stimulated formation of the AppDNA intermediate (by at least a factor of 6, calculated from the extent of total product formation as a function of input LigC3), while suppressing formation of the ligated 24-mer (compare Figures 2C-Mn2+ and 2A). This effect was evident in the presence of manganese or magnesium (Figure 2C).
The quaternary structures of AtuLigC2 and AtuLigC3 were examined by zonal velocity sedimentation in 15–30% glycerol gradients. Marker proteins catalase (native size 248 kDa), BSA (66 kDa) and cytochrome c (12 kDa) were included as internal standards in the gradients. The ligase activity profiles for AtuLigC2 and AtuLigC3 comprised single peak components sedimenting between BSA and cytochrome c (Supplementary Figure S2), which paralleled the abundance of the AtuLigC2 and AtuLigC3 polypeptides (not shown). A plot of the S values of the three standards versus fraction number yielded a straight line. We calculated S values of 3.4 for AtuLigC2 and 3.4 for AtuLigC3 by interpolation to the internal standard curves. The results are consistent with a monomeric quaternary structure for AtuLigC2 and AtuLigC3. In summary, these experiments show that Agrobacterium LigC2 and LigC3 resemble M. tuberculosis LigC in that they have feeble DNA nick-sealing activity in the presence of ATP.
Agrobacterium LigD paralogs
LigD1 and LigD2 are 771-aa and 884-aa polypeptides composed of a central ligase domain flanked by putative N-terminal phosphoesterase (PE) and C-terminal polymerase (POL) domains. The ligase domains include all of the canonical nucleotidyltransferase motifs (Supplementary Figure S3). We produced AtuLigD1 and AtuLigD2 in E. coli as His10-AtuLigC fusions and purified them from soluble extracts by nickel-agarose chromatography. SDS-PAGE revealed that the LigD1 and LigD2 preparations contained major polypeptides of 90 and 102 kDa, corresponding to the respective His-tagged LigD proteins (Figure 1). AtuLigD1 and AtuLigD2 migrated slightly faster and slower, respectively, during SDS-PAGE than did Pseudomonas LigD (840-aa; ∼97 kDa by SDS-PAGE).
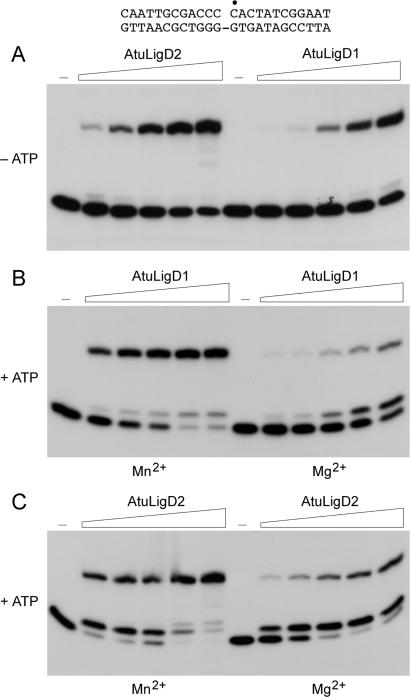
AtuLigD1 and AtuLigD2 catalyzed nick sealing in the absence of added ATP, with little accumulation of AppDNA (Figure 3A). From the slopes of the titration curves, we estimated that ligase-AMP comprised 5 and 27% of the AtuLigD1 and AtuLigD2 preparations, respectively. ATP-independent nick sealing by AtuLigD1 and AtuLigD2 required a divalent cation cofactor, which could be manganese, magnesium or cobalt. Calcium, copper and zinc did not support activity (data not shown). Figure 3B shows that inclusion of ATP stimulated the manganese-dependent activity of AtuLigD1 by a factor of 8 (calculated from the extent of total product formation as a function of input LigD1), with relatively little trapping of the AppDNA intermediate. In contrast, when magnesium was the divalent cation, the enzyme was 6-fold less reactive overall (as judged by consumption of substrate) and AppDNA was the majority product (Figure 3B, Mg2+ versus Mn2+). ATP stimulated the manganese-dependent reaction of AtuLigD2 with nicked DNA by a factor of 8, with AppDNA accumulating at lower enzyme concentrations and ligated 24-mer predominating at higher levels of input LigD2 (Figure 3C). ATP trapping of the AppDNA intermediate during the LigD2 reaction was more pronounced in the presence of magnesium (Figure 3C).
Figure 3.
Nick-sealing activity of AtuLigD1 and AtuLigD2. (A) Reaction mixtures (20 µl) containing either 50 mM Tris-HCl (pH 7.5) (AtuLigD1) or 50 mM Tris-acetate (pH 6.0) (AtuLigD2), 5 mM DTT, 5 mM MnCl2, 1 pmol 32P-labeled nicked 24-mer DNA substrate (shown at the top), and increasing amounts of AtuLigD1 (63, 125, 250, 500 and 1000 ng) or AtuLigD2 (31, 63, 125, 250, 500 ng) were incubated for 20 min at 37°C. (B) Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2 (left panel) or MgCl2 (right panel), 250 µM ATP, 1 pmol 32P-labeled nicked 24-mer DNA substrate, and increasing amounts of AtuLigD1 (63, 125, 250, 500, 1000 ng) were incubated at 37°C for 20 min. (C) Reaction mixtures (20 µl) containing 50 mM Tris-acetate (pH 6.0), 5 mM DTT, 5 mM MnCl2 (left panel) or MgCl2 (right panel), 250 µM ATP, 1 pmol 32P-labeled nicked 24-mer DNA substrate, and increasing amounts of AtuLigD2 (31, 63, 125, 250, 500 ng) were incubated at 37°C for 20 min. Enzyme was omitted from control reaction mixtures (lanes –).
AtuLigD1 and AtuLigD2 were sedimented in 15–30% glycerol gradients with internal markers catalase, BSA and cytochrome c (Supplementary Figure S4). The nick-sealing activity profiles for AtuLigD1 and AtuLigD2 comprised single peaks that coincided with the AtuLigD1 and AtuLigD2 polypeptides. We calculated S values of 4.5 for AtuLigD1 and 5.5 for AtuLigD2 by interpolation to the internal standard curves. These results are consistent with monomeric quaternary structures for AtuLigD1 and AtuLigD2. (The S value of 4.5 for AtuLigD1 is slightly lower than expected for a globular protein of 90 kDa, suggesting that it might have an elongated shape.)
Activities of the POL domain of Agrobacterium LigD2
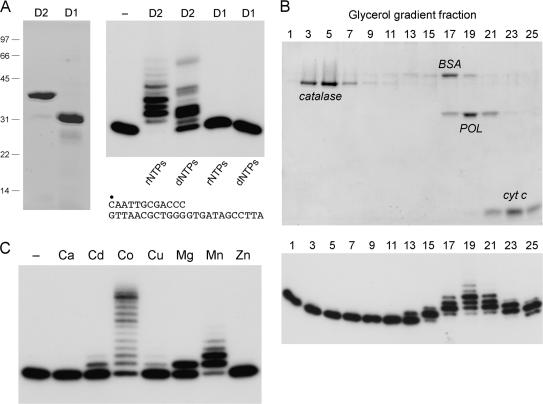
The primary structure of the C-terminal domain of AtuLigD2 from aa 569 to 884 is similar throughout its length to the POL domain of Pseudomonas LigD (aa 533–840) (Figure S3). In contrast, the C-terminal module of AtuLigD1 is truncated by 63-aa compared to AtuLigD2 (Figure S3). To assess what activities, if any, are associated with the AtuLigD2 and AtuLigD1 POL domains, we produced AtuLigD2-(569–884) and AtuLigD1-(536–771) in E. coli as His10 fusions and purified them from soluble extracts by Ni-agarose chromatography. SDS-PAGE showed that the preparations were highly enriched with respect to the recombinant D2 and D1 POL domains (Figure 4A, left panel). The LigD2 POL domain was capable of template-directed DNA or RNA synthesis, as gauged by its ability to extend a primer-template composed of a 5′ 32P-labeled 12-mer DNA strand annealed to a complementary 24-mer strand (Figure 4A, right panel). In contrast, the LigD1 POL domain displayed no polymerase activity with rNTP or dNTP substrates (Figure 4A, right panel). A mixing experiment revealed that primer extension by LigD2 POL was unaffected by addition of an equal amount of LigD1 POL (not shown); thus the inactivity of the LigD1 POL protein was not caused by an inhibitor in the preparation. We surmise that the C-terminal truncation of AtuLigD1 compromises its POL function. Thus, AtuLigD2 and AtuLigD1 are not functionally equivalent.
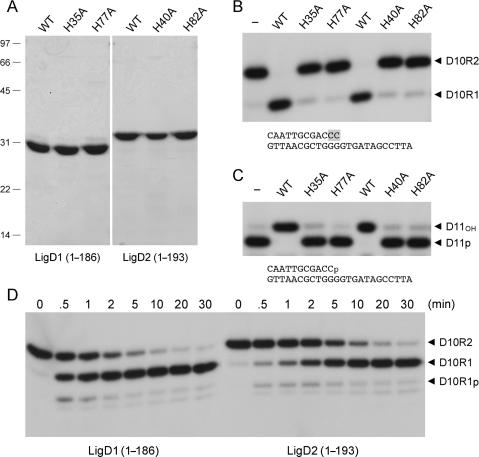
Figure 4.
Recombinant LigD2 and LigD1 POL domains. (A) Aliquots (7 µg) of the Ni-agarose preparations of the POL domains of AtuLigD2 and AtuLigD1 were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown in the left panel. Polymerase reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 100 µM rNTPs or dNTPs as specified, 50 nM 5′ 32P-labeled 12-mer/24-mer DNA primer-template (depicted at bottom) and 1 µg of LigD2 or LigD1 POL as specified were incubated at 37°C for 20 min. The products were analyzed by denaturing PAGE. An autoradiograph of the gel is shown in the right panel. POL was omitted from the control reaction in lane −. (B) Glycerol gradient sedimentation was performed as described under Experimental Procedures. Aliquots (15 µl) of the odd-numbered fractions were analyzed by SDS-PAGE (top panel). The catalase, BSA, LigD2 POL and cytochrome c polypeptides are indicated. Polymerase activity was gauged by incubating 20 µl reaction mixtures containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 100 µM rNTPs, 1 pmol 5′ 32P-labeled 12-mer/24-mer DNA primer-template and 2 µl of the indicated gradient fractions at 37°C for 20 min. PAGE analysis of the primer extension products is shown in the bottom panel. (C) Metal specificity. Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 7.5), 100 µM rNTPs, 50 nM 5′ 32P-labeled 12-mer/24-mer DNA primer-template, 156 nM LigD2 POL, and either no divalent cation (–) or 5 mM Ca2+, Cd2+, Co2+, Cu2+, Mg2+, Mn2+ or Zn2+ were incubated at 37°C for 20 min.
The native size of the active LigD2 POL domain was gauged by glycerol gradient sedimentation. The 38 kDa LigD2 POL polypeptide sedimented as a discrete component (peaking in fraction 19) between the BSA and cytochrome c markers (Figure 4B, top panel), coincident with the peak of the templated ribonucleotide addition activity (Figure 4B, bottom panel). An S value of 3.7 for LigD2 POL was calculated by interpolation to the internal standard curve. These results are consistent with a monomeric quaternary structure for LigD2 POL.
Templated ribonucleotide addition by the POL domain required a divalent cation cofactor (Figure 4C). Testing various metals at 5 mM concentration showed that activity, expressed as the percent of the input primer that had been extended, was optimal with manganese (89% extended) or cobalt (80% extended). Whereas most of the products formed in manganese were elongated by one or two nucleotides, the cobalt-directed products comprised a ladder extending to the end of the template strand (Figure 4C). Magnesium was less effective (43% of primer extended) and the reaction was limited to a single step of rNMP addition (Figure 4C). Cadmium and copper were weak activators of the polymerase; calcium and zinc were ineffective (Figure 4C). Manganese, cobalt and magnesium displayed relative activities and product distributions similar to that seen in Figure 6C over a concentration range of 0.3–10 mM (not shown).
Figure 6.
End-healing activities of the AtuLigD1 and AtuLigD2 PE domains. (A) Aliquots (5 µg) of the Ni-agarose preparations of the wild-type (WT) and mutated versions of the N-terminal PE domains of AtuLigD1 and AtuLigD2 were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Reaction mixtures (10 µl) containing 50 mM Tris-acetate (pH 6.0), 5 mM DTT, 0.5 mM MnCl2, 50 nM pmol 32P-labeled D10R2 primer-template (shown at bottom, with ribonucleotides highlighted in shaded boxes), and 4 µM WT or mutant PE domain as specified were incubated at 37°C for 20 min. The products were resolved by PAGE and visualized by autoradiography. The labeled species corresponding to the D10R2 substrate and the D10R1 end-product are indicated by arrowheads on the right. (C) Reaction mixtures (10 µl) containing 50 mM Tris-acetate (pH 6.0), 5 mM DTT, 0.5 mM MnCl2, 50 nM 32P-labeled D11p primer-template (shown at bottom) and 4 µM WT or mutant PE domain as specified were incubated at 37°C for 20 min. The products were analyzed by PAGE and visualized by autoradiography. The labeled species corresponding to the D11p substrate and the D11OH product are indicated by arrowheads on the right. (D) Reaction mixtures (90 µl) containing 50 mM Tris-acetate (pH 6.0), 5 mM DTT, 0.5 mM MnCl2, 100 nM 32P-labeled D10R2 primer-template and 4 µM AtuLigD1 or AtuLigD2 PE as specified were incubated at 37°C. Aliquots (10 µl) were withdrawn at the times specified above the lanes and quenched immediately with EDTA/formamide. The products were resolved by PAGE and visualized by autoradiography. The labeled species corresponding to the D10R2 substrate, the D10R1p intermediate and the D10R1 end-product are indicated by arrowheads on the right.
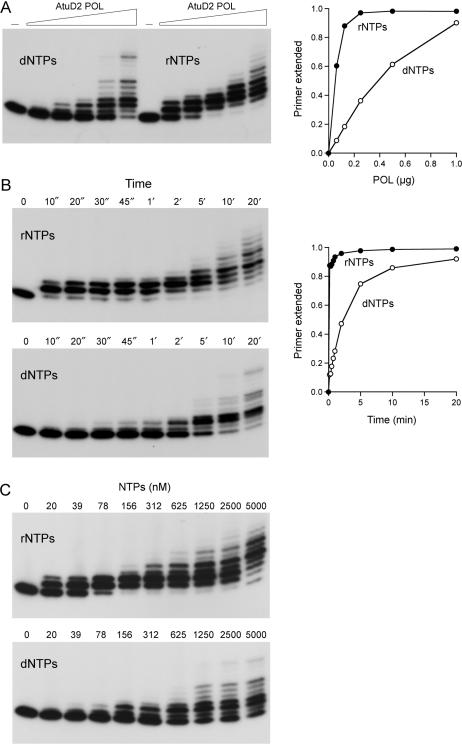
Further characterization of the POL activity is shown in Figure 5. When provided with manganese and a mixture of the four dNTPs or rNTPs (100 µM of each nucleotide), increasing concentrations of input POL catalyzed progressive elongation of the 12-mer primer to yield a ladder of products 13–18 nt long (Figure 5A, left panel). Very little of the primer was elongated to the end of the template strand, even under conditions of POL excess when all of the primer had been extended. The efficiency of the extension reaction was quantified as the percent of the input 12-mer primer strand that was elongated by at least one nucleotide. A plot of primer utilization versus input POL showed that ribonucleotide substrates were preferred over deoxynucleotides by a factor of 8 (Figure 5A, right panel).
Figure 5.
Preferential utilization of ribonucleotides by LigD2 POL. (A) Enzyme titration. Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 50 nM 5′ 32P-labeled 12-mer/24-mer DNA primer-template, 100 µM rNTPs or dNTPs as specified, and increasing amounts of LigD2 POL (62, 125, 250, 500 and 1000 ng, from left to right in each titration series, corresponding to 78, 156, 312, 625 and 1250 nM POL) were incubated at 37°C for 20 min. PAGE analysis of the primer extension products is shown in the left panel. POL was omitted from the control reaction in lane –. The fraction of input primer extended by at least one nucleotide is plotted as a function of input POL in the right panel. (B) Kinetics of primer extension. Reaction mixtures (220 µl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 50 nM 5′ 32P-labeled 12-mer/24-mer DNA primer-template, 100 µM rNTPs or dNTPs as specified, and 1.25 µM LigD2 POL were incubated at 37°C. Aliquots (20 µl) were withdrawn at the times specified and quenched immediately with EDTA/formamide. PAGE analysis of the primer extension products is shown in the left panels. The fraction of input primer extended by at least one nucleotide is plotted as a function of time in the right panel. (C) NTP concentration dependence. Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 50 nM 5′ 32P-labeled 12-mer/24-mer DNA primer-template, 1.25 µM LigD2 POL and rNTPs or dNTPs as specified were incubated at 37°C for 20 min.
A kinetic analysis of primer extension by excess POL in the presence of 100 µM rNTPs or dNTPs is shown in Figure 5B. The first cycle of ribonucleotide addition was rapid, with nearly all of the 12-mer primer elongated to 13-mer (with some 14-mer) at 10 s, the earliest timepoint in the experiment. We estimated from the 10 s datum an apparent rate constant of ≥0.21 s−1 for the first rNMP incorporation step. It was apparent that the second cycle of templated ribonucleotide addition was much slower, insofar as the 13-mer product formed within 10 s was gradually converted to 14-mer or longer product over the subsequent 5 min. By quantifying the flux through the 13-mer species (not shown), we estimated a rate constant of 0.016 s−1 for the second cycle of rNMP incorporation. The 13-fold rate decrement after the first cycle of ribonucleotide addition by AtuLigD2 POL agrees well with the findings for Pseudomonas LigD POL (30), which becomes progressively less active with each ribonucleotide added at the 3′ primer terminus (31). The kinetics of dNMP incorporation by AtuLigD2 POL were much slower than extension with rNTPs (Figure 5B). The first cycle of dNMP addition displayed a pseudo-first order pattern with an apparent rate constant of 0.0062 s−1. Thus, by comparing the rate constants, LigD2 POL is at least 34-fold faster at incorporating a ribonucleotide than a deoxynucleotide during the first reaction cycle at a DNA primer end.
The effect of limiting the rNTP pool is shown in Figure 5C (top panel), where we see that LigD2 POL was remarkably efficient at scavenging ribonucleotide substrate and adding it to the primer-template at stoichiometric ratios of rNTP to primer. For example, virtually all of the primer was elongated at least once during the 20-min reaction at a 3:1 ratio of rNTP to primer-template (156:50 nM). At limiting rNTP concentrations—20 and 39 nM—the yields of extended primer were 19 and 33 nM, respectively. LigD2 POL was ∼8-fold less effective at scavenging dNTPs at the same limiting concentrations (Figure 5C bottom panel).
3′ End-healing activities of the Agrobacterium LigD1 and LigD2 PE domains
Pseudomonas LigD (PaeLigD) has a 3′-ribonuclease/3′-phosphatase activity, whereby it resects a short tract of 3′-ribonucleotides on a primer-template substrate to the point at which the primer strand has a single 3′-ribonucleotide remaining (31–33). The failure to digest beyond this point reflects a requirement for a 2′-OH group on the penultimate nucleoside of the primer strand. The ribonucleotide resection activity resides within the 187-aa N-terminal PE domain and is the result of at least two component steps: (i) the 3′-terminal nucleoside is first removed to yield a primer strand with a ribonucleoside 3′-PO4 terminus; (ii) the 3′-PO4 is hydrolyzed to a 3′-OH. The 3′-ribonuclease and 3′-phosphatase activities are both dependent on manganese. The PaeLigD PE domain also catalyzes hydrolysis of the 3′-PO4 of an all-DNA primer-template substrate (32).
The primary structures of the N-terminal segments of AtuLigD1 and AtuLigD2 resemble that of the PaeLigD PE domain (Supplementary Figure S3), which has no apparent structural or mechanistic similarity to any previously characterized nucleases or 3′-phosphatases. Extensive mutational analysis of the PaeLigD PE domain has identified an ensemble of side chain functional groups that are essential for phosphoesterase activity, which is conserved in the two Agrobacterium LigD proteins (32,33). In order to probe whether the AtuLigD paralogs possess an end-healing function, we produced the N-terminal PE domains of AtuLigD1 (aa 1–186) and AtuLigD2 (aa 1–193) in E. coli as His10 fusions and purified them from soluble bacterial lysates by Ni-agarose chromatography. SDS-PAGE analysis verified that the preparations were enriched to the same extent with respect to the LigD1 and LigD2 PE polypeptides, which migrated anomalously at ∼30 kDa (predicted size 24 kDa) and ∼33 kDa (predicted size 25 kDa), respectively (Figure 6A). This electrophoretic anomaly echoes the properties of the Pseudomonas PE domain (32). We also produced and purified two mutated versions of each of the Atu PE domains, in which the conserved counterparts of essential PaeLigD residues His42 and His84 were substituted by alanine. The purities of the AtuLigD1 H35A and H77A PE proteins and the AtuLigD2 H40A and H82A PE mutants were comparable to the respective wild-type versions (Figure 8A).
Figure 8.
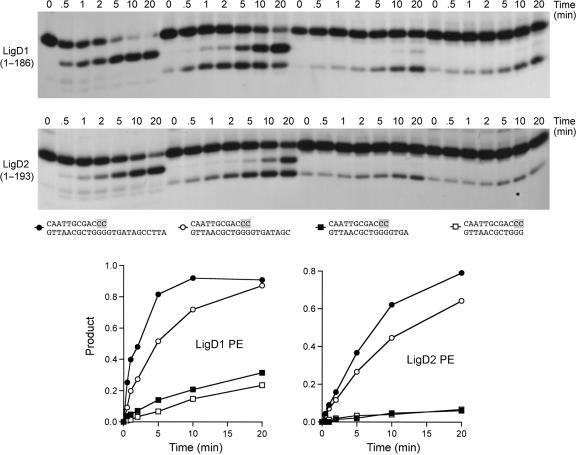
Effect of the template strand 5′ tail on AtuLigD PE activity. Reaction mixtures (80 µl) containing 50 mM Tris-acetate (pH 6.0), 5 mM DTT, 0.5 mM MnCl2, 50 nM 32P-labeled D10R2 primer-templates with variable 5′ tails and 4 µM AtuPE1 or AtuPE2 were incubated at 37°C. Aliquots (10 µl) were withdrawn at the times specified and quenched immediately with EDTA/formamide. The products were resolved by PAGE. The structures of the primer-templates are depicted below the autoradiograms, with ribonucleotides highlighted in shaded boxes. The kinetic profiles of the reactions were quantified by scanning the gel. Phosphodiesterase activity, expressed as the fraction of radiolabeled material in the D10R1-p plus D10R1 products, is plotted as a function of time. The substrate symbols are as depicted below the autoradiograms.
The PE proteins were reacted with a 5′ 32P-labeled D10R2 primer-template composed of a 12-mer primer strand with two terminal ribonucleotides and a 24-mer DNA template strand (Figure 6B). The wild-type AtuLigD PE domains converted virtually all of the input labeled strand to a more rapidly migrating end-product, D10R1 (Figure 6B). This species migrated identically to the D10R1 product generated by the Pseudomonas LigD PE domain (not shown). A kinetic analysis of the ribonucleotide resection reaction in enzyme excess showed the transient appearance of small amounts of D10R1p at early times (Figure 6D), suggesting that the Atu PE domains remove the terminal ribonucleotide in a two-step pathway similar to that described for the Pseudomonas LigD PE domain (31). The 3′-phosphomonoesterase activity of the AtuLigD PE domains was demonstrated directly by reaction with a 5′ 32P-labeled D11p primer-template composed of a 3′-phosphate-terminated 11-mer primer strand annealed to a 24-mer DNA strand (Figure 6C). The wild-type AtuLigD PE domains converted the input D11p strand to a more slowly migrating 3′-OH end-product, D11OH (Figure 6C). The 3′-ribonucleotide resection and polynucleotide 3′-phosphatase activities of the AtuLigD1 and AtuLigD2 PE domains were abolished by each of the histidine-to-alanine mutations (Figure 6B and C). These results verify that the 3′ ribonuclease and 3′ phosphomonoesterase activities are intrinsic to the AtuLigD PE domains and that they rely on the same essential histidines as the Pseudomonas homolog.
The quaternary structures of the PE domains were examined by zonal velocity sedimentation in a 15–30% glycerol gradient with marker proteins catalase, BSA and cytochrome c included as internal standards. After centrifugation, the polypeptide compositions of the odd-numbered gradient fractions were analyzed by SDS-PAGE and aliquots of the fractions were assayed for shortening of the D10R2 primer-template. The LigD1 and LigD2 PE proteins sedimented as a discrete peaks between BSA and cytochrome c, coincident with the 3′ ribonuclease activity (not shown). We infer that the isolated PE domains are monomers.
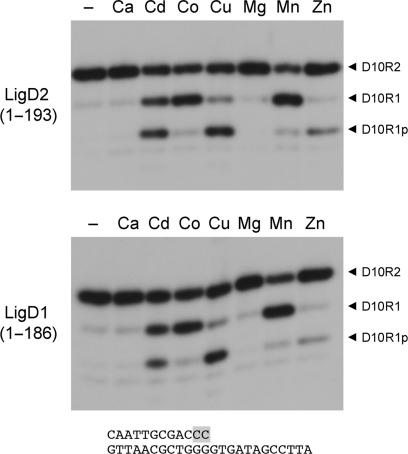
The 3′ ribonucleotide resection activity of the AtuLigD PE domain was optimal at pH 5.5–8.0 in 50 mM Tris-acetate or Tris-HCl buffer (data not shown) and was strictly dependent on a divalent cation, preferably manganese or cobalt (Figure 7). A manganese titration experiment showed that the 3′ ribonuclease activity was optimal at 0.2–5 mM MnCl2 (not shown). Magnesium and calcium were ineffective at 0.5 mM concentration. Cadmium, copper and zinc (0.5 mM) were capable of sustaining progressively reduced activity that resulted in the formation of the initial D10R1p product, little of which was converted to the D10R1OH end-product in the presence of copper or zinc (Figure 7).
Figure 7.
Divalent cation specificity of the 3′-ribonuclease activity of AtuLigD PE domains. Reaction mixtures (10 µl) containing 50 mM Tris-acetate (pH 6.0), 100 nM 32P-labeled D10R2 primer-template, 0.5 mM divalent cation as specified, and either 1 µM AtuLigD1 PE (bottom panel) or 2 µM AtuLigD2 PE (top panel) were incubated at 37°C for 20 min.
The experiments in Figure 8 probed the contribution of the 5′ single-strand tail of the primer-template to the ribonucleotide resection reactions. We annealed the labeled D10R2 primer to a series of incrementally shortened template strands to form substrates with 12-, 8- or 4-nt 5′ tails attached to identical 12-bp duplex segments, and a substrate consisting only of the 12-bp duplex with no single-strand tail. We examined the kinetics of the reaction of the AtuLigD1 PE domain with this set of primer-templates under conditions of enzyme excess (Figure 8, top panel). As noted above, the labeled D10R2 strand of the 12-mer/24-mer substrate was converted to a 3′-OH end-product, D10R1, with low levels of the 3′-phosphorylated species, D10R1-p, appearing transiently at 0.5–1 min. The extent of substrate decay and the distribution of the reaction products were strongly affected by shortening of the 5′ tail (Figure 8, top panel). The primer-template containing an 8-nt 5′ tail was consumed at 44% of the rate of the ‘standard’ DNA with the 12-mer tail (Figure 8, bottom left panel) and there was a clear delay in the conversion of the D10R1-p intermediate to the D10R1 end-product, i.e. the intermediate persisted for the entire time course and exceeded the level of the end-product for the first 5 min of the reaction (Figure 8, top panel). Further shortening of the tail to 4 nt slowed the rate of substrate consumption to 7% of the standard primer-template (Figure 8, bottom panel) and there was scant formation of the D10R1 end-product. The reaction rate on the blunt duplex was 4% of the rate on the standard primer-template. Similar effects of tail shortening on the rate of resection and the distribution of the D10R1-p and D10R1 products were observed for the AtuLigD2 PE domain (Figure 8, middle and lower panels).
Ku stimulates repair of double-strand breaks by Atu ligases C2, C3, and D2
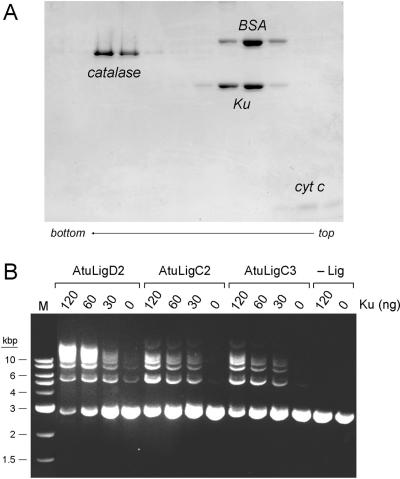
The DNA-binding protein Ku is critical for NHEJ-mediated repair of linear plasmid DNA in mycobacteria (17). Mycobacterial Ku interacts physically and functionally with mycobacterial LigD (17–19,35). In particular, Ku stimulates the joining of linear DNA fragments by mycobacterial LigD (18). In light of the genetic evidence that LigC functions in a Ku-dependent NHEJ pathway in vivo (17), it was of interest to query whether Ku might affect also the strand-sealing activity of LigC in vitro. The abundance of LigC and LigD enzymes in Agrobacterium affords a unique opportunity to address the Ku-ligase connections. However, the situation is complicated by the multiple Ku isoforms in the Agrobacterium proteome. Mycobacteria specify a single Ku protein, which has a homodimeric quaternary structure (18). Eukaryal Ku is a heterodimer of separately encoded subunits (39). The homomeric and heteromeric association properties of the three Agrobacterium Ku polypeptides are not known and the combinatorial possibilities are daunting. Our initial efforts to produce and purify recombinant His-tagged versions of the individual AtuKu proteins were not fruitful, e.g. Ku1 and Ku2 were not expressed in E. coli. Consequently, we produced and purified the single Ku protein encoded by P. aeruginosa (PaeKu; locus tag PA2150; Genbank accession NC_002516). PaeKu was chosen as a stand-in for Agrobacterium Ku in light of the shared domain organization and biochemical activities of Pseudomonas LigD and Agrobacterium LigD2. Analysis of the quaternary structure of recombinant PaeKu by zonal velocity sedimentation in a glycerol gradient revealed it to be a single discrete component that overlapped the ‘heavy’ side of the BSA marker (Figure 9A). We surmise that PaeKu is a homodimer of the recombinant 35 kDa Ku polypeptide.
Figure 9.
Ku stimulation of double-strand break repair by Atu ligases. (A) Homodimeric quaternary structure of Ku. Glycerol gradient sedimentation of PaeKu was performed as described under Experimental Procedures. Aliquots (15 µl) of odd-numbered gradient fractions were analyzed by SDS-PAGE. The catalase, BSA, Ku and cytochrome c polypeptides are indicated. (B) Plasmid end joining. Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 0.25 mM ATP, 0.2 µg BamHI-digested pUC19 DNA (120 fmol plasmid; 240 fmol double-strand breaks), either AtuLigD2 (250 ng), AtuLigC2 (75 ng; 1.9 pmol) or AtuLigC3 (125 ng; 3.1 pmol), and PaeKu as indicated were incubated for 2 h at 37°C. The reactions were quenched by adjusting the mixtures to 0.2% SDS and 6 mM EDTA. The products were resolved by electrophoresis through a 1% agarose gel in TBE containing 0.05% ethidium bromide. A photograph of the gel under UV illumination is shown. The positions and sizes (kbp) of linear duplex DNA markers (lane M) are indicated on the left. The products of ligation are linear concatemers. A closed monomer circle migrates ahead of the linear monomer; a nicked monomer circle migrates between the linear monomer and linear dimer.
To approximate an NHEJ-like double-strand break repair scenario in vitro, we tested the ability of Atu ligases to join linear pUC19 plasmid DNA that had been digested with BamHI. The reaction mixtures contained ∼120 fmol plasmid DNA, corresponding to 240 fmol double-strand breaks. Whereas addition of a molar excess of AtuLigC2 (1.9 pmol) or AtuLigC3 (3.1 pmol) over the available DNA ends resulted in little end joining, the inclusion of Ku clearly stimulated end joining, as evinced by the appearance of a ladder of linear concatemers, comprising dimer, trimer, tetramer and pentamer products (Figure 9B). The Ku-mediated stimulation of break repair by LigC2 and LigC3 was proportional to the amount of Ku added in the range of 30, 60 and 120 ng, corresponding to 0.43, 0.86 and 1.7 pmol of Ku homodimer. Ku by itself (120 ng) had no strand-joining activity (Figure 9B). The absence of monomer circles among the reaction products signifies that Ku promotes intermolecular repair events by LigC2 and LigC3. In contrast, reaction of T4 DNA ligase with the same linear DNA yielded a significant amount of monomer circle along with linear concatemers (not shown). Ku also strongly stimulated plasmid end joining by AtuLigD2 (Figure 9B). The amount of LigD2 added (250 ng) sufficed to join a small fraction of the input DNA, generating mostly linear dimer and trimer products. Supplementation with increasing amounts of Ku resulted in reaction of nearly all of the input DNA and its conversion to higher order concatamers (tetramer or greater) (Figure 9B). In contrast, Ku had no apparent effect on plasmid end joining by AtuLigD1 (data not shown). These experiments provide the first evidence that Ku directly affects the double-strand break repair activity of a LigC-type enzyme and they implicate LigD2, LigC2 and LigC3 as likely agents of Ku-dependent NHEJ in Agrobacterium.
DISCUSSION
The menu of DNA ligases in the Agrobacterium proteome is especially rich, owing to the proliferation of paralogs within the LigC and LigD clades, which is accompanied by an expansion of paralogs of Ku. The complexity of the bacterial NHEJ apparatus now ranges from the relatively simple state found in P. aeruginosa (which has a single Ku, a single LigD and no LigC), to progressively more complex forms in M. tuberculosis (single Ku, single LigD, single LigC), M. smegmatis (single Ku, single LigD, two LigCs) and A. tumefaciens (three Ku paralogs, two LigDs, three LigCs). Understanding the meaning (if any) of the larger complement of putative NHEJ ligases in Agrobacterium (and other plant-associated bacteria such as Mesorhizobium loti and Sinorhizobium meliloti) requires a systematic dissection of their biochemical properties, focused on the following questions: (i) Do the putative ligases really have strand-joining activity? (ii) What other catalytic functions are associated with the ligases, particularly the LigD paralogs? (iii) How do the paralogs compare biochemically to each other? (iv) How do they compare to putative orthologs from other species? (v) Are they affected by Ku?
The present study addressed these issues and thereby illuminates shared features within the LigC and LigD clades, as well as remarkable differences. The key findings are: (i) Not all putative ligases have demonstrable strand-joining activity, e.g. AtuLigC1; (ii) Atu ligases C2, C3, D1 and D2 are relatively feeble at sealing singly nicked DNA duplexes, a characteristic shared with mycobacterial LigC and LigD (16); (iii) AtuLigD1 and AtuLigD2 have an intrinsic 3′ end-healing function similar to that of Pseudomonas LigD; (iv) AtuLigD1 and AtuLigD2 are not functionally equivalent, because only LigD2 has an intrinsic polymerase activity; (v) AtuLigD2 POL prefers ribonucleotide substrates; (vi) The capacity of Atu LigC2, C3, and D2 to seal double-strand breaks in vitro (a model NHEJ-like reaction) is enhanced strongly by Ku.
The Ku stimulation of plasmid end joining by AtuLigC is significant, in that it provides the first biochemical support for the genetic evidence in mycobacteria of a Ku/LigC-driven pathway of NHEJ (17). It is remarkable that Agrobacterium LigC2 and LigC3 are stimulated by Ku derived from Pseudomonas, which does not have a LigC protein. We presume that Ku assists in approximating the duplex ends for sealing by the bacterial ligase, although the molecular details remain obscure. Clearly, the functional interaction of LigC and Ku extends across species lines and does not require ancillary structural modules flanking the core ligase domain. This differs from the LigD–Ku interaction, where the POL domain is implicated in binding to Ku (35). If such an interaction is indeed critical for Ku function in the LigD-mediated NHEJ pathway, then it also appears to cross species lines, because we find that PaeKu stimulates sealing of broken plasmid ends by AtuLigD2 in vitro. The observation that PaeKu does not stimulate plasmid joining by AtuLigD1, which lacks the complete POL domain, provides additional support for the idea that Ku–LigD interactions occur via POL. However, we do not exclude an alternative scenario in which AtuLigD1 interacts specifically with one of the AtuKu paralogs (e.g. AtuKu1, which is encoded in a putative operon adjacent to AtuLigD1). Ultimately, it will be of interest to test whether LigD and Ku proteins from different bacterial species can be exchanged in vivo.
The polymerase activity of LigD is a major contributor to the mutagenic character of bacterial NHEJ at blunt and 5′-overhang DNA breaks in vivo (17,29). Whereas the single LigD enzymes encoded by mycobacteria and P. aeruginosa have catalytically active POL domains, the situation is more complex in Agrobacterium. The domain order of AtuLigD1 and AtuLigD2 (PE-LIG-POL) mimics that of PaeLigD, yet only AtuLigD2 has an intrinsic polymerase activity. The C-terminal POL domain of AtuLigD2 comprises a complete homolog of the PaeLigD POL domain, whereas the POL domain of AtuLigD1 is truncated at its C-terminus. The equivalent of the missing segment of AtuLigD1 POL constitutes the ‘back’ surface of PaeLigD POL, away from the active site (29). As PaeLigD POL exemplifies a minimized version of the archaeal-eukaryal polymerase-primase family (29), it is reasonable that the C-terminal deletion in AtuLigD1 results in loss of activity, most likely because the fold of the residual POL domain is compromised, notwithstanding that the domain contains most of the catalytic residues in the POL active site. The presence of a rump POL domain in AtuLigD1 suggests that LigD1 and LigD2 evolved by duplication of a single full-length ancestral LigD gene, followed by deletion of the C-terminal segment of the LigD1 POL module.
The salient feature of AtuLigD2 POL is its much higher rate of templated ribonucleotide addition at a DNA primer terminus compared to deoxyribonucleotide addition. This property is shared with PaeLigD POL (30) and is likely to be relevant to NHEJ in vivo. NHEJ provides the means to repair double-strand breaks in the absence of a sister chromatid homolog, i.e. in quiescent cells that have a single copy of the bacterial genome. In the absence of ongoing DNA replication, dNTP pools are likely to be limiting because ribonucleotide reductase is cell-cycle regulated (40); thus the shared capacity of LigD POL enzymes to efficiently scavenge and preferentially incorporate ribonucleotides could be advantageous for bacterial survival during quiescence, by providing a temporary ‘ribo patch’ at the 3′-OH side of the repair junction.
The present study underscores that the 3′ end-healing functions of the PE domain of bacterial LigD are conserved across species (e.g. Pseudomonas and Agrobacterium) and among LigD paralogs within a species (AtuLigD1 and AtuLigD2). The Agrobacterium PE domains each catalyze two phosphoesterase reactions: (i) a phosphodiesterase-type resection of a diribonucleotide end of a primer-template to leave a single ribonucleotide and (ii) a phosphomonoesterase reaction that removes a 3′-PO4 from either a terminal ribo or deoxyribonucleotide of a primer-template to leave a 3′-OH. The phosphodiesterase activity of the AtuLigD PE domains is apparently limited to resecting an RNA 3′ terminus, insofar as we observe no 3′ nucleotide resection activity on an all-DNA primer-template and the resection reaction of the AtuLigD PE proteins halts once the primer has a single 3′ ribonucleoside. The ribonucleotide resection activity of the AtuLigD PE domains is strongly stimulated by the 5′ single-strand tail of the complementary DNA strand. These features are shared with the PaeLigD PE domain (31–33). In contrast, it has been reported that M. tuberculosis LigD has an intrinsic 3′ exonuclease activity on DNA phosphodiesters (19). Although the observed DNA exonuclease of full-length MtuLigD was suppressed by a mutation in the PE segment of the protein (20), it was surprising that no DNA exonuclease activity was detected for the isolated PE domain (35). We have purified the PE domain of MtuLigD and find that it performs a manganese-dependent 3′ ribonucleotide resection reaction on a 3′ diribonucleotide-containing primer-template and that it removes the 3′-PO4 from an all-DNA primer-template (Supplementary Figure S5). Thus, the PE domain of M. tuberculosis LigD has the activities ascribed to the homologous modules of Agrobacterium and Pseudomonas LigD. We surmise that 3′ ribonuclease and 3′ phosphatase reactions are the relevant repair functions of the PE domain that are conserved in all LigD proteins characterized to date.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENT
This work was supported by NIH grant GM63611. SS is an American Cancer Society Research Professor. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM63611.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lehman IR. DNA ligase: structure, mechanism, and function. Science. 1974;186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson A, Day J, Bowater R. Bacterial DNA ligases. Mol. Microbiol. 2001;40:1241–1248. doi: 10.1046/j.1365-2958.2001.02479.x. [DOI] [PubMed] [Google Scholar]
- 3.Singleton MR, Håkansson K, Timson DJ, Wigley DB. Structure of the adenylation domain of an NAD+-dependent DNA ligase. Structure. 1999;7:35–42. doi: 10.1016/s0969-2126(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 4.Lee JY, Chang C, Song HK, Moon J, Yang J, Kim HK, Kwon ST, Suh SW. Crystal structure of NAD+-dependent DNA ligase: modular architecture and functional implications. EMBO J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gajiwala K, Pinko C. Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure. 2004;12:1449–1459. doi: 10.1016/j.str.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 6.Srivastiva SK, Tripathi RP, Ramachandran R. NAD+-dependent DNA ligase (Rv3014c) from Mycobacterium tuberculosis: crystal structure of the adenylylation domain and identification of novel inhibitors. J. Biol. Chem. 2005;280:30273–30281. doi: 10.1074/jbc.M503780200. [DOI] [PubMed] [Google Scholar]
- 7.Sriskanda V, Shuman S. Conserved residues in domain Ia are required for the reaction of Escherichia coli DNA ligase with NAD+ J. Biol. Chem. 2002;277:9685–9700. doi: 10.1074/jbc.M111164200. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman MM, Hicks ML, Gellert M. Genetics and function of DNA ligase in Escherichia coli. J. Mol. Biol. 1973;77:531–547. doi: 10.1016/0022-2836(73)90221-0. [DOI] [PubMed] [Google Scholar]
- 9.Konrad EB, Modrich P, Lehman IR. Genetic and enzymatic characterization of a conditional lethal mutant of Escherichia coli K12 with a temperature-sensitive DNA ligase. J. Mol. Biol. 1973;77:519–529. doi: 10.1016/0022-2836(73)90220-9. [DOI] [PubMed] [Google Scholar]
- 10.Park UE, Olivera BM, Hughes KT, Roth JR, Hillyard DR. DNA ligase and the pyridine cycle in Salmonella typhimurium. J. Bacteriol. 1989;171:2173–2180. doi: 10.1128/jb.171.4.2173-2180.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petit MA, Ehrlich SD. The NAD-dependent ligase encoded by yerG is an essential gene of Bacillus subtilis. Nucleic Acids Res. 2000;28:4642–4648. doi: 10.1093/nar/28.23.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaczmarek FS, Zaniewski RP, Gootz TD, Danley DE, Mansour MN, Griffor M, Kamath AV, Cronan M, Mueller J, et al. Cloning and functional characterization of an NAD+-dependent DNA ligase from Staphylococcus aureus. J. Bacteriol. 2001;183:3016–3024. doi: 10.1128/JB.183.10.3016-3024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 14.Cheng C, Shuman S. Characterization of an ATP-dependent DNA ligase encoded by Haemophilus influenzae. Nucleic Acids Res. 1997;25:1369–1375. doi: 10.1093/nar/25.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magnet S, Blanchard JS. Mechanistic and kinetic study of the ATP-dependent DNA ligase of Neisseria meningitidis. Biochemistry. 2004;43:710–717. doi: 10.1021/bi0355387. [DOI] [PubMed] [Google Scholar]
- 16.Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 2004;279:20594–20606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- 17.Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS. Mechanism of nonhomologous end-joining in mycobacteria: a low fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- 18.Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 19.Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, et al. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 20.Korycka-Machala M, Brzostek A, Rozalska S, Rumijowska-Galewicz A, Dziedzic R, Bowater R, Dziadek J. Distinct DNA repair pathways involving RecA and nonhomologous end joining in Mycobacterium smegmatis. FEBS Microbiol. Lett. 2006;258:83–91. doi: 10.1111/j.1574-6968.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sata T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 23.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 24.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, et al. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science. 2001;294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 25.Gallego ME, Bleuyard JY, Daoudal-Cotterell S, Jallut N, White CI. Ku80 plays a role in non-homologous recombinant but is not required for T-DNA integration in Arabidopsis. Plant J. 2003;35:557–565. doi: 10.1046/j.1365-313x.2003.01827.x. [DOI] [PubMed] [Google Scholar]
- 26.Van Attikum H, Bundock P, Overmeer RM, Lee LY, Gelvin SB, Hooykaas PJJ. The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 2003;31:4247–4255. doi: 10.1093/nar/gkg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H, Shuman S. A primer-dependent polymerase function of Pseudomonas aeruginosa ATP-dependent DNA ligase (LigD) J. Biol. Chem. 2005;280:418–427. doi: 10.1074/jbc.M410110200. [DOI] [PubMed] [Google Scholar]
- 29.Zhu H, Nandakumar J, Aniukwu J, Wang LK, Glickman MS, Lima CD, Shuman S. Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D. Proc. Natl Acad. Sci. USA. 2006;103:1711–1716. doi: 10.1073/pnas.0509083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yakovleva L, Shuman S. Nucleotide misincorporation, 3'-mismatch extension, and responses to abasic sites and DNA adducts by the polymerase component of bacterial DNA ligase D. J. Biol. Chem. 2006;281:25026–25040. doi: 10.1074/jbc.M603302200. [DOI] [PubMed] [Google Scholar]
- 31.Zhu H, Shuman S. Novel 3′-ribonuclease and 3′-phosphatase activities of the bacterial non-homologous end-joining protein, DNA ligase D. J. Biol. Chem. 2005;280:25973–25981. doi: 10.1074/jbc.M504002200. [DOI] [PubMed] [Google Scholar]
- 32.Zhu H, Shuman S. Essential constituents of the 3′-phosphoesterase domain of bacterial DNA ligase D, a nonhomologous end-joining enzyme. J. Biol. Chem. 2005;280:33707–33715. doi: 10.1074/jbc.M506838200. [DOI] [PubMed] [Google Scholar]
- 33.Zhu H, Shuman S. Substrate specificity and structure-function analysis of the 3′-phosphoesterase component of the bacterial NHEJ protein, DNA Ligase D. J. Biol. Chem. 2006;281:13873–13881. doi: 10.1074/jbc.M600055200. [DOI] [PubMed] [Google Scholar]
- 34.Akey D, Martins A, Aniukwu J, Glickman MS, Shuman S, Berger JM. Crystal structure and nonhomologous end joining function of the ligase domain of Mycobacterium DNA ligase D. J. Biol. Chem. 2006;281:13412–13423. doi: 10.1074/jbc.M513550200. [DOI] [PubMed] [Google Scholar]
- 35.Pitcher RS, Tonkin LM, Green AJ, Doherty AJ. Domain structure of a NHEJ repair ligase from Mycobacterium tuberculosis. J. Mol. Biol. 2005;351:531–544. doi: 10.1016/j.jmb.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 36.Nair GR, Liu Z, Binns AN. Reexamining the role of the accessory plasmid pATC58 in the virulence of Agrobacterium tumefaciens strains C58. Plant Physiol. 2003;133:989–999. doi: 10.1104/pp.103.030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shuman S. Vaccinia DNA ligase: specificity, fidelity, and inhibition. Biochemistry. 1995;34:16138–16147. doi: 10.1021/bi00049a029. [DOI] [PubMed] [Google Scholar]
- 38.Sriskanda V, Shuman S. Mutational analysis of Chlorella virus DNA ligase: catalytic roles of domain I and motif VI. Nucleic Acids Res. 1998;26:4618–4625. doi: 10.1093/nar/26.20.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 40.Gon S, Camara JE, Klungsøyr HK, Crooke E, Skarstad K, Beckwith J. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006;25:1137–1147. doi: 10.1038/sj.emboj.7600990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.