Abstract
Much effort has long been devoted to unraveling the coordinated cellular response to genotoxic insults. In view of the difficulty of obtaining human biological samples of homogeneous origin, I have established a set of stable human clones where one DNA repair gene has been stably silenced by means of RNA interference. I used pEBVsiRNA plasmids that greatly enhance long-term gene silencing in human cells. My older clones reached >500 days in culture. Knock-down HeLa clones maintained a gene silencing phenotype for an extended period in culture, demonstrating that I was able to mimic cells from cancer-prone syndromes. I have silenced >20 genes acting as sensors/transducers (ATM, ATR, Rad50, NBS1, MRE11, PARG and KIN17), or of different DNA repair pathways. In HeLa cells, I have switched off the expression of genes involved in nucleotide excision repair (XPA, XPC, hHR23A, hHR23B, CSA and CSB), nonhomologous end-joining (DNA-PKcs, XRCC4 and Ligase IV), homologous recombination repair (Rad51 and Rad54), or base excision repair (Ogg1 and Ligase III). These cells displayed the expected DNA repair phenotype. We could envisage untangling the complex network between the different DNA repair pathways. In this study, no viral vehicles, with their attendant ethical and safety concerns, were used.
INTRODUCTION
A large body of evidence demonstrates that impaired responses to genotoxic insults are involved in cancer development. After a DNA insult, a coordinated cellular response takes place, including damage recognition, signal-transduction pathways (with both sensor and transducer proteins) and the response to the DNA damage per se (cell cycle checkpoints, DNA repair and apoptosis) (1). These interwoven responses insure genome integrity. As a consequence, mutations in genes encoding proteins involved in the main DNA repair pathways, such as nucleotide excision repair (NER), translesion synthesis (TLS), mismatch repair (MMR), nonhomologous end-joining (NHEJ) or base excision repair (BER) may cause chromosome and genetic instability, telomere defects and cancer predisposition (2–5).
Fewer than 20 DNA repair disorders have been identified so far. Besides the well-known xeroderma pigmentosum (XP), Cockayne's syndrome (CS) and trichothiodystrophy (TTD) syndromes, which affect the two branches of the NER pathway (global genome repair versus transcription-coupled repair), mutations in other DNA repair pathways have also been identified. For instance, mutations in some components of the NHEJ have been observed for ligase IV (LIG4 syndrome), Artemis (RS-SCID, human radiosensitive severe combined immunodeficiency syndrome), DNA-PKcs (M059J glioblastoma cells) and Cernunnos (6). Also, chromosomal instability associated with impaired RecQ helicase functions is associated with Bloom syndrome (BS protein), Werner syndrome (WS; WR protein) and Rothmund–Thompson syndrome (RTS) (7–9). Mutations are also found in different proteins that act as sensor proteins for DNA lesions and/or chromatin alterations, or transducer proteins for conveying the damage signal to downstream effector proteins. These hereditary mutations have been found in several syndromes and diseases, such as the Li-Fraumeni syndrome ( p53 gene), ataxia-telangiectasia (AT; ATM gene), ATR-Seckel syndrome (ATR; ATR gene), AT-like disease (ATLD, MRE11 gene), Nijmegen breakage syndrome (NBS, NBS1 gene) and Fanconi anemia (FA).
It turns out that cells arising from these patients represent the major source of DNA repair-deficient human cells (10). However, the major limitation is the large genetic variation in the human population. As a consequence, to study the relationship between the main DNA repair pathways I envisaged establishing a set of isogenic human cell lines in which one specific gene is silenced by the RNA interference method. Because RNAi is an endogenous natural pathway, its power as a gene-silencing tool is greater than that of other approaches.
Most studies describing RNA interference in cultured human cells are based on transient transfections of siRNA duplexes or plasmids, or transient infection of virus-expressing siRNA constructs. In the strategy based on siRNA-expressing plasmids, use is made of integrative plasmids that could lose their siRNA cassette during selection pressure. An elevated number of siRNA-based plasmids per cell or an elevated viral titer may saturate the RNAi machinery (RISC). As a consequence, overexpression of siRNA through either siRNA duplexes or integrative vectors may lead to mistaken conclusions in transient experiments. Hence, long-term maintenance of gene silencing is riddled with pitfalls.
To overcome these shortcomings, I have designed episomal plasmids driving the in cellulo siRNA synthesis. I have harnessed the efficiency of the siRNA approach with Epstein–Barr virus (EBV)-derived vectors in order to establish cell lines carrying a few plasmid copies per cell. For many years, pEBV-based plasmids have been used to efficiently modify human cell genotypes (11,12). These plasmids bear the latent replication origin of the EBV virus (OriP) and allow the expression of only one viral protein, EBV nuclear antigen-1 (EBNA-1). The main features of these vectors have been recently reviewed (13). EBNA-1 tethers EBV episomes to metaphase chromosomes, providing the basis for their nuclear retention and their successful segregation at mitosis (14,15). Interestingly, the presence of EBNA-1 does not interfere with the differentiation of human cells (16). EBV DNA replication is semi-conservative and is coordinated with genomic replication (17–19). OriP appears to be regulated like human endogenous DNA replication. Since these vectors behave like endogenous transcription units, it was expected that they would tightly regulate siRNA expression over a long period of time.
So far, I have developed and characterized >200 replicative pEBVsiRNA plasmids for their ability to impose efficient, specific and very long-term gene silencing in different human cell lines. In terms of DNA repair, my main aim was to establish a set of isogenic cell lines in which the expression of one specific DNA repair gene is abrogated. Here >40 genes were targeted, and my older clones reached >500 days in culture. These older clones maintained impressive gene silencing. This approach allows us to validate the choice of an efficient siRNA sequence that remains efficient in cells even several months following transfection. This also affords us the opportunity to compare short- and long-term effects of RNAi. Here, I present new clones that will help us to untangle the relationships between the different DNA repair pathways.
MATERIALS AND METHODS
Design of pEBVsiRNA vectors
The crucial step in RNAi is the unfailing recognition of the cognate mRNA by the siRNA sequence. Since Tuschl and colleagues described the initial design paradigm for efficient 21 nt siRNA (20,21), many rational designs have been developed, based on a better understanding of RNAi biochemistry (22,23). I adopted the algorithm developed by Sonnhammer and collaborators (24). All my siRNA sequences targeted the open reading frame of a specific gene.
Cloning was performed using my pEBVsiRNA-LacZ’ vectors carrying either a hygromycin B or puromycin resistance cassette (pBD751 and pBD899, respectively) (13). These vectors carry the H1 RNA polymerase III promoter (RNA pol III) to drive the transcription of short hairpin RNA (shRNA), which gives rise to siRNA-like molecules in vivo. These cloning plasmids allow blue/white screening of bacterial colonies carrying a recombinant pEBVsiRNA plasmid with the DNA coding for a shRNA sequence. Hence, shRNA cloning reaches 100% efficiency, thus reducing the time of selection and characterization of recombinant plasmids. For each gene, 2–4 vectors were assessed for both short- and long-term silencing (several months). In each case, I selected the best vector according to three criteria: (i) highly efficient silencing of the gene of interest, (ii) maintenance of this gene silencing over time and (iii) accuracy of the expected phenotype (loss-of-function) when this phenotype is known.
Cell culture, treatment and flow cytometry
HeLa (cervical adenocarcinoma) cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin and 100 μg/ml streptomycin, under 5% CO2. HeLa cells were transfected using 2 μl of LipofectAMINE2000 (Invitrogen) with 2 μg of DNA. Transfected cells were propagated in culture in the presence of hygromycin B (125 μg/ml; Invitrogen). After transfection of HeLa cells, 12 clones per gene were propagated in culture, and growing clones were analyzed for loss-of-function. Thereafter, only one clone was selected and added to my battery of knock-down clones (other clones being frozen). Among the 40 genes targeted, 20 are listed in Table 1. The other targeted genes have also been successfully silenced for a long period in culture, and the new knock-down clones have to be further characterized (data not shown). Among them, I have targeted the PARP2, PARG, BRCA1, ERCC1, XRCC1, TRF2 or cdc25b genes.
Table 1.
Efficient pEBVsiRNA vectors for long-term silencing
| Protein | pEBVsiRNA plasmid | siRNA sequence | Position (nt) | |
|---|---|---|---|---|
| 1 | XPC (Q01831) | pBD634 (pEBVsiXPC-267) | 5′-GGATGAAGCCCTCAGCGAT-3′ | 267–285 |
| 2 | XPA (P23025) | pBD695 (pEBVsiXPA-587) | 5′-GTCAAGAAGCATTAGAAGA-3′ | 587–605 |
| 3 | XRCC4 (Q13426) | pBD694 (pEBVsiXRCC4-674) | 5′-GAGATCCAGTCTATGATGA-3′ | 674–692 |
| 4 | DNA-PKcs (P78527) | pBD743 (pEBVsiDNAPK-5980) | 5′-GTTGAGGTTCCTATGGAAA-3′ | 5980–5998 |
| 5 | Rad50 (Q92878) | pBD872 (pEBVsiRad50-274) | 5′-GAACTTATAGCTGTGCAAA-3′ | 274–292 |
| 6 | MRE11A (P49959) | pBD930 (pEBVsiMRE11-464) | 5′-GTTCAATGTCTGTGGAGAA-3′ | 464–482 |
| 7 | NBS1 (O60934) | pBD932 (pEBVsiNBS1-160) | 5′-GTAACCAACCTGAGTCAAA-3′ | 160–178 |
| 8 | Rad51 (Q06609) | pBD864 (pEBVsiRad51-114) | 5′-GAAGAAATTGGAAGAAGCT-3′ | 114–132 |
| 9 | Rad54 (Q92698) | pBD912 (pEBVsiRad54-987) | 5′-GAATGATCTGCTTGAGTAT-3′ | 987–1005 |
| 10 | hHR23A (P54725) | pBD805 (pEBVsiR23A-491) | 5′-AGACGATGCTGACGGAGAT-3′ | 491–509 |
| 11 | hHR23B (P54727) | pBD804 (pEBVsiR23B-1123) | 5′-GCATTAGGATTTCCTGAAG -3′ | 1123–1141 |
| 12 | ATM (Q13315) | pBD935 (pEBVsiATM-2594) | 5′-GTGTTAGTGATGCAAACGA-3′ | 2594–2612 |
| 13 | ATR (Q13535) | pBD962 (pEBVsiATR-854) | 5′-CTGACCAATTGAAACTCTA-3′ | 854–872 |
| 14 | CSA (Q13216) | pBD918 (pEBVsiCSA-191) | 5′-CAGATGGTGTGATTGTACT-3′ | 191–209 |
| 15 | CSB (Q03468) | pBD921 (pEBVsiCSB-1358) | 5′-GAGATGATGGAGATGAAGA-3′ | 1358–1376 |
| 16 | Ligase III (P49916) | pBD941 (pEBVsiLigIII-164) | 5′-GCATGTTTGAGAAACTAGA-3′ | 164–182 |
| 17 | Ligase IV (P49917) | pBD940 (pEBVsiLigIV-1939) | 5′-CCTAACCTTACTAACGTTA-3′ | 1939–1957 |
| 18 | Ogg1 (O15527) | pBD784 (pEBVsiOgg1-177) | 5′-AGCGGATCAAGTATGGACA-3′ | 177–195 |
| 19 | Kin17 (O60870) | pBD674 (pEBVsiK180) | 5′-TCCTCAGCAGTTTATGGAT-3′ | 180–198 |
| 20 | PARG (Q86W56) | pBD813 (pEBVsiPARG-2325) | 5′-GGATCACAATGAATGTCTA-3′ | 2325–2343 |
| 21 | Ku70 (P12956) | pBD699 (pEBVsiKu70-156R) | 5′-GAGTGAAGATGAGTTGACA-3′ | 156–174 |
For each gene, 2–4 vectors were constructed. Puromycin plasmids were systematically constructed for all genes tested in order to allow the establishment of double knock-down cells. The siRNA sequence and its position in the ORF are indicated (with ATG as the first nucleotide). It is noteworthy that only Ku70 gene silencing leads to cell death after ∼10 days of culture.
For clonogenic growth, only established long-term clones were used. Cells were plated at 500 cells per 6-cm-diameter dish 24 h before treatments. Growing clones were fixed with 4% paraformaldehyde, and stained with methylene blue (in 30% methanol) after 14 days of culture. Clones containing >50 cells were counted. Each point represents the mean of three culture dishes. Experiments were performed at least twice. Colony formation was normalized as a percentage of the control.
For acute treatments, 400 000 cells from established clones were seeded per 6-cm-diameter dish 2 days before treatments. Cells were treated with UVC 10 J/m2, γ rays 6 Gy (137Cs γ-ray source; dose rate of 1.9 Gy/min; IBL 637 CisBio International, Bedford, MA, USA) or etoposide (Vépéside-Sandoz; 25 μM for 1.5 h) and harvested 24 h later. Flow cytometry analysis was described elsewhere (25). Briefly, adherent cells were collected by trypsinization, washed with PBS and fixed in 75% ethanol at 4°C for at least 24 h. Cells were washed twice in PBS and nuclear DNA was stained with propidium iodide (Sigma, St. Louis, MO, USA; 4 μg/ml) in the presence of RNase (Sigma; 10 μg/ml) in PBS for at least 30 min. Stained cells were analyzed on a FACScalibur (Becton Dickinson, Franklin Lakes, NJ, USA) using CellQuest software. Here, ∼10 000 cells gated as single cells using FL2A/FL2W scatter were analyzed.
Western blot, immunostaining and in vitro NHEJ assay
Procedures were described elsewhere (26). For in vitro NHEJ, whole-cell extracts and DNA substrates were prepared as described elsewhere (27–29).
RESULTS
Silencing of genes of the NER pathway
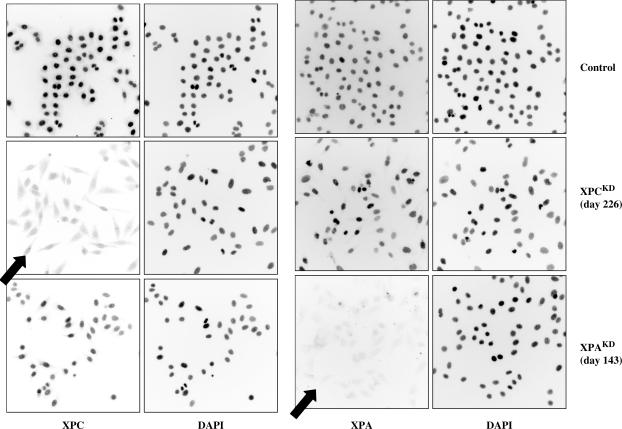
I targeted genes of the main pre- and post-replicative DNA repair pathways. Preliminary experiments were performed to silence the XPA and XPC genes in order to circumvent the transcription-coupled (TC) and global genome (GG) NER pathways. XPA is involved in both TC- and GG-NER pathways and XPC is only required for GG-NER. Numerous knock-down clones were characterized, and all of them displayed very low or nearly undetectable amounts of either XPA or XPC protein (Figure 1). Several XPAKD and XPCKD clones have been recently described in detail (25,29). As expected, XPAKD and XPCKD clones exhibited an ‘XP’ phenotype with great sensitivity to UVC and an impaired unscheduled DNA repair synthesis (UDS) after UVC irradiation (25). Strikingly, in HeLa or MCF-7 cells, XPCKD cells displayed major growth disadvantages in comparison with their XPAKD counterparts. This could be correlated with the critical interactions of XPC (C-terminal portion) with hHRad23B, centrin 2 or TFIIH (30).
Figure 1.
Immunocytochemical staining of knock-down HeLa clones. Long-term silenced clones were propagated in culture and analyzed (one representative field is shown for each clone; Magnification ×100).
I have now enlarged this approach by focusing on two other genes tightly associated with the XPC pathway, hHR23A and hHR23B. Two interpretations have been proposed: (i) hHR23A&B may stabilize the XPC protein by protecting it from 26S proteasome-dependent protein degradation (31), and (ii) hHR23B may be a key event in the displacement of XPC from damaged DNA by the XPA–RPA complex (32). I found that hHR23B knock-down dramatically hampered HeLa cell growth, as did XPC gene silencing. In contrast, the shutting down of the hHR23A gene expression did not significantly affect cell growth (data not shown).
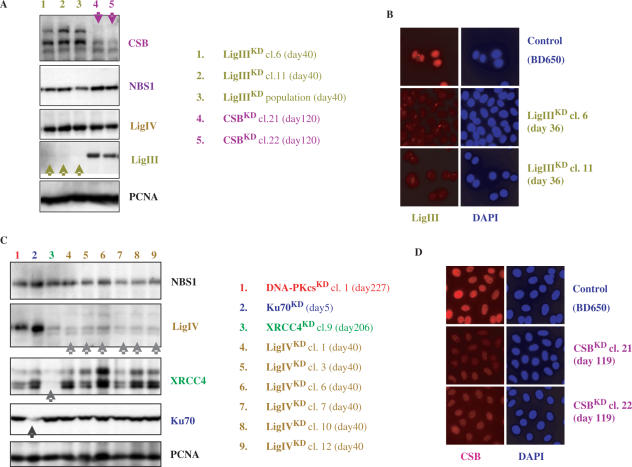
I also focused my attention on the CSA and CSB genes (Cockayne's syndrome genes). Figure 2 shows the remaining CSB protein content observed in two CSBKD HeLa clones 120 days after transfection. The human CSB (rad26 homolog) belongs to a large complex that transiently interacts with the transcription machinery to check whether RNA synthesis occurs faithfully (for review 33). As a SWI/SNF chromatin-remodeling factor, CSB affects the chromatin conformation of both active genes and those found throughout the genome. CSB participates in TC-NER in human cells, but presumably in other DNA repair mechanisms too, because it was found tightly associated with other DNA repair proteins, such as Ogg1 and PARP1. While CSA and CSB nullizygous mice are not viable (34), I found that all isolated CSAKD and CSBKD cells maintained a residual amount of the targeted protein, suggesting that these proteins could be essential for cell survival in the absence of genotoxic injuries.
Figure 2.
Protein analysis of long-term LigIIIKD, LigIVKD, XRCC4KD and CSBKD clones.
Silencing of genes of other DNA repair pathways (e.g. NHEJ versus HR)
The repair of DNA double-strand breaks (DSBs) is critical for the maintenance of genomic stability. In eukaryotes, two pathways deal with such DNA lesions to ensure faithful DNA repair: the NHEJ and the homologous recombination (HR) pathways. Although NHEJ can function throughout the cell cycle, it predominates in G0 and G1 phases. In contrast, HR acts in S and G2 phases.
The NHEJ pathway includes at least seven factors: Ku70, Ku80, DNA-PK(cs), Artemis, Xrcc4 and DNA ligase IV (Lig IV), and recently Cernunnos (6). The Ku70–Ku80 heterodimer binds to DNA ends and recruits the PI-3K-like kinase DNA-PKcs (catalytic subunit of the DNA-dependent protein kinase). DNA-PKcs phosphorylates, associates with, and activates the Artemis endonuclease, which is required to resolve DNA hairpin structures. One of the targets of DNA-PKcs is the XRCC4 protein, which associates with DNA-Lig IV. The XRCC4/Lig IV complex is responsible for the final ligation step (for review 35). To impair the NHEJ pathway, I targeted Ku70, XRCC4, DNA-PKcs and Lig IV.
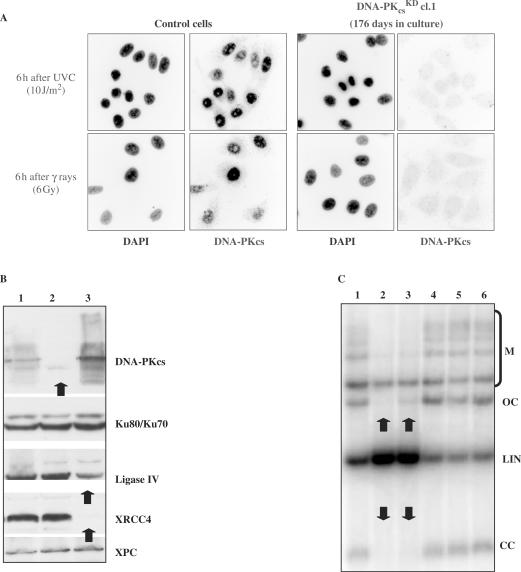
All attempts to silence the Ku70 gene with different vectors (e.g. pBD699 plasmid; Figure 2C) led to cell death after ∼15 days of culture (in the presence of hygromycin B), further indicating that the Ku70 protein is an essential factor in human cells. This contrasts with Ku70 knock-out mice, which are viable, but display severe defects in DSBR, growth and B cell development (for review 34). In contrast, I successfully established XRCC4KD (pBD694), DNA-PKcsKD (pBD743) and more recently Lig IVKD (pBD940) cells for a very long period of time. The protein content of knock-down HeLa clones was monitored constantly, as shown in Figures 2 and 3. The establishment of XRCC4KD and DNA-PKcsKD clones demonstrates that the cells were sensitive to ionizing radiation and DSB-inducing agents. The marked reduction in either XRCC4 or DNA-PKcs protein levels triggers a failure in DSB rejoining activity, as shown by an in vitro NHEJ assay. In vitro NHEJ activities were assessed using whole-cell extracts and a wide range of substrates (29). In these conditions, NHEJ activities were reduced by 70–80% in DNA-PKcsKD and XRCC4KD cells, depending on DNA end configuration after cutting with different restriction enzymes: cohesive ends, blunt ends, 3′ end/3′ end, 5′ end/blunt end, 3′ end/blunt, 5′ end/3′ end (Figure 3). As controls, KIN17KD clones were used. This study indicated that the reduction in the XRCC4 protein level abrogated the re-circularization of different linear substrates, suggesting that XRCC4 is a limiting factor in NHEJ. The establishment of XRCC4KD HeLa clones is interesting because no human cell line lacking this protein has been identified to date. Only Artemis, Lig IV and Cernunnos are known V(D)J/NHEJ factors deficient in human heritable diseases. In parallel to DNA-PKcsKD and XRCC4KD, I have recently isolated different Lig IVKD clones (Figure 2). We observed that Lig IV deficiency lowered in vitro NHEJ activities to a lesser extent than did DNA-PKcsKD and XRCC4KD (data not shown). My stable knock-down clones may allow us to answer numerous exciting questions. For instance, there are at least two NHEJ complementary sub-pathways with different kinetics, the high-speed classical D-NHEJ (strictly dependent on DNA-PK and XRCC4 protein) and a low-speed B-NHEJ (Backup; also termed microhomology-based end-joining pathway). These two sub-pathways seem to contribute to the major rescue of IR-induced DSBs in higher eukaryotes (36,37). It appeared that in vitro NHEJ activity was tightly dependent on both XRCC4 and DNA-PKcs proteins. Beside, XRCC4 protein may be a limiting factor in NHEJ, but not DNA-PKcs, because XRCC4KD cells displayed no detectable re-circularization activity (‘OC’ and ‘CC’ products). On the other hand, most of the building blocks of B-NHEJ are unknown; DNA ligase III (Lig III) has been identified as one (38). Lig III participates in the last step of the BER pathway by sealing the remaining single-strand break. Lig III is stabilized in vivo by its tight interaction with XRCC1 (for review 39). I have now established different Lig IIIKD clones (two clones are depicted in Figure 2A, lanes 1 and 2) that will allow us to investigate this issue. Interestingly, Lig IIIKD cells were easily obtained with a nearly undetectable protein level, as shown by western blot and immunocytochemical staining. Furthermore, a cell population continues to maintain an undetectable Lig III protein level 40 days after transfection, strongly suggesting that the loss of this protein did not alter the viability and growth of HeLa cells in the absence of genotoxic injuries (Figure 2A, lane 3).
Figure 3.
Functional analysis of NHEJ-deficient HeLa cells. Protein contents were analyzed (A) by immunocytochemical staining (DNA PKcsKD cells at day 176 after UVC or γ ray irradiation) or (B) by western blotting. 1: Control (pBD650); 2: DNA PKcs KD cl.1 (pBD743; day 94); 3: XRCC4KD cl.9 (pBD694; day 94). (C) NHEJ activities were measured using an in vitro NHEJ assay. Agarose gel separation of NHEJ products was followed by Southern blotting. Reaction products are marked on the right side: M: linear multimers (inter-molecular joining); OC: open circles (intra-molecular joining); LIN: input linear substrate; CC: covalently closed circles (intra-molecular joining). 1: Control (pBD650); 2: XRCC4KD cl.9 (pBD694; day 75). 3: DNA PKcsKD cl.1 (pBD743; day 75); 4: Control (pBD650); 5: KIN17KD cl.6 (pBD674; day 81). 6: KIN17KD cl.12 (pBD674; day 81).
I also switched off the expression of genes of the Rad52 epistasis group, such as Rad51, Rad52 and Rad54, in order to abrogate a part of HR activity. Here, I present results from Rad51KD and Rad54KD clones. Rad51 gene silencing induced cell death and few clones with a reduced Rad51 protein level could be expanded in mass culture. Preliminary observations indicate that Rad51KD cells display an expected phenotype, as discussed below (Figures 5 and 6).
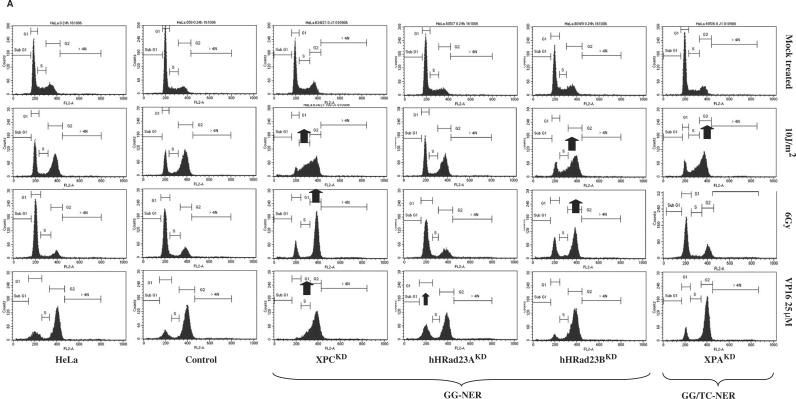
Figure 5.
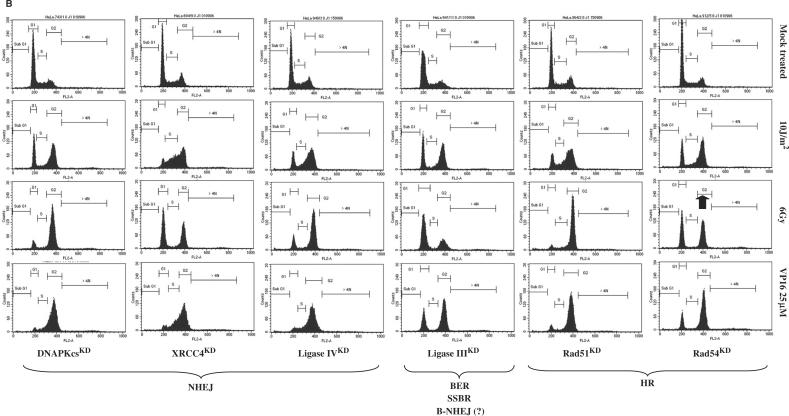
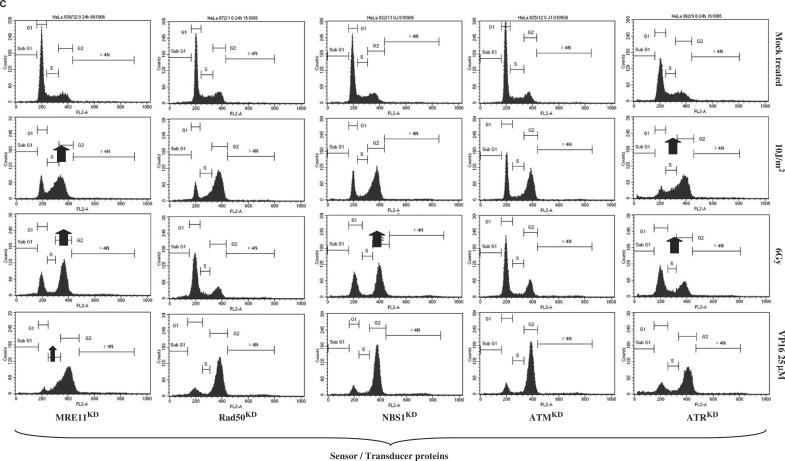
(A) Cell cycle progression of stable knock-down HeLa clones after UVC (10 J/m2), γ rays (6 Gy) or etoposide treatment (VP16; 25 μM for 1.5 h): GG-NER and TC-NER pathways. Exponentially growing cells were treated and 24 h later adherent cells were collected, washed and fixed in 75% ethanol at 4°C for at least 24 h. Cells were stained with propidium iodide (4 μg/ml) in the presence of RNase (10 μg/ml) for at least 30 min. Stained cells were analyzed on a FACScalibur (Becton Dickinson) using CellQuest software. Here, ∼10 000 cells gated as single cells using FL2A/FL2W scatter were analyzed. Each clone was analyzed at least three times. Long-term HeLa clones: Control (BD650, day115), XPCKD (BD634/21; day 333), hHR23AKD (BD805/7; day 158), hHR23BKD (BD804/9; day 191), XPAKD (BD695/6; day 422). Arrows indicate a striking point highlighting intolerance to DNA damage. (B) NHEJ and HR pathways. Long-term HeLa clones: DNAPKcsKD (BD743/1; day 321), XRCC4KD (BD694/9; day 235), LigIVKD (BD940/3; day 103), LigIIIKD (BD941/11; day 82), Rad51KD (BD864/2; day 107), Rad54KD (BD912/5; day 148). (C) Signaling pathways Long-term HeLa clones: MRE11KD (BD930/32; day 61), Rad50KD (BD872/1; day 158), NBS1KD (BD932/11; day 108), ATMKD (BD935/12; day 111), ATRKD (BD962/9; day 56).
Figure 6.
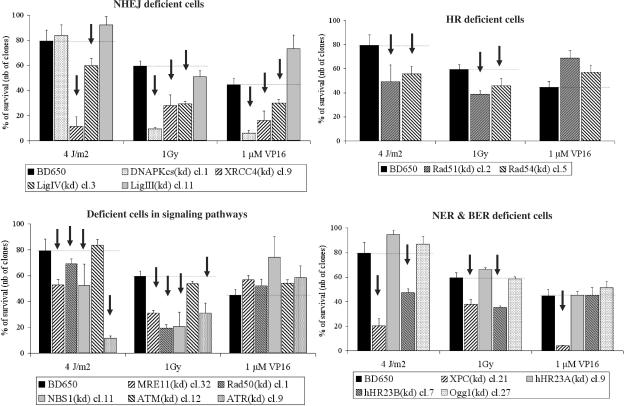
Clonogenic cell survival assays after UVC (4 J/m2), γ rays (1 Gy) or etoposide treatment (VP16; 1 μM for 1.5 h). Established long-term clones were seeded at 500 cells per 6-cm diameter-dish 24 h before treatment. Here, ∼14 days later, cells were fixed and stained. Each point represents the mean of two independent experiments; each experiment represents three culture dishes per point. SD are indicated. Age of cells at the time of the last clonogenic cell survival assay: Control (pBD650; day 238); XPCKD cell cl.21(day 456); hHR23AKD cl.7 (day 236); hHR23BKD cl.9 (day 272); DNA PKcsKD cl. 1 (day 365); XRCC4KD cl.9 (day 205); Ligase IVKD cl.3 (day 176); Ligase IIIKD cl.11 (day 178); Rad51KD cl.2 (day 193); Rad54KD cl.5 (day 322); MRE11KD cl.32 (day 177); Rad50KD cl.1 (day 175); NBS1KD cl.11 (day 176); ATMKD cl.12 (day 302); ATRKD cl.9 (day 196); Ogg1KD cl.27 (day 232). Arrows indicate a striking point highlighting intolerance to DNA damage.
Silencing of the component of the MRN complex
Another interesting field of investigation is the sensor/transducer pathway, which is critical for genomic stability. The MRN complex (MRE11, Rad50 and NBS1) plays an essential role in the intracellular signaling pathway activated after DNA damage, in particular in the cellular response to stalled replication forks. MRN acts as a DNA damage sensor, continuously localized at sites of unrepaired DNA damage. MRN also participates in other essential mechanisms, such as DNA replication, cell cycle checkpoints and telomere maintenance. Nbs1 binds to γH2AX in the vicinity of DSBs, and recruits Rad50 and Mre11. While Mre11 has both nuclease and DNA-unwinding activities, Rad50 adopts a ‘V-like’ conformation to act as a bridge to hold together the broken ends of DSBs and prevent extensive degradation of the DNA (for review see 1,40). Targeting of MRN components is essential to understand the role of the MRN complex in the NHEJ and HR pathways. MRE11, Rad50 and NBS1 are essential genes in vertebrates, which limits experimental work. Experiments are usually performed with cells from patients displaying hypomorphic mutations in either MRE11 (ATLD disease) or NBS1 (NBS syndrome). Recently, one patient with two germline mutations in RAD50 has been identified (for review see 41). For instance, most human NBS cells express an NH2-terminally truncated Nbs1 protein that contains an intact Mre11-binding domain (for review see 42). The latter cells may maintain several biological activities associated with the MRN complex. In mouse models, the assessment of MRN functions has also been hampered by the lethal phenotype of null Mre11, Rad50 and Nbs1 mutants in cultured cells and in vivo (for review see 43). Only mouse models carrying Rad50S alleles or mimicking the mutations identified in human NBS and A-TLD patients have been characterized (for review see 44).
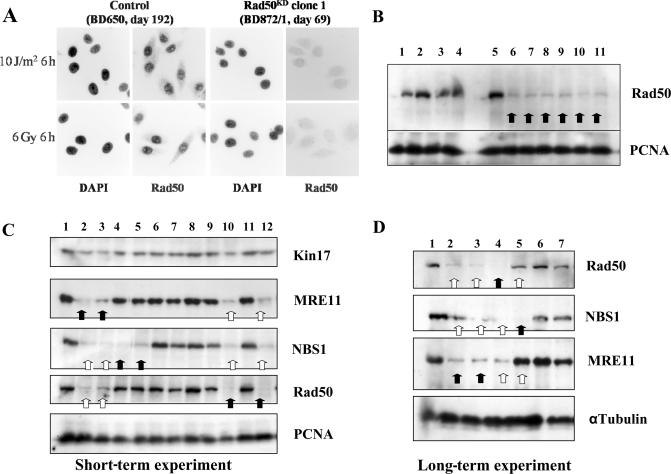
I have isolated stable knock-down HeLa clones for the three components of the MRN complex. In a HeLa genetic background, all of my Rad50KD clones exhibited a very low Rad50 protein level, even 6, 45, 69, 76, 120 or 149 days after transfection, with impressive stability over time (Figure 4). Hence, the reduced Rad50 protein level did not significantly impair cell growth. Interestingly, in all experiments performed, the lowered Rad50 protein level in Rad50KD clones triggered a marked fall in both Mre11 and Nbs1 protein levels. MRE11 gene silencing induced a severe decrease in the protein level of the two other components of the MRN complex (Rad50 and Nbs1). In contrast, the nearly undetectable level of the Nbs1 protein in NBS1KD cells did not induce any change in Mre11 or Rad50 protein levels. This result was observed with different pEBVsiRNA vectors per gene and in either short- or very long-term gene silencing experiments (Figure 4C and D).
Figure 4.
Short- and long-term gene silencing for MRN components. (A and B) 45, 69 and 76 days after transfection, Rad50KD HeLa clones were analyzed by immunocytochemical staining (magnification ×315) or western blot. 1: Control (pBD650; day 189); 2: XPCKD cl. 21 (pBD634; day 376); 3: XPAKD clone 6 (pBD695; day 436); 4: XRCC4KD clone 9 (pBD694; day 107); 5: Control (pBD650; day 31); 6: Rad50KD clone 1 (pBD872; day 76); 7: Rad50KD clone 1 (day 45); 8: Rad50KD clone 2 (day 45); 9: Rad50KD clone 3 (day 45); 10: Rad50KD clone 4 (day 45); 11: Rad50KD clone 5 (day 45). (C) Transient gene silencing: 6 days after transfection, MRE11KD (two pEBVsiRNA plasmids), Rad50KD (one plasmid) and NBS1KD (two plasmids) cell populations were analyzed by western blot. 1: Control (pBD650; day 6); 2: MRE11KD (pBD930; day 6); 3: MRE11KD (pBD931; day 6); 4: NBS1KD (pBD932; day 6); 5: NBS1KD (pBD934; day 6); 6: ATMKD (pBD935; day 6); 7: ATMKD (pBD936; day 6); 8: ATMKD (pBD937; day 6); 9: PARGKD (pBD812; day 6); 10: Rad50KD (pBD872; day 6); 11: Control (pBD650; day 6); 12: Rad50KD (day 120). (D) Long-term gene silencing: Clones were picked up from cell populations described in the panel ‘C’ and propagated for several months in culture. 1: Control (pBD650; day 238); 2: MRE11KD cell population (pBD930; day 33); 3: MRE11KD clone 32 (pBD930; day 132); 4: Rad50KD clone 1 (pBD872; day 149); 5: NBS1KD clone 11 (pBD932; day 172); 6: DNA PKcsKD clone 1 (pBD743; day 391); 7: Ligase IIIKD clone 11 (pBD941; day 180).
This observation suggests that the trimeric complex MRN supports different interactions between each building block. Rad50 and Mre11 are essential for the maintenance and stability of the MRN complex, certainly in the vicinity of DSBs. The loss of either Rad50 or Mre11 triggers destabilization of the whole MRN complex and the disappearance of the other components. This was already observed in a patient exhibiting a truncated mutation in the MRE11 gene associated with reduced expression of three members of the MRN (45). Interestingly, the loss of Nbs1 did not interfere with the stability of either Rad50 or Mre11 proteins, as previously mentioned with mouse models (for review see 43). These compelling findings show that my stable knock-down cell model has the hallmarks of MRN stability.
Other sensor/transducer proteins
Besides the MRN complex, I targeted other essential proteins involved in the pathway signaling the presence of DNA damage, such as the nuclear protein kinases ataxia telangiectasia mutated (ATM) and ataxia telangiectasia- and Rad3-related (ATR). ATM and ATR belong to a conserved family of proteins termed the ‘phosphatidylinositol 3-kinase-like protein kinases’ (PIKKs). PIKKs are conserved from yeast to mammals, and respond to various stresses by phosphorylating substrates in the appropriate pathways. The chief transducer of the DSB signal is ATM (for review see 46). Another member of this family of kinases is DNA-PKcs, which has been described above for its gene silencing. Cells from AT patients (or ATM-nullizygous mice) are sensitive to ionizing radiation and other agents that induce DSBs. AT cells fail to activate the IR-induced G1/S or G2/M checkpoints, and exhibit radioresistant DNA synthesis, which is indicative of an impaired S-phase checkpoint (for review see 47). ATR mediates responses to a broad spectrum of genotoxic stimuli, including DNA replication inhibitors (e.g. hydroxyurea), UV radiation, IR and agents such as cis-platinum that induce DNA interstrand cross-links.
I am currently assessing loss-of-function in ATMKD and ATRKD clones. In my ATMKD cells (clone BD935/12; 70 days after transfection), there was no appearance of phosphorylated ATMSer-1981 protein after induction of DNA damage, and, consequently, p53Ser-15 protein was absent (J. Bouley, personal communication). Interestingly, phosphorylation of p53Ser-15 is necessary for an IR-induced G1/S arrest via transcriptional induction of the cyclin-dependent kinase inhibitor p21 (47). ATM gene silencing did not significantly affect HeLa cell growth, even after several months of culture. In contrast, ATRKD cells displayed an elevated rate of spontaneous cell death. This was in agreement with previous reports showing that while ATM−/− mice are viable, ATR-deficient mice die early during embryogenesis. Similarly, conditional knock-out of ATR gene function in human cells leads to a loss of cellular viability.
Interconnecting DNA repair processes
In an effort to reveal the interconnecting features of DNA repair pathways, I treated my stable clones with genotoxic agents that inflict major damage on DNA, triggering defects in the cell cycle progression. I postulated that defects in a specific DNA repair pathway could be easily and rapidly assessed by means of flow cytometry. I have already applied this method to XPAKD and XPCKD cells (25). I used as genotoxic agents UVC, γ rays or etoposide (VP16), for the following reasons: (i) UVC-induced DNA damage blocks DNA replication and transcription, (ii) γ rays induce DSBs, which are a major threat to genomic stability and (iii) etoposide is a ‘topoisomerase II (topo II) poison’, which converts topo II into a DNA-damaging enzyme by disrupting the cleavage-religation equilibrium; VP16 induces accumulation of DSBs, activation of DNA damage sensors, cell cycle arrest, and initiation of apoptosis or repair (for review see 48).
All of my stable clones were cultivated, treated and analyzed using the same procedure at least three times. Briefly, cells were treated with UVC (10 J/m2), γ rays (6 Gy) or etoposide (25 μM for 1.5 h in the culture medium). Twenty-four hours later, cells were trypsinized, fixed with EtOH (70%), and stored at +4°C before flow cytometry analysis. Not all doses used induced lethality in control cells at the time of the analysis. Only adherent cells were analyzed by means of flow cytometry. Here, I show some of these results. It was noteworthy that I observed no marked differences in cell cycle progression in response to genotoxic insults as a function of the age of the cells in culture. Afterwards, data from flow cytometry analyses were constantly checked with classical clonogenic cell survival assays. In this manner, I could assess the sensitivity of my clones to both low and high doses of genotoxic agents after a short or long period in culture.
The presence of pEBV plasmids in HeLa cells for several months did not modify cell behavior toward genotoxic insults. Only a slight increase in the percentage of cells in G2–M phase after ionizing radiation was observed in control BD650 cells (21%), in comparison with mock transfected HeLa cells (13%), which was certainly due to the presence of hygromycin B.
UVC irradiation
UVC irradiation (10 J/m2) of NER-proficient cells (HeLa cells or control BD650 cells) induces strong G2–M phase arrest, suggesting that UVC lesions are removed without hampering progression of DNA replication. However, the high UVC dose used here leads to the accumulation of damaged cells in G2 phase (Figure 5A).
As expected, highly UVC-sensitive XPAKD and XPCKD cells were arrested in early and middle S phase, showing that remaining UVC lesions block DNA replication. We have previously reported that UVC lesions were not removed in our NER-deficient cells, which was a typical feature of XP cells. In our previous study, we characterized the XP phenotype of these knock-down clones (25). Strikingly, while hHR23AKD cells were not blocked in S phase after UVC irradiation, many hHR23BKD cells were hindered in getting out of S phase (Figure 5A). This suggested the presence of unrepaired UVC-induced DNA damage in hHR23BKD cells. Therefore, hHR23BKD cells seemed to behave like XP cells. This was confirmed by clonogenic survival assays, where hHR23BKD cells displayed a significant sensitivity to UVC, in contrast to hHR23AKD cells, which strongly tolerated UVC irradiation (Figure 6). This also suggested that hHR23A and hHR23B displayed diverse biological functions in humans. In contrast, mHR23A and mHR23B appeared to have redundant roles in NER. Interestingly, inactivation of both genes caused embryonic lethality, but was compatible with cellular viability (49).
UVC-induced stalled replication could trigger an early commitment to recombination repair pathways because at later times after treatment arrested forks are degraded into double-strand breaks (50). This alternative pathway was reinforced when the p53 protein was inactivated, as observed in HeLa cells, where the HPV18-E6 protein may degrade a part of the newly synthesized p53 protein. As a consequence, I expected to observe an impaired cellular response to UVC in some recombination repair-deficient cells. XRCC4KD and Lig IVKD on the one hand, and Rad51KD cells on the other, seemed to interfere with the response to UVC-induced DNA damage (Figure 5B). This was confirmed by clonogenic cell survival assays (Figure 6). Hence, in a HeLa genetic background, XRCC4, Lig IV and Rad51 could be essential partners or limiting factors or both in the commitment of DNA repair processes after UVC irradiation. These observations have to be verified in other cell lines. In contrast, this relationship between NER and DSBR pathways seemed to be independent of DNA PKcs.
Since the MRN complex plays an essential role at sites of blocked replication, it was expected that MRN-deficient cells would encounter major difficulties after UVC irradiation. MRE11KD cells, but not Rad50KD and NBS1KD ones, displayed an impaired response to high doses of UVC irradiation, since many cells remained stalled in S phase 24 h after 10 J/m2 irradiation, as did XP cells (Figure 5C). However, survival assays revealed that all MRN-deficient cells (MRE11KD, Rad50KD and NBS1KD) displayed enhanced sensitivity to UVC (Figure 6).
While ATMKD cells failed to elicit impaired progression of cells in the cell cycle after UVC irradiation, ATRKD cells exhibited great intolerance to UVC-induced DNA damage (Figure 5C). Survival assays reinforced this observation by showing that ATRKD cells were very sensitive to UVC (Figure 6). More generally, major growth disadvantages and elevated spontaneous cell death were observed in ATRKD HeLa cells in comparison with ATMKD ones.
γ Ray irradiation
Ionizing radiation triggered a slight G1 phase arrest, with moderate G2 phase blockage in HeLa cells (Figure 5A). DNA-PKcsKD, XRCC4KD and Lig IVKD cells, which are deficient in the major DSB-repair mechanism (NHEJ), displayed marked G2–M arrest 24 h after ionizing radiation (Figure 5B). In particular, most DNA-PKcsKD cells were blocked in G2–M phase 24 h after 6 Gy. This accumulation of damaged cells demonstrated intolerance to DSB-inducing agents in DNA-PKcsKD, XRCC4KD and Lig IVKD cells, which was confirmed by clonogenic cell survival assays (Figure 6). A similar result was observed with Rad51KD and Rad54KD cells, most of which were arrested in G2–M phase (Figure 5B). However, clonogenic cell survival assays revealed that HR-deficient cells were less sensitive to ionizing radiation than their NHEJ counterparts (Figure 6). In contrast, Lig IIIKD cells remained insensitive to γ rays, suggesting a minor role of this protein in the cellular response to ionizing radiation. It was noteworthy that the hallmark of Lig III-deficient cells was enhanced sensitivity to cross-linking agents such as mitomycin C.
Interestingly, MRE11KD and NBS1KD cells elicited intolerance to a high dose of γ rays with marked G2–M phase blockage (Figure 5C). In clonogenic cell survival assays, all MRN-deficient cells appeared to be sensitive to a low dose of γ rays (Figure 6).
Strikingly, XPCKD and hHR23BKD cells displayed intolerance to γ rays, in contrast to XPAKD and hHR23AKD cells. Interestingly, XPCKD cells displayed a moderate sensitivity in survival assays after 1 Gy irradiation (Figure 6). This suggested an indirect role in a regulatory process. We have recently showed that the long-term inactivation of the XPC gene induced important changes in the DNA repair potential by affecting the NER process, but also other repair pathways. We have found that XPC deficiency in HeLa cells could be correlated with a decrease in end-joining efficiency (NHEJ DNA repair) (29)
Twenty-four hours after irradiation, ATMKD and ATRKD cells failed to display an impaired cell cycle progression. However, clonogenic survival assays revealed that while ATRKD cells displayed an enhanced sensitivity after a low dose of ionizing radiation (1 Gy) (Figure 6), ATMKD cells exhibited a very low radiosensitivity. I could not rule out that the remaining ATM proteins were enough to ensure their biological functions.
Etoposide treatment
Cells were treated with etoposide (VP16) for 1.5 h and thereafter culture mediums were changed to allow recovery after DNA damage. In these conditions, the treatment was reversible and normal cells were able to circumvent etoposide-induced DNA lesions (e.g. DSBs) in order to progress through S phase. Twenty-four hours after the beginning of the treatment, most normal cells (mock-treated HeLa cells and BD650) accumulated in G2–M phase in order to restore their genomic integrity (Figure 5A).
In NHEJ-deficient cells (DNA-PKcsKD, XRCC4KD and Lig IV), etoposide induced persistent arrest during the whole S phase, suggesting the presence of unrepaired (or unrepairable) DNA damage (Figure 5B). This was confirmed by clonogenic survival assays (Figure 6). In contrast, Rad51KD and Rad54KD cells appeared to be insensitive to this treatment using my two separate approaches (Figures 5B and 6). This may suggest that the NHEJ pathway is the main DNA repair process required during cell recovery after etoposide treatment and withdrawal in HeLa cells. No difference was observed between control and MRN-deficient cells after etoposide treatment (Figures 5C and 6). The integrity of the MRN complex seemed not to be a prerequisite for repair of etoposide-induced DNA damage.
Interestingly, XPCKD HeLa cells, but not their XPA counterparts, displayed intolerance toward etoposide (Figures 5A and 6). This reproducible result was discussed in a recent report (29). It appears that beside its canonical function in the early steps of the NER, the XPC protein could be essential in the coordination of other recovery pathways, such as those involved in the repair of etoposide-induced DNA damage. Hence, my set of cell lines helps us to emphasize new relationships between different DNA repair processes.
DISCUSSION
Long-term gene silencing opens up new areas in the field of DNA repair. Because evolution has retained highly sophisticated DNA repair processes that prevent most of the damage inflicted on the genome, it is necessary to unravel the connections between these pathways in a genetically homogeneous cell model.
For the first time, I present stable clones silenced for genes acting as sensors/transducers (ATM, ATR, Rad50, NBS1 and MRE11), or genes of different DNA repair pathways (XPA, XPC, hHR23A, hHR23B, CSA, CSB, DNA-PKcs, XRCC4, Ligase IV, Rad51, Rad54 or Ligase III). Other HeLa clones have also been established, such as PARGKD (in culture for 113 days) and Ogg1KD (in culture for 180 days), which display a spectacular loss-of-function (G. de Murcia and A. Campalans, personal communications). I have also constructed and characterized new efficient pEBVsiRNA plasmids directed against PARP2, BRCA1, ERCC1, XRCC1, cdc25b or TRF2 and new clones are now under investigation (data not shown). All of these clones are being maintained in culture for further analysis.
My approach is based on new siRNA-expressing plasmids termed pEBVsiRNA. The intrinsic features of these plasmids, such as their nuclear retention, their high stability and their tethering to endogenous chromosomes, greatly improve short- and long-term gene silencing in human cells (for review see 13). Because the transactivating function of the oriP/EBNA1 complex is essential for activation of promoters carried by the plasmid, the reverse position of the shRNA cartridge related to EBV components could enhance the long-term transcription of shRNA sequences (25). This transactivating process involves EBNA1-induced changes in the chromatin structure, including DNA looping and nucleosome destabilization (51). In these conditions, 2 weeks after transfection (and hygromycin B selection) of pEBVsiRNA plasmids, ∼100% of transfected cells were silenced. This was the case for numerous genes tested, such as XPA, hHR23A, MRE11, Lig III, Lig IV or Rad50. As a consequence, these cell populations are easily propagated for >1 month and maintained an impressive gene silencing. Selected clones elicited this marked gene silencing for an undetermined period in culture, even after several freeze–thaw cycles. Interestingly, all clones described here are still being maintained in culture, after several months or >1 year for my older clones (XPAKD, XPCKD or DNA-PKcsKD).
I initially developed these new plasmids to compare short- and long-term gene silencing in DNA repair. I thought that transient transfection assays could either amplify or mask the real effects of specific gene silencing, due to an overproduction of siRNA in cultured cells, which could be due to (i) high concentrations of siRNA duplexes, (ii) very high numbers of copies per cell of an integrative plasmid or (iii) elevated virus titers. I targeted genes of the main DNA repair processes (NHEJ, NER, HR and BER) and of crucial DNA damage signaling pathways (ATM, ATR and MRN). Stable HeLa clones were established for all targeted genes. We could then assess the short- and long-term effects of specific DNA repair deficiency on different biological endpoints, such as chromosomal and telomeric abnormalities. Experiments are under way to compare genomic stability and telomere maintenance as a function of the loss of one specific DNA repair pathway (e.g. NHEJ versus HR), after short- and long-term gene silencing. The high efficiency of pEBVsiRNA vectors in different cell lines affords us the opportunity to rapidly obtain new knock-down clones that will be stable over time, in particular for certain genes where to date no mutant lines are available. Most of these clones display very low or undetectable levels of the protein of interest, as shown by western blot or immunocytochemical staining. However, the major drawback of the RNAi approach is underestimation of the biological role of a protein, because in theory a residual amount of the targeted protein remains. This raises two questions: (i) is the residual protein sufficient to ensure its biological functions, and (ii) can we detect a loss-of-function in knock-down clones? At present, my results reveal that knock-down clones exhibited the expected phenotype with an associated loss-of-function. For instance, XPAKD and XPCKD cells displayed an impaired UDS with an enhanced UVC sensitivity (25), XRCC4KD, LigIVKD and DNA-PKcsKD cells failed to accurately join DNA ends during NHEJ and were highly sensitive to DSB-generating agents (29). The cellular response of knock-down clones to diverse genotoxic injuries seems to be in agreement with the expected phenotype (Figures 5 and 6). Another point to be considered is the existence of redundancy in some functions in mammalian cells. For instance, Rad54B protein could compensate for the silencing of Rad54 (52).
My data also raise some intriguing and provocative questions. For instance, (i) do XPC and hHR23B proteins participate in the cellular response to ionizing radiation? (ii) does Lig III protein participate in the low-speed B-NHEJ? (iii) is the NHEJ pathway required for cellular recovery after etoposide-induced DNA damage? and (iv) are MRN proteins required during HR or NHEJ or both? Many other interesting questions emerge from these data, and the aforementioned stable knock-down clones afford us the opportunity to answer them.
Because my approach is based on long-term culture, I cannot exclude the possibility that suppression of gene expression over a long period may provoke compensatory cellular responses. In culture, an ‘adaptive period’ may mask the true biological consequences of specific gene silencing. However, this is also true during the multistage process leading to tumorigenesis, where a normal cell encounters serial genetic changes, including initiation, clonal expansion, pre-malignant lesions and malignant progression, before acquiring a tumoral phenotype. This adaptive period is also present in other strategies to switch off gene expression, as in knock-out mouse models where clonal cell expansion is required. It is therefore necessary to characterize the expected phenotype in knock-down cells during the culture, and also to compare short- and long-term experiments. In my hands, long-term silencing of genes of the NER or NHEJ pathways, mediated by pEBVsiRNA vectors, always gave the expected phenotypic modifications.
To conclude, the combination of RNAi technology with pEBV-derived vectors offers an exceptional opportunity to rapidly create a set of stable knock-down clones covering various fields of genetics. We could mimic human diseases in a comparable genetic background. This approach may have several applications in drug screening, and in the early stages of testing new therapies. Now I have to develop my approach in other human cells, in particular normal cells such as stem cells. A recent study reinforces my strategy by demonstrating that the pEBV vector could effectively enhance RNA interference efficiency in human stem cells (hES) (53). Abrogation of DNA repair genes in hES could offer new opportunities, because genetic modifications of stem cells have great therapeutic potential. Another fundamental aspect of this work is the validation of a specific siRNA sequence for several weeks or months in culture. This could ensure unfailing gene silencing before the beginning of time-consuming experiments, such as assays on hES.
ACKNOWLEDGEMENTS
My thanks go to J. Délic and S. Marcand for critical reading of this manuscript, and to E. Despras for performing in vitro NHEJ assays. In accordance with the Quality Management Program of my department, this work is reported on custom laboratory books numbered CEA/LGR/0760, 3090, 3191, 4658, 4659, 4660, 6093, 6094 and 7231. I thank ‘Electricité de France (EDF)’ for its financial support. Funding to pay the Open Access publication charges for this article was provided by EDF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhang Y, Zhou J, Lim CU. The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res. 2006;16:45–54. doi: 10.1038/sj.cr.7310007. [DOI] [PubMed] [Google Scholar]
- 2.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 3.Hoeijmakers JH. Nucleotide excision repair. II: from yeast to mammals. Trends Genet. 1993;9:211–217. doi: 10.1016/0168-9525(93)90121-w. [DOI] [PubMed] [Google Scholar]
- 4.Giglia-Mari G, Coin F, Ranish JA, Hoogstraten D, Theil A, Wijgers N, Jaspers NG, Raams A, Argentini M, et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 2004;36:714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 5.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 6.Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP. Cernunnos interacts with the XRCC4 × DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J. Biol. Chem. 2006;281:13857–13860. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
- 7.Duker NJ. Chromosome breakage syndromes and cancer. Am. J. Med. Genet. 2002;115:125–129. doi: 10.1002/ajmg.10688. [DOI] [PubMed] [Google Scholar]
- 8.O’Driscoll M, Gennery AR, Seidel J, Concannon P, Jeggo PA. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair (Amst.) 2004;3:1227–1235. doi: 10.1016/j.dnarep.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 9.O’Driscoll M, Jeggo PA. The role of double-strand break repair—insights from human genetics. Nat. Rev. Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 10.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 11.Biard DS, Cordier A, Sarasin A. Establishment of a human cell line for the detection of demethylating agents. Exp. Cell Res. 1992;200:263–271. doi: 10.1016/0014-4827(92)90172-5. [DOI] [PubMed] [Google Scholar]
- 12.Calos MP. The potential of extrachromosomal replicating vectors for gene therapy. Trends Genet. 1996;12:463–466. doi: 10.1016/0168-9525(96)40049-x. [DOI] [PubMed] [Google Scholar]
- 13.Biard DSF, Angulo JF. Long term RNA interference: toward a set of isogenic human cells deficient in DNA repair genes. In: Landseer BR, editor. New Research on DNA Repair. Vol. 7. Hauppauge: Nova Science Publishers, Inc; 2007. (in press) [Google Scholar]
- 14.Kapoor P, Frappier L. EBNA1 partitions Epstein-Barr virus plasmids in yeast cells by attaching to human EBNA1-binding protein 2 on mitotic chromosomes. J. Virol. 2003;77:6946–6956. doi: 10.1128/JVI.77.12.6946-6956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas JC. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Wilson A, Alber S, Ma Z, Tang ZL, Satoh E, Mazda O, Watkins S, Huang L, et al. Prolonged gene expression in mouse lung endothelial cells following transfection with Epstein-Barr virus-based episomal plasmid. Gene. Ther. 2003;10:822–826. doi: 10.1038/sj.gt.3301958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106:287–296. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- 18.Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 2003;116:3971–3984. doi: 10.1242/jcs.00708. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Chau CM, Deng Z, Shiekhattar R, Spindler MP, Schepers A, Lieberman PM. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 2005;24:1406–1417. doi: 10.1038/sj.emboj.7600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 21.Tuschl T. RNA interference and small interfering RNAs. Chembiochem. 2001;2:239–245. doi: 10.1002/1439-7633(20010401)2:4<239::AID-CBIC239>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 22.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 24.Chalk AM, Wahlestedt C, Sonnhammer EL. Improved and automated prediction of effective siRNA. Biochem. Biophys. Res. Commun. 2004;319:264–274. doi: 10.1016/j.bbrc.2004.04.181. [DOI] [PubMed] [Google Scholar]
- 25.Biard DS, Despras E, Sarasin A, Angulo JF. Development of new EBV-based vectors for stable expression of small interfering RNA to mimick human syndromes: application to NER gene silencing. Mol. Cancer Res. 2005;3:519–529. doi: 10.1158/1541-7786.MCR-05-0044. [DOI] [PubMed] [Google Scholar]
- 26.Biard DS, Miccoli L, Despras E, Frobert Y, Creminon C, Angulo JF. Ionizing radiation triggers chromatin-bound kin17 complex formation in human cells. J. Biol. Chem. 2002;277:19156–19165. doi: 10.1074/jbc.M200321200. [DOI] [PubMed] [Google Scholar]
- 27.Pfeiffer P, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 28.Pfeiffer P, Feldmann E, Odersky A, Kuhfittig-Kulle S, Goedecke W. Analysis of DNA double-strand break repair by nonhomologous end joining in cell-free extracts from mammalian cells. Methods Mol. Biol. 2005;291:351–371. doi: 10.1385/1-59259-840-4:351. [DOI] [PubMed] [Google Scholar]
- 29.Despras E, Pfeiffer P, Salles B, Calsou P, Kulle SK, Angulon JF, Biard DSF. Long-term XPC silencing affects DNA double-strand break repair in HeLa cells. Cancer Res. 2007;67:2526–2534. doi: 10.1158/0008-5472.CAN-06-3371. [DOI] [PubMed] [Google Scholar]
- 30.Bunick CG, Miller MR, Fuller BE, Fanning E, Chazin WJ. Biochemical and structural domain analysis of xeroderma pigmentosum complementation group C protein. Biochemistry. 2006;45:14965–14979. doi: 10.1021/bi061370o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lommel L, Ortolan T, Chen L, Madura K, Sweder KS. Proteolysis of a nucleotide excision repair protein by the 26 S proteasome. Curr. Genet. 2002;42:9–20. doi: 10.1007/s00294-002-0332-9. [DOI] [PubMed] [Google Scholar]
- 32.You JS, Wang M, Lee SH. Biochemical analysis of the damage recognition process in nucleotide excision repair. J. Biol. Chem. 2003;278:7476–7485. doi: 10.1074/jbc.M210603200. [DOI] [PubMed] [Google Scholar]
- 33.Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–436. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage (Version 6) DNA Repair (Amst.) 2004;3:1617–1638. doi: 10.1016/j.dnarep.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 35.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 36.Verkaik NS, Esveldt-van Lange RE, van Heemst D, Bruggenwirth HT, Hoeijmakers JH, Zdzienicka MZ, van Gent DC. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 37.Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 2004;104:14–20. doi: 10.1159/000077461. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 39.Petermann E, Keil C, Oei SL. Roles of DNA ligase III and XRCC1 in regulating the switch between short patch and long patch BER. DNA Repair (Amst.) 2006;5:544–555. doi: 10.1016/j.dnarep.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Lim CU, Li JJ, Cai L, Zhang Y. The role of NBS1 in the modulation of PIKK family proteins ATM and ATR in the cellular response to DNA damage. Cancer Lett. 2006;243:9–15. doi: 10.1016/j.canlet.2006.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gennery AR. Primary immunodeficiency syndromes associated with defective DNA double-strand break repair. Br. Med. Bull. 2006:71–85. doi: 10.1093/bmb/ldl006. [DOI] [PubMed] [Google Scholar]
- 42.Lee JH, Lim DS. Dual role of Nbs1 in the ataxia telangiectasia mutated-dependent DNA damage response. FEBS J. 2006;273:1630–1636. doi: 10.1111/j.1742-4658.2006.05191.x. [DOI] [PubMed] [Google Scholar]
- 43.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst.) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 44.Usui T, Petrini JH, Morales M. Rad50S alleles of the Mre11 complex: questions answered and questions raised. Exp. Cell. Res. 2006;312:2694–2699. doi: 10.1016/j.yexcr.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 45.Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, Ottini L, Crescenzi M, Martinotti S, et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3:248–254. doi: 10.1093/embo-reports/kvf044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Fabbro M, Savage K, Hobson K, Deans AJ, Powell SN, McArthur GA, Khanna KK. BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J. Biol. Chem. 2004;279:31251–31258. doi: 10.1074/jbc.M405372200. [DOI] [PubMed] [Google Scholar]
- 48.Sung PA, Libura J, Richardson C. Etoposide and illegitimate DNA double-strand break repair in the generation of MLL translocations: new insights and new questions. DNA Repair (Amst.) 2006;5:1109–1118. doi: 10.1016/j.dnarep.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 49.Ng JM, Vermeulen W, van der Horst GT, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JH. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Limoli CL, Giedzinski E, Cleaver JE. Alternative recombination pathways in UV-irradiated XP variant cells. Oncogene. 2005;24:3708–3714. doi: 10.1038/sj.onc.1208515. [DOI] [PubMed] [Google Scholar]
- 51.Zetterberg H, Borestrom C, Nilsson T, Rymo L. Multiple EBNA1-binding sites within oriPI are required for EBNA1-dependent transactivation of the Epstein-Barr virus C promoter. Int. J. Oncol. 2004;25:693–696. [PubMed] [Google Scholar]
- 52.Wesoly J, Agarwal S, Sigurdsson S, Bussen W, Van Komen S, Qin J, van Steeg H, van Benthem J, Wassenaar E, et al. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell Biol. 2006;26:976–989. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren C, Zhao M, Yang X, Li D, Jiang X, Wang L, Shan W, Yang H, Zhou L, et al. Establishment and applications of Epstein-Barr virus-based episomal vectors in human embryonic stem cells. Stem Cells. 2006;24:1338–1347. doi: 10.1634/stemcells.2005-0338. [DOI] [PubMed] [Google Scholar]