Abstract
Metallic nanoparticles of gold functionalized with oligonucleotides conventionally use a terminal thiol modification and have been used in a wide range of applications. Although readily available, the oligonucleotide–nanoparticle conjugates prepared in this way suffer from a lack of stability when exposed to a variety of small molecules or elevated temperatures. If silver is used in place of gold then this lack of stability is even more pronounced. In this study we report the synthesis of highly stabilized oligonucleotide–nanoparticle conjugates using a simple oligonucleotide modification. A modified solid support was used to generate 3′-thioctic acid modified oligonucleotides by treatment with an N-hydroxysuccimidyl ester of thioctic acid. Unusually, both gold and silver nanoparticles have been investigated in this study and show that these disulphide-modified oligonucleotide probes offer significant improvements in nanoparticle stability when treated with dithiothreitol (DTT) compared with monothiol analogues. This is a significant advance in oligonucleotide–nanoparticle conjugate stability and for the first time allows silver nanoparticles to be prepared that are more stable than standard gold-thiol functionalized nanoparticles. This opens up the possibility of using silver nanoparticles functionalized with oligonucleotides as an alternative to gold.
INTRODUCTION
The area of molecular diagnostics is at the forefront of modern bioanalytical research and a sector of growing importance is the interface with materials science on the nanoscale and the conjugation of metallic nanoparticles with biomolecules (1–5). The ability to detect DNA sequences in an highly selective manner for the rapid detection of genetic mutations and disease states is of ever growing interest and central to a number of new approaches to disease management. Specific DNA sequences can be detected by hybridization to complementary oligonucleotide probes conjugated with nanoparticle substrates in a number of different ways (1). Gold nanoparticles have been most widely used in this manner with oligonucleotide adsorption typically achieved through thiol modification, following Brust's observation that alkylthiols stabilize gold colloidal solutions (6–8). Bioanalytical probes based on those observations have been shown to be capable of discriminating between fully complementary and single base mismatched sequences (9). This approach relies on the hybridization-induced reversible aggregation of the nanoparticles resulting in a distinctive red-shifting of the plasmon of the nanoparticles. The nanoparticles have unusually high extinction coefficients in the visible region of the spectrum which makes them easy to visualize colourimetrically, by eye or instrumentation, when a change in the plasmon relating to hybridization takes place. A particular disadvantage of the thiol adsorption strategy to immobilize the oligonucleotide probe on the gold is its lability under certain conditions such as prolonged or cycled elevated temperatures, high NaCl concentrations and treatment with biological buffer additives e.g. dithiothreitol (DTT) or mercaptoethanol (10). Upon thiol desorption, irreversible aggregation occurs and the probe system is rendered inactive. To minimize these undesirable aggregation events, structurally complex multiple thiol linker systems have been investigated but were only ever reported as being used with gold nanoparticles (10,11).
Thioctic acid is an inexpensive, commercially available and structurally simple disulphide species. Herein we report its preliminary employment as a linker molecule for oligonucleotide nanoparticle functionalization of greater stability than standard thiol analogues. Thioctic acid has received great attention in recent years in the area of gold and silver nanoparticle functionalization for a variety of applications (12–18), ranging from the immobilization of tetrathiofulvalenes as cation sensors on gold electrodes (12) and the investigation of metal ion chelation (13), to the probing of nanoparticle surface adsorption by nitroxide-modified thioctic ester spin labelled probes (14). Indeed, it has also been used to immobilize molecules for bioanalytical applications; carbohydrates for non-specific protein interactions (15), antibodies (16) and transition metal complexes for protein capture (17). From an oligonucleotide perspective thioctic acid oligonucleotide templates have been generated by attachment to gold nanoparticles via mid-sequence amino-modified sites (18) and via multiple-step post-synthetic modification (19), however, there was no data presented on the subsequent improved properties of the oligonucleotide–nanoparticle conjugates (18). Until now, the stability of thioctic acid functionalized oligonucleotide–nanoparticle conjugates has not been reported and their ultimate application to oligonucleotide sequence analysis has not been realized.
INSTRUMENTATION
1H NMR and 13C NMR were recorded on a Brüker DPX 400 MHz spectrometer with the appropriate solvent peak as reference. J values are quoted in Hertz. Mass spectrometry was carried out as a service by Swansea EPSRC mass spectrometry centre. Preparative reversed phase HPLC was carried out on a Dionex UVD170U detector fitted with a P680 pump through a Phenomenex Clarity column. Desalting HPLC was carried out on a Biocad Sprint HPLC using a HiTrap (5 ml) size exclusion column. Oligonucleotides were synthesized on an MerMade 6 Nucleic Acid Synthesiser, unless otherwise stated. ZipTipC18 purification pipette tips were purchased from Millipore Corp. Oligonucleotide synthesis reagents were purchased from Link Technologies.
MATERIALS AND METHODS
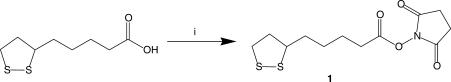
1-{[5-(1,2-dithiolan-3-yl)pantanoyl]oxy}-2,5-pyrrolidinedione, 1
N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC.HCl) (1.840 g, 9.6 mmol, 1.2 eq) was dissolved in DCM (anhydrous, 20 ml), which, 1.7 ml diisopropylethylamine (DIPEA) (anhydrous, 9.6 mmol, 1.2 eq) was added and stirred for 10 min. To the solution N-hydroxysuccinimide (1.290 g, 11.2 mmol, 1.4 eq) was added and the reaction mixture stirred in an ice bath. Thioctic acid (1.643 g, 8.0 mmol, 1.0 eq) was dissolved in anhydrous DCM (10 ml) and added to the reaction over 5 min and stirred overnight. The reaction mixture was washed with HCl (5% (v/v), 50 ml × 2) and water (distilled, 50 ml). The organic layer was dried over Na2SO4. Solvents were concentrated in vacuo to yield 2.236 g of ester 1, as a yellow, solid. The residue was dry-loaded on Na2SO4 and the product purified by flash column chromatography giving the product in 83% yield. δH(400 MHz; DMSO) 1.4–1.7 (6H, m, CH2 × 3), 1.84–1.93 (1H, m, CH), 2.38–2.46 (1H, m, CH), 2.68 (2H, t, J 7.2, CH2), 2.81 (4H, s, succinimidyl CH2 × 2), 3.12–3.21 (2H, m, CH2), 3.56–3.65 (1H, m, CH). δC(400 MHz; DMSO) 24.415, 25.830, 28.026, 30.425, 34.217, 38.485, 56.011, 169.287, 170.612. M/Z 321.0937 ([M+NH4+] C12H17NO4S2 requires, 321.0937).
Synthesis of 3′-disulphide-modified oligonucleotides
The NHS ester, 1, was added to the 3′-end of oligonucleotides (5′- FAM CAT TGA AGC TTC (Pr1), 5′- FAM CAT TGA AGC TTC TTT TTT TTT T (Pr2), 5′-FAM CAT TGA AGC TTC AAA AAA AAA A (Pr3) and 5′- ATC CTG AAT GCG AAA AAA AAA A (Pr4)) using 3′-amino-modifier C7 CPG solid supports (1 μmol). The Fmoc protecting group was removed from the 3′-amino group by treatment with 20% (v/v) piperidine/MeCN for 30 min. The NHS ester, 1 (20 mg in 1 ml of MeCN) was applied to the column overnight then washed with MeCN (3 × 1 ml) before being used as the solid support for oligonucleotide synthesis. The oligonucleotide sequence was prepared on a MerMade 6 Nucleic Acid Synthesiser with the FAM introduced as a commercially available terminal phosphoramidite. The oligonucleotides were cleaved from the CPG supports by incubation in 1 ml of conc. NH4OH for 3 days at room temperature. The ammonium hydroxide was removed in vacuo (room temperature) and re-dissolved in H2O (1 ml, distilled) and purified by reversed-phase HPLC on a Dionex UVD170U detector fitted with a P680 pump through a Phenomenex Clarity column. Buffer A: TEAA (0.1 M, pH 7), Buffer B: CH3CN; T = 0, 95% TEAA: 5% MeCN, with a gradient of 1% min−1 B over 15 min and held at 20% B for 5 min at a flow rate of 1 ml/min. After HPLC purification the fractions were collated and concentrated in vacuo (room temperature) and redissolved in H2O (1 ml, distilled), they were then freeze-dried and redissolved in H2O (1 ml, distilled) and stored at 4°C. HPLC analysis of 3′-thioctic acid, 5′-FAM-modified oligonucleotides indicated modification had occurred. Confirmation of this was achieved by reference to MALDI mass spectrometry of a known thioctic acid modified sequence, M/Z 3977.6 ([MALDI] C15H28NO3S2-ATGCTCAACTCT requires, 3977.4).
Synthesis of 3′-thiol-modified oligonucleotide probes
Sequences were prepared on a MerMade 6 Nucleic Acid Synthesiser with commercially available 3′-thiol-modified universal support (propyl) and FAM introduced at the 5′-end by a commercially available phosphoramidite. The oligonucleotides were cleaved from the CPG supports by incubation in 1 ml of conc. NH4OH for 3 days at room temperature. The ammonium hydroxide was removed in vacuo (room temperature) and re-dissolved in H2O (1 ml, distilled) and purified by reversed-phase HPLC on a Dionex UVD170U detector fitted with a P680 pump through a Phenomenex Clarity column. Buffer A: TEAA (0.1 M, pH 7), Buffer B: CH3CN; T = 0, 95% TEAA: 5% MeCN, with a gradient of 1% min−1 B over 15 min and held at 20% B for 5 min at a flow rate of 1 ml/min. After HPLC purification, the fractions were collated and concentrated in vacuo (room temperature) and redissolved in H2O (1 ml, distilled), they were then freeze-dried and then redissolved in H2O (1 ml, distilled) and stored at 4°C.
Functionalization of nanoparticles with oligonucleotides
Thirteen nanometre gold nanoparticles were prepared via citrate reduction of HAuCl4. Thirty seven nanometre silver nanoparticles were prepared via citrate reduction of AgNO3.
Disulphide-modified oligonucleotides
Desalted oligonucleotide (5.71 nmol) was added to Au (3 ml, 17 nM) or Ag (3 ml, 1.0 nM) colloid in an autoclaved glass vial. After 18 h, phosphate (NaH2PO4/Na2HPO4) buffer (60 mM) was added to 10 mM final concentration. 2 M NaCl was then added at intervals, increasing the salt concentration by 0.05 M ncrements to a final concentration of 0.3 M. The oligonucleotide–gold conjugates were stored at 4°C.
Thiol-modified oligonucleotides
Desalted oligonucleotide (5.71 nmol) was added to Au (3 ml 17 nM) or Ag (3 ml, 1.0 nM) colloid in an autoclaved glass vial. After 18 h, phosphate (NaH2PO4/Na2HPO4) buffer (60 mM) was added to 10 mM final concentration. 2 M NaCl was then added at intervals, increasing the salt concentration by 0.05 M (final concentration) increments to a final concentration of 0.3 M. The oligonucleotide–gold conjugates were stored at 4°C.
Pre-treated thiol-modified oligonucleotides
An aliquot of 1 ml of desalted oligonucleotide (10 µM) in Na2HPO4 (pH 8.5) was shaken with DTT (0.1 M) for 30 min. DTT was removed by size exclusion chromatography though a Hi-Trap desalt column and 1 ml of the oligonucleotide collected directly onto 3 ml of Au or Ag colloid.
RESULTS
Synthesis of thioctic-modified oligonucleotides
A number of options were considered for the attachment of thioctic acid to oligonucleotides. Ultimately, the synthesis of an active ester of the thioctic acid was determined as being the most versatile approach to functionalizing the termini of oligonucleotides. Primary amine groups are readily added to the 3′- or 5′-termini of oligonucleotides and will react with the active ester of thioctic acid either on a solid support or in solution to provide the disulphide functionalized oligonucleotide. In addition the thioctic acid was shown to be stable to the conditions used for oligonucleotide synthesis and deprotection indicating its compatibility with routine oligonucleotide synthesis. The N-hydroxysuccinimidyl ester, 1, of thioctic acid was prepared in good yield by the esterification of the carboxylic acid with hydroxysuccinimide using EDC (Scheme 1). This active ester was then used in two different approaches.
Scheme 1.
(i) EDC, NHS, 0°C, DCM, 83%.
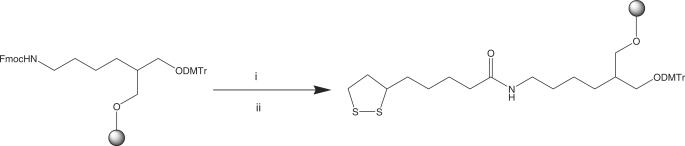
Modification at the 3′-terminus was achieved by using an amino-modified solid support. Initially, the Fmoc protecting group was removed using standard piperidine deprotection to give the free amine on the CPG. The active ester was added to the CPG in acetonitrile and left overnight (Scheme 2). Following washing with acetonitrile, the thioctic acid solid support was used with standard phosphoramidite chemistry.
Scheme 2.
(i) 20% Piperidine/MeCN (v/v), 30 min and (ii) 1, MeCN, overnight.
Three sequences were generated with the thioctic acid modification at the 3′-terminus and a FAM label at the 5′-terminus: 5′- FAM CAT TGA AGC TTC (Probe 1), 5′- FAM CAT TGA AGC TTC TTT TTT TTT T (Probe 2) and 5′-FAM CAT TGA AGC TTC AAA AAA AAA A (Probe 3), respectively. The sequences chosen were designed to assess whether the presence of spacer bases (adenine or thymine) would affect stability and/or surface coverage of the Au or Ag conjugates. Preparing both disulphide (DX) and thiol (TX) analogues allows for comparison of surface coverage and assessment of stability of the nanoparticle–oligonucleotide conjugates (where X denotes the Probe sequence as indicated above). In addition, thiol conjugates were prepared that omitted the treatment of DTT prior to conjugation (TTX). This is usually carried out to ensure that the thiol-modified oligonucleotide is in a mono-thiol form, rather than disulphide, when applied to the nanoparticles.
Nanoparticle functionalization and stability studies
Gold nanoparticle stability
Immobilization of the disulphide-modified oligonucleotide sequences, D1, D2 and D3 on citrate-reduced gold nanoparticles (20) was carried out using standard protocols with incremental elevation of NaCl concentration. In earlier investigations, thioctic acid oligonucleotides were added to gold nanoparticles in water and after 18 h phosphate buffer added followed by aliquots of NaCl over 5 days to give a final salt concentration of 0.3 M. If the salt concentration is raised too rapidly then the nanoparticles can precipitate if a thiolated oligonucleotide is used. The NaCl concentration of the disulphide-modified sequences, while still raised incrementally, was condensed to a considerably shorter timescale of 2 days, whilst maintaining nanoparticle stability on citrate-reduced gold and silver nanoparticles. The final conditions of the conjugate materials were 10 mM phosphate, pH 7, 0.3 M NaCl (0.3 M PBS). Thiol-modified oligonucleotides (1 ml, 10 µM) were subjected, as standard, to treatment with DTT, 0.1 M, phosphate at pH 8.5 for 30 min before desalting directly onto the nanoparticles, generating probes T1, T2 and T3. A set of oligonucleotide–nanoparticle conjugates were also prepared without this DTT treatment prior to addition of oligonucleotide, probes TT1, TT2 and TT3. Both sets of thiol-modified oligonucleotides were treated to a ‘softer’ salt-ageing process and were complete in 5 days.
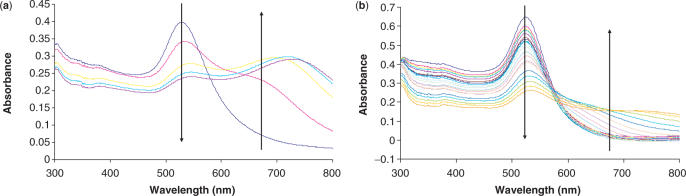
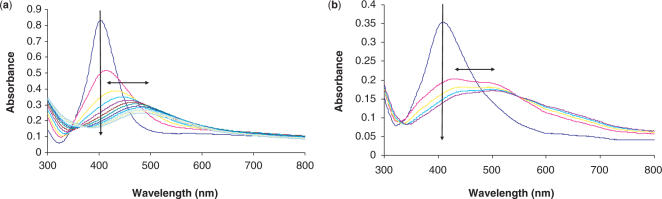
Following literature precedent, the effect of the thioctic acid linker moiety on the stability of oligonucleotide–nanoparticle conjugates, was assessed by treating the conjugates with DTT (10 mM final concentration) at 40°C (10,22). Typical UV–Vis spectra taken periodically for both thiolated (T3 at 1 min intervals) and disulphide-modified oligonucleotide (D1 at 10 min intervals) gold conjugates are shown in Figure 1. Figure 1a shows the rapid degradation of the conjugates typical of thiolated oligonucleotides. Even from these ‘busy’ plots, the enhancement in stability of the conjugates can be seen when employing the disulphide modification, as each entry signifies another 10 min interval cf. 1 min for the thiol plot.
Figure 1.
UV–Vis spectra showing degradation upon treatment with 10 mM DTT of (a) T3–Au conjugates at 1 min intervals and (b) D1–Au conjugates at 10 min intervals. Arrows added to highlight the ‘movement’ of the spectra with time.
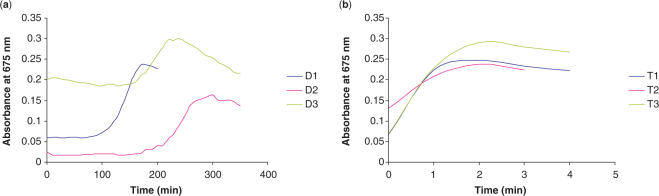
Monitoring the disappearance of the plasmon band at 520 nm and the appearance of one between 600 and 700 nm indicates the progressive aggregation event when gold nanoparticles are used. Absorption spectra were taken at periodic intervals. Plotting absorbance at a particular wavelength versus time for each experiment clearly shows the stability of the system. This was carried out for the D1–Au, D2–Au, D3–Au, TT1–Au, TT2–Au, TT3–Au, T1–Au, T2–Au and T3–Au conjugates. Figure 2 shows the absorbance versus time at the ‘significant’ 675 nm plasmon wavelength for each of these samples. The DX results (Figure 2a) are plotted together to highlight the effect of spacer bases as are TX (Figure 2b). Note that TTX–Au conjugates show the same profile as those in Figure 2b.
Figure 2.
Monitoring the emergence of the plasmon at 675 nm against time gives an indication of the durability of the oligonucleotide conjugates when subject to the same conditions. Figures (a) and (b) show the plots of the DX–Au and TX–Au conjugates at 675 nm, respectively. These results are pronounced when the timescale over which they were acquired are considered.
The half-life (time taken for absorbance at 675 nm to reach half the value for complete aggregation) was calculated for each conjugate (Table 1) for ease of comparison.
Table 1.
Half-lives for Au–oligonucleotide conjugates
The disparate stabilities of the thiol and disulphide systems are striking when comparing the half-lives of the conjugate systems. All of the Au-monothiol systems have reached the half-way point of complete aggregation in 1 min or less. Compare this with the thioctic acid disulphide system and the enhanced stability is clear with half-lives of 140, 195 and 245 min. It should be noted (as shown in Figure 2a) that DTT-induced aggregation event is not linear, a long period of sustained stability is followed by a sharp increase in absorbance. With the disulphide conjugates (Figures 2a), aggregation begins at 90 min for D1 (no spacers) or 2 h incubation for D2 and D3 (polyT and polyA spacers, respectively) and is considerable by 3 h. This is an excellent enhancement when compared with the monothiol results (Figure 2b) which show complete aggregation in <5 min. The emergence of the 675 nm peak is indicative of nanoparticle aggregation, however, by 300 min in the disulphide system there is a loss of signal altogether; this feature can be attributed to the aggregate species growing in size and eventually settling out of the colloidal solution leaving a colourless mother liquor.
In similar studies where a steroidal disulphide was used Mirkin and co-workers state that the structurally complex steroidal disulphide system begins aggregation after 2 h (10). This, he argues, is due to a hydrophobic ‘screening’ effect of the gold surface as well as the formation of a chelate structure. Indeed the dihydrothioctic acid, the reduced dithiol, form of thioctic acid is known to bind through both sulphur atoms (22), and form stable monolayers on gold surfaces, and this now appears to be the driving factor in stability enhancement as similar results have been obtained in this non-steroidal system. It is worthy to note that Mirkin achieved further enhancement of stability when employing a complex trithiol head group (which maintained optical stability for 10 h) (10).
Comparison of thiol and disulphide surface coverage
Whilst there is no doubt that the result of employing disulphide attachment of oligonucleotides to nanoparticles is enhanced stability of the ‘conjugate’ species cf. monothiol, one cannot immediately make the argument that this is due to the disulphide–gold interaction being greater than the thiol–gold one. There are many factors which influence the stability of oligonucleotide conjugates and surface coverage plays an important role.
For this reason the surface coverage of each of the conjugate systems was assessed (Table 2). This was carried out by a method devised by Mirkin et al. (23) Triplicates of the FAM-modified oligonucleotide conjugates were treated with DTT (10 mM) and left to completely aggregate. After which they were centrifuged to ensure that all nanoparticulate sediment was separated from the supernatant. Aliquots of the supernatant were taken, again in triplicate, and fluorescence measured. These fluorescence values when correlated with a standard calibration curve gave a concentration for FAM-labelled oligonucleotide in solution. This FAM-oligonucleotide concentration divided by the starting molar concentration of nanoparticle gives a value of oligonucleotides per nanoparticle and dividing by the surface area of the sphere gives normalized surface coverage values which are easily converted to pmol·cm−2.
Table 2.
Surface coverage data for oligonucleotide–nanoparticle conjugates
| Probe name | Surface coverage (pmol·cm−2) | Standard deviation | Probe name | Surface coverage (pmol·cm−2) | Standard deviation |
|---|---|---|---|---|---|
| D1–Au | 7.4 | 0.3 | D1–Ag | 21.1 | 1.3 |
| D2–Au | 59.9 | 6.7 | D2–Ag | 331 | 8.3 |
| D3–Au | 12.6 | 0.8 | D3–Ag | 105.7 | 0.9 |
| TT1–Au | 17.5 | 0.5 | TT1–Ag | 144.7 | 14.0 |
| TT2–Au | 12.0 | 0.3 | TT2–Ag | 38.1 | 6.5 |
| TT3–Au | 21.1 | 1.2 | TT3–Ag | 76.1 | 2.2 |
| T1–Au | 12.6 | 0.6 | T1–Ag | 31.2 | 1.5 |
| T2–Au | 21.1 | 1.2 | T2–Ag | 64.4 | 3.0 |
| T3–Au | 12.2 | 0.7 | T3–Ag | 123.1 | 15.1 |
Interestingly, it was found that D1 actually has less oligonucleotide surface coverage than both TT1 and T1 7.4 ± 0.3 pmol·cm−2 cf. 17.5 ± 0.5 pmol·cm−2 and 12.6 ± 0.6 pmol·cm−2, respectively. This shows that with no spacer bases the surface coverage by thioctic acid modified oligonucleotide–Au nanoparticle conjugates is less than that of the standard thiol conjugates. In turn, the surface coverage of standard thiol conjugates is found to be less than those that are not treated with DTT prior to conjugation (when the oligonucleotides are added directly to the nanoparticles without treating with DTT and purifying with size exclusion chromatography). This indicates that the conjugation process can be improved when using alkyl-thiolated oligonucleotides as there is no trade-off in conjugate stability or surface coverage as a result of omitting reduction by DTT.
With the polyT spacer, D2, there is greater surface coverage of the Au nanoparticle by the oligonucleotide, 59.9 ± 6.7 pmol·cm−2 compared with 12.0 ± 0.3 pmol·cm−2 and 21.1 ± 1.2 pmol·cm−2 for the TT2 and T2 samples. Therefore, it is difficult to argue that the enhanced stability was due to an increased surface coverage since in the previous case, without spacers, there is decreased surface coverage and the enhanced conjugate stability is still observed.
Indeed, moving to the polyA spacer, D3, the surface coverage is in line with the thiol systems at 12.6 ± 0.8 pmol·cm−2 cf. 21.1 ± 1.2 pmol·cm−2 and 12.2 ± 0.7 pmol·cm−2 for TT3 and T3. Again, as this disulphide conjugate has the same surface coverage as the standard monothiol the enhanced stability cannot be attributed to surface coverage effects.
Whilst these surface coverage results are variable depending upon whether spacer bases are present and indeed which spacers are utilized, it can clearly be seen that the enhanced stability of the conjugate systems cannot be determined by surface coverage. If that was the influencing factor we would expect to see discrepancies in the stability between the disulphide examples in line with surface coverage. That is, D1 would be the least stable, D3 would be of intermediate stability and D2 would be the most stable. This is not observed as D3 and D2 show very similar stability profiles, despite a reasonably large difference in surface coverage.
Silver nanoparticle–oligonucleotide conjugate stability
To demonstrate the versatility of the thioctic acid modified oligonucleotides, silver nanoparticles were used in a similar study. Silver nanoparticles are considerably less stable than gold and as a consequence have been subject to less success in DNA sensing primarily due to the lack of robust surface chemistry. A limited number of studies have been reported but use homo-oligonucleotides or direct hybridization approaches (24–26). Figure 3 shows the degradation of thioctic acid- and thiol-terminated oligonucleotide sequences immobilized on 37 nm citrate-reduced silver nanoparticles. Figure 3 again shows the progressive red-shift of absorbance as a result of aggregation. The thiol system taken at 1 min intervals is clearly less stable than the disulphide which was monitored at 10 min intervals.
Figure 3.
UV–Vis spectra taken for (a) D1–Ag conjugates at 10 min intervals and (b) T3–Ag conjugates at 1 min intervals again clearly shows enhanced stability on the conjugates when thioctic acid tethering to the nanoparticles is employed.
It should be noted that with oligonucleotide–Ag conjugates rather than a new peak appearing at red-shifted wavelength due to a change in plasmon on the nanoparticle surface, the plasmon broadens and there is a loss of absorbance at 407 nm. For that reason it is less informative to plot an ‘emergence’ profile for the silver conjugates and their stability was assessed by reference to the 407 nm peak. For ease of comparison the ‘half-lives’ of the conjugate systems were calculated, taken as the time required for half the total absorbance change to occur at 407 nm, the results are shown in Table 3.
Table 3.
Half-lives for Ag–oligonucleotide conjugates
| Probe | t½ (min) |
|---|---|
| D1–Ag | 15 |
| D2–Ag | 25 |
| D3–Ag | 30 |
| TT1–Ag | 0.5 |
| TT2–Ag | 0.5 |
| TT3–Ag | 0.75 |
| T1–Ag | 0.5 |
| T2–Ag | 0.5 |
| T3–Ag | 0.5 |
With both the thiol and disulphide–Ag conjugate systems aggregation commences immediately upon treatment with DTT (10 mM). There is a marked difference in the rate of aggregation, however, with all of the thiol systems having a half-life of less than a minute, compared with the disulphide examples which have 15–30 min half-lives. Whilst it is difficult to compare gold and silver conjugate systems, we can say that the overall ‘conjugate’ stability displayed by the silver–disulphide systems is more stable (with respect to DTT-induced aggregation) than the ‘standard’ thiol–gold system (see Figure 2).
Due consideration must be given to the surface coverage of the conjugates as a high surface coverage could explain enhanced stability. As with the gold conjugates the surface coverage data is variable depending upon whether there are spacer bases and what those spacer bases are. For example, the surface coverage of oligonucleotide on Ag nanoparticles for D1–Ag conjugates was found to be 21.1 ± 1.3 pmol·cm−2, compared with TT1–Ag which have a greater surface coverage, 144.7 ± 14 pmol·cm−2. Even T1–Ag has a greater surface coverage than the disulphide species at 31.2 ± 1.5 pmol·cm−2, albeit less than the standard thiol sample.
Once again, the disparity in surface coverage does not impact the conjugate stability since we have seen that the disulphide systems are by far more stable than both of the thiol conjugates and yet has lower surface coverage. This surface-coverage-independent stability is also observed with the polyT and polyA sequence conjugates (refer to Table 2).
Comparing the results for stability of gold and silver ‘conjugates’ it can be seen that as anticipated, the disulphide does not stabilize the silver nanoparticles to the same extent as gold, due to weaker thiol–silver interactions. Surface coverage effects can again be dismissed as the stabilizing factor when comparing samples D2–Au (with a surface coverage of 59.9 ± 6.7 pmol·cm−2) and T2–Ag (with a surface coverage of 64.4 ± 3.0 pmol·cm−2) and yet there are vastly differing stabilities. Similarly, D1–Ag (surface coverage of 21.1 ± 1.3 pmol·cm−2) is considerably less stable than D3–Au (surface coverage of 12.6 ± 0.8 pmol·cm−2) despite their surface coverages being quite similar. It is worthy to note, however, that the disulphide on silver remains more stable than the ‘standard’ monothiol linker systems on gold and this is in spite of similar surface coverages in some cases e.g. D1–Ag (surface coverage of 21.1 ± 1.3 pmol·cm−2) and TT3–Au (surface coverage 21.1 ± 1.2 pmol·cm−2). This is highly significant as it now allows oligonucleotide–silver nanoparticle conjugates to be exploited in a similar manner to gold nanoparticles.
Silver–oligonucleotide conjugate hybridization
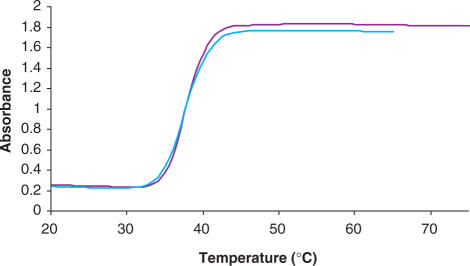
As shown in Figure 4, hybridization between the disulphide-immobilized oligonucleotide and a fully complementary sequence induces a characteristically sharp Tm melting profile (monitored at 413 nm). This is a particularly exciting result since the hybridization was not carried out in the normal ‘sandwich’ fashion. Instead silver nanoparticles conjugated with a thioctic acid modified sequence, 5′- ATC CTG AAT GCG AAA AAA AAA A MOD 3′ (D4) were hybridized with the ‘unconjugated’ complement. This has been observed previously with gold nanoparticles, but not silver (27–29). This shows the efficacy of the thioctic acid terminated oligonucleotide–Ag conjugates and their potential for employment in DNA detection. Furthermore, investigations are underway to establish oligonucleotide–Ag nanoparticle conjugates for DNA detection purposes in a manner similar to that of the established gold analogues. This is particularly attractive as silver nanoparticles have a higher molar extinction coefficient than gold.
Figure 4.
The melting profiles of functionalized silver nanoparticles hybridized with a fully complementary sequence (413 nm).
In conclusion, this data demonstrates the applicability of thioctic acid to simple modification of oligonucleotides, producing enhanced performance with respect to oligonucleotide–nanoparticle ‘conjugate’ stability. This offers a routine way forward for those requiring more robust surface attachment chemistry for oligonucleotides on gold or silver nanoparticles and in particular provides surface chemistry for silver nanoparticles that allows their use in a wide variety of conditions previously unthinkable.
ACKNOWLEDGEMENT
The authors acknowledge the support of the EPSRC to J.A.D. and the Analytical Chemistry Trust Fund through the award of their Analytical Grand-Prix to D.G. Funding to pay the Open Access Publication charges for this article was provided by the EPSRC.
REFERENCES
- 1.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem.Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 2.Yguerabide J, Yguerabide EE. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications – I. Theory. Anal. Biochem. 1998;262:137–156. doi: 10.1006/abio.1998.2759. [DOI] [PubMed] [Google Scholar]
- 3.Yguerabide J, Yguerabide EE. Light-scattering submicroscopicparticles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications – II. Experimental characterization. Anal. Biochem. 1998;262:157–176. doi: 10.1006/abio.1998.2760. [DOI] [PubMed] [Google Scholar]
- 4.Niemeyer CM. Nanoparticles, proteins, and nucleic acids: Biotechnology meets materials science. Angew. Chem. Int. Ed. 2001;40:4128–4156. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 5.Verma A, Rotello V. Surface recognition of biomacromolecules using nanoparticle receptors. Chem. Commun. 2005;20:303–312. doi: 10.1039/b410889b. [DOI] [PubMed] [Google Scholar]
- 6.Brust M, Kiely CJ. Some recent advances in nanostructure preparation from gold and silver particles: a short topical review. Colloids Surf. A Physicochem. Eng. Asp. 2002;202:175–186. [Google Scholar]
- 7.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. Synthesis of thiol-derivatized gold nanoparticles in a 2-phase liquid-liquid system. J. Chem. Soc. Chem. Commun. 1994;7:801–802. [Google Scholar]
- 8.Brust M, Fink J, Bethell D, Schiffrin DJ, Kiely C. Synthesis and reactions of functionalized gold nanoparticles. J. Chem. Soc. Chem. Commun. 1995;16:1655–1656. [Google Scholar]
- 9.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-pot colorimetric differentiation of polynucleotides with singlebase imperfections using gold nanoparticle probes. J. Am. Chem. Soc. 1998;120:1959–1964. [Google Scholar]
- 10.Li Z, Jin R, Mirkin CA, Letsinger RL. Multiple thiol-anchor capped DNA-gold nanoparticle conjugates. Nucleic Acids Res. 2002;30:1558–1562. doi: 10.1093/nar/30.7.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letsinger RL, Elghanian R, Viswanadham G, Mirkin CA. Use of a steroid cyclic disulphide anchor in constructing gold nanoparticle-oligonucleotide conjugates. Bioconjugate Chem. 2000;11:289–291. doi: 10.1021/bc990152n. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Liu S, Echegoyen L. Remarkably stable self-assembled monolayers of new crown-ether annelated tetrathiafulvalene derivatives and their cation recognition properties. Chem. Commun. 1999;16:1493–1494. [Google Scholar]
- 13.Berchmans S, Thomas PJ, Rao CNR. Novel effects of metal ion chelation on the properties of lipoic acid-capped Ag and An nanoparticles. J. Phys. Chem. B. 2002;106:4647–4651. [Google Scholar]
- 14.Checkik V, Wellsted HJ, Korte A, Gilbert BC, Caldararu H, Ionita P, Caragheorgheopol A. Spin-labelled Au nanoparticles. FaradayDiscuss. 2004;125:279–291. doi: 10.1039/b302730a. [DOI] [PubMed] [Google Scholar]
- 15.Karamanska R, Mukhopadhyay B, Russell DA, Field RA. Thioctic acid amides: convenient tethers for achieving low nonspecific protein binding to carbohydrates presented on gold surfaces. Chem. Commun. 2005;26:3334–3336. doi: 10.1039/b503843j. [DOI] [PubMed] [Google Scholar]
- 16.Chang J-Y, Wu H, Chen H, Ling Y-C, Tan W. Oriented assembly of Au nanorods using biorecognition system. Chem. Commun. 2005;8:1092–1094. doi: 10.1039/b414059a. [DOI] [PubMed] [Google Scholar]
- 17.Abad JM, Mertens SFM, Pita M, Fernández VM, Schiffrin DJ. Functionalization of thioctic acid-capped gold nanoparticles for specific immobilization of histidine-tagged proteins. J. Am. Chem. Soc. 2004;127:5689–5694. doi: 10.1021/ja042717i. [DOI] [PubMed] [Google Scholar]
- 18.Stevenson KA, Muralidharan G, Maya L, Wells JC, Barhen J, Thundat T. Covalent attachment of gold nanoparticles to DNA templates. J. Nanosci. Nanotech. 2002;2:397–404. doi: 10.1166/jnn.2002.110. [DOI] [PubMed] [Google Scholar]
- 19.Harrison JG, Balasubramanian S. A convenient systhetic route to oligonucleotide conjugates. 1997;7:1041–1046. [Google Scholar]
- 20.Grabar KC, Freeman RG, Hommer MB, Natan MJ. Preparation and characterization of au colloid monolayers. Anal. Chem. 1995;67:735–743. [Google Scholar]
- 21.Roux S, Garcia B, Bridot J-L, Salomé M, Marquette C, Lemelle L, Gillet P, Blum L, Perriat P, et al. Synthesis, characterization of dihydrolipoic acid capped gold nanoparticles, and functionalization by the electroluminescent luminol. Langmuir. 2005;21:2526–2536. doi: 10.1021/la048082i. [DOI] [PubMed] [Google Scholar]
- 22.Garcia B, Salomé M, Lemelle L, Bridot J-L, Gillet P, Perriat P, Roux S, Tillement O. Sulfur K-edge XANES study of dihydrolipoic acid capped gold nanoparticles: dihydrolipoc acid is bound by both sulphur ends. Chem. Commun. 2005;3:369–371. doi: 10.1039/b411231h. [DOI] [PubMed] [Google Scholar]
- 23.Demers LM, Mirkin CA, Mucic RC, Reynolds III RA, Letsinger RL, Elghanian R, Viswanadham G. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal. Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 24.Cai H, Xu Y, Zhu N, He P, Fang Y. An electrochemical DNA hybridization detection assay based on a silver nanoparticle label. Analyst. 2002;127:803–808. doi: 10.1039/b200555g. [DOI] [PubMed] [Google Scholar]
- 25.Tokareva I, Hutter E. Hybridization of oligonucleotide-modified silver and gold nanoparticles in aqueous dispersions and on gold films. J. Am. Chem. Soc. 2004;126:15784–15789. doi: 10.1021/ja046779k. [DOI] [PubMed] [Google Scholar]
- 26.Vidal BC, Jr, Deivaraj TC, Yang J, Too H-P, Chow G-M, Gan LM, Lee JY. Stability and hybridization-driven aggregation of silver nanoparticle-oligonucleotide conjugates. New J. Chem. 2005;29:812–816. [Google Scholar]
- 27.Sato K, Hosokawa K, Maeda M. Rapid aggregation of gold nanoparticles induced by non-cross-linking DNA hybridisation. J. Am. Chem. Soc. 2003;125:8102–8103. doi: 10.1021/ja034876s. [DOI] [PubMed] [Google Scholar]
- 28.Sato K, Hosokawa K, Maeda M. Non-cross-linking gold nanoparticle aggregation as a detection method for single-base substitutions. Nucleic Acids Res. 2005;33:e4. doi: 10.1093/nar/gni007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato K, Hosokawa K, Maeda M. Colorimetric biosensors based on DNA-nanoparticle conjugates. Anal. Sci. 2007;23:17–20. doi: 10.2116/analsci.23.17. [DOI] [PubMed] [Google Scholar]