Abstract
Calmodulin (CaM) is involved in defense responses in plants. In soybean (Glycine max), transcription of calmodulin isoform 4 (GmCaM4) is rapidly induced within 30 min after pathogen stimulation, but regulation of the GmCaM4 gene in response to pathogen is poorly understood. Here, we used the yeast one-hybrid system to isolate two cDNA clones encoding proteins that bind to a 30-nt A/T-rich sequence in the GmCaM4 promoter, a region that contains two repeats of a conserved homeodomain binding site, ATTA. The two proteins, GmZF-HD1 and GmZF-HD2, belong to the zinc finger homeodomain (ZF-HD) transcription factor family. Domain deletion analysis showed that a homeodomain motif can bind to the 30-nt GmCaM4 promoter sequence, whereas the two zinc finger domains cannot. Critically, the formation of super-shifted complexes by an anti-GmZF-HD1 antibody incubated with nuclear extracts from pathogen-treated cells suggests that the interaction between GmZF-HD1 and two homeodomain binding site repeats is regulated by pathogen stimulation. Finally, a transient expression assay with Arabidopsis protoplasts confirmed that GmZF-HD1 can activate the expression of GmCaM4 by specifically interacting with the two repeats. These results suggest that the GmZF-HD1 and –2 proteins function as ZF-HD transcription factors to activate GmCaM4 gene expression in response to pathogen.
INTRODUCTION
In plants, cytosolic-free calcium level (Ca2+) plays pivotal roles as an intracellular second messenger in response to a variety of stimuli, including light, phytohormones, oxidative stress, drought, cold and pathogens (1,2). One of the earliest events in response to pathogen attack is an increase in the [Ca2+]cyt, with complex changes in its amplitude, frequency and duration (3,4). A Ca2+ influx has been shown to be essential for the activation of defense-related genes, phytoalexin biosynthesis and hypersensitive cell death (5). Ca2+ signals are sensed by intracellular Ca2+-binding proteins and transduced by downstream effector proteins that regulate cellular processes (6,7).
Calmodulin (CaM), one of the best characterized Ca2+-binding proteins, contains four helix–loop–helix Ca2+-binding motifs referred to as EF hands. Ca2+-bound CaM transduces Ca2+ signals by modulating the activity of numerous diverse CaM-binding proteins such as metabolic enzymes, transcription factors, ion channels, protein kinases/phosphatases and structural proteins, which generates physiological responses to various stimuli (8,9). Mammalian cells have only a few CaM genes encoding one or a few isoforms. In contrast, plants possess a large repertoire of CaM and CaM-like genes that encode several CaM isoforms (7,8). In all plants examined, CaM genes, even those encoding the same isoform, are differentially expressed in response to various external stimuli such as touch, heat shock, cold, light, auxin and pathogens (10).
We previously cloned five CaM isoforms (GmCaM1–5) from soybean (Glycine max). The conventional isoforms GmCaM1, −2 and −3 share more than 96% identity with mammalian CaM, while the divergent isoforms GmCaM4 and −5 share only 78% identity to GmCaM1; these are the most divergent isoforms reported thus far in the plant and animal kingdoms (11,12). The cellular level of GmCaM4 rapidly and dramatically rises in response to specific stimuli such as pathogen exposure and salinity (13). Furthermore, ectopic expression of GmCaM4 or GmCaM5 in tobacco and Arabidopsis confers increased pathogen resistance through the formation of spontaneous hypersensitive response-associated lesions (even in the absence of pathogens), which is not mediated by elevated levels of salicylic acid but by increased levels of systemic acquired resistance gene expression (14). In addition, these transgenic plants exhibit salt tolerance and can accumulate high levels of proline (15). These results demonstrate that Ca2+ signaling mediated by specific CaM isoforms contributes to pathogen and salt stress resistance in plants.
Homeodomain proteins in the homeobox gene family play important roles as transcription factors in plant, animal and fungal development (16). Mutant analyses of monocot and dicot plants showed that leaf development involves the down-regulation of meristem-specific homeobox genes such as the knotted-like homeobox (knox) genes (17,18). Ectopic expression of these genes leads to altered cell fates in the leaf, suggesting a pivotal function in early leaf development (19). However, it is not well established that homeodomain proteins are involved in plant stress signaling. Pathogenesis-related homeodomain proteins from parsley and pathogenesis-related homeodomain proteins from Arabidopsis (PRHP and PRHA), members of the PHD finger subfamily, were isolated on the basis of their interaction with a 125-nt region within the pr2 promoter, which is rapidly stimulated by fungal or bacterial elicitors (20).
We are interested in understanding how plants recognize biological and environmental stresses such as pathogens and salinity, and how signals are transduced to activate transcription of the GmCaM4 gene in response to such stresses. For this purpose, it is crucial to identify cis-acting elements and trans-acting factors that regulate GmCaM4 expression. We previously identified two regions (−1286 to −1065 and −858 to −728) in the GmCaM4 promoter that are bound by proteins induced by pathogen or NaCl exposure (13). The GT-1 cis-acting element in the −858 to −728 bp region of the GmCaM4 promoter is partially involved in GmCaM4 expression by interacting with a GT-1 like transcription factor in response to pathogen and salt stresses (13). However, cis-acting element(s) in the −1286 to −1065 region of the GmCaM4 promoter and their DNA binding proteins remained to be identified and characterized.
Here, we report the presence of two repeats of a conserved homeodomain binding site, ATTA, within the −1286 to −1065 region of the GmCaM4 promoter. We used the yeast one-hybrid system to isolate two transcriptional regulators, GmZF-HD1 and GmZF-HD2, which specifically bind to a 30-nt A/T-rich cis-acting element within the −1207 to −1128 region. Supershift and transient expression assays confirmed that GmZF-HD1 is a component of DNA-nuclear proteins complexes formed in vitro and that it functions as an in vivo transcriptional regulator of pathogen-responsive expression of the GmCaM4 gene.
MATERIALS AND METHODS
Plant cell culture, bacterial and yeast strains
Soybean suspension cells (Glycine max L. cv. Williams 82; W82) were cultured in MS medium supplemented with 0.75-mg l−1 benzyl adenine and maintained at 25°C in the dark with shaking at 130 r.p.m. Pseudomonas syringae pv glycinea carrying avrA (Psg. avrA) was grown and manipulated as previously described (21). The Escherichia coli strain XL-1 Blue MRF’ (Stratagene La Jolla, CA, USA) was used for cloning. Glutathione S-Transferase (GST) fusion proteins were expressed in E. coli BL21 (pLys S) DE3. The yeast strain YM4271 (MATα, ura3-52, his3-200, ade2-101, lys2-801, leu2-3, 112, trp1-901, tyr1-501, gal4-Δ512, gal80-Δ538, ade5::hisG) was used as a host for transformation with a reporter vector in the yeast one-hybrid screen (Clontech).
Isolation of GmZF-HD1 and GmZF-HD2 cDNAs by yeast one-hybrid screening
Yeast one-hybrid screening was performed using the MATCHMAKER one-hybrid system (Clontech). To make a target reporter construct, four tandem repeats of the A2 region of the GmCaM4 promoter (positions −1207 to −1128 bp) containing putative cis-acting elements were inserted into the SmaI and XhoI sites of pLacZi and the SmaI site of pHisi. The two expression plasmids were simultaneously transformed into YM4271. Cells from a 300-ml culture were transformed with soybean cDNA libraries and plated on synthetic minimal medium containing 20 mM 3-aminotriazole (3-AT) but lacking His and Leu. The soybean cDNA library was constructed using the yeast expression vector pAD-GAL4-2.1 (Stratagene) and RNA isolated from apical and elongating regions (0.3–1.3 cm from the cotyledon) of 4-day-old etiolated soybean hypocotyl tissue. After incubation at 30°C for five days, positive colonies were cultured in YPD and selected on medium containing 20 mM, 45 mM or 60 mM 3-AT but lacking His and Leu. Colonies were then transferred to filter paper and tested for β-galactosidase activity. Plasmids were extracted from positive colonies, amplified in E. coli, and purified for sequencing.
Preparation of soybean nuclear extracts and electrophoretic mobility shift assay (EMSA)
Nuclear protein extracts were prepared as described (22) from soybean suspension culture cells (W82). Aliquots were taken for determining protein concentrations with the Bradford protein assay kit (Bio-Rad), and aliquots of the extracts were frozen in tubes at −70°C. EMSA was performed as described (13,23) using [32P]-labeled DNA probes. Soybean nuclear extracts or GST fusion proteins were pre-incubated in a binding buffer (20 mM HEPES, pH 7.9, 0.5 mM DTT, 0.1 mM EDTA, 2 μg poly dI/dC, 50 mM KCl, 15% glycerol) for 10 min at room temperature and then incubated with 40 Kcpm of end-labeled probes for 20 min. The resulting protein–DNA complexes were electrophoresed in non-denaturing 8% polyacrylamide gels in 0.5 X TBE buffer. The gels were dried and exposed to X-ray films.
DNase I footprinting assay
The DNase I footprinting assay was performed as described (24,25) using a 118 bp DNA fragment containing −1207 to −1128 bp of the GmCaM4 promoter and pGEM-T Easy vector (Promega) sequences between the SphI and SpeI sites. The fragment was labeled with α-32P isotope at the SpeI site. A DNA-nuclear protein binding mixture (100 μl) containing 10 mM HEPES pH 7.9, 60 mM KCl, 1 mM EDTA pH 8.0, 7% glycerol and 1 mM DTT was pre-incubated for 20 min at room temperature, one unit of DNase I cofactor solution (50 μl) was added, and incubation was continued for 3 min at room temperature. Stop solution (100 μl) with 100 μg of proteinase K was immediately added and incubation was continued for 30 min at 55°C. Total reaction mixtures were extracted with Tris-saturated phenol, and products were precipitated by adding 25 μl 3 M sodium acetate, pH 5.2, and 500 μl 100% ethanol. The final pellet was re-suspended in sequencing loading buffer (0.1 M NaOH:formamide [1:2], 0.1% bromophenol blue, 0.1% xylene cyanol), and the mixture was heated at 100°C for 2 min, chilled on ice, and loaded onto 8% polyacrylamide-urea DNA sequencing gels.
Expression of GST::GmZF-HD1 and −2 fusion proteins in E. coli
Full-length GmZF-HD1 and GmZF-HD2 cDNA clones were cloned into the BamHI and XhoI sites of the expression vector pGEX-2T-linkerI (Amersham Pharmacia Biotech), respectively. The resulting constructs were confirmed by sequencing and introduced into E. coli BL21 (pLys S) DE3, and GST fusion proteins were expressed and purified using glutathione–agarose beads according to the manufacturer's instruction (Amersham Pharmacia Biotech).
RNA gel blot analysis
Various tissues harvested from 4-day etiolated soybean (Glycine max L. cv. Williams) seedlings and W82 cells collected on filter papers by vacuum filtration were used for isolation of total RNA. RNA gel blot analysis was carried out as previously described (26). Total RNA (20 μg each sample) prepared from apical hypocotyls, elongating hypocotyls, mature hypocotyls, plumule, seed, root and pathogen-treated soybean suspension cells (Psg. avrA) was separated on 1.5% formaldehyde/agarose gels, transferred onto a nylon membrane (Hybond N+, Amersham), and hybridized with the GmZF-HD1 cDNA probe. To verify equal loading, rRNA was visualized by staining with ethidium bromide. Hybridization and washing were performed under high stringency conditions.
Antibody preparation and supershift assay
A GmZF-HD1 antibody was raised against the GST::GmZF-HD1 protein in rabbits (Animal Culture Facility, Gyeongsang National University). A protein A-Sepharose column was used to separate the IgG fraction of the rabbit serum. After adjusting the pH of the crude serum to 8.0 by adding 1/10 volume of 1.0 mM Tris–HCl (pH 8.0), the antibody solution was washed extensively with 10 column vol of 100 mM Tris–HCl (pH 8.0) and in turn 10 mM Tris–HCl (pH 8.0). The antibody was eluted with 0.1 M glycine–HCl (pH 2.5) and immediately neutralized with 0.1 vol of 1 M Tris–HCl (pH 8.0). The supershift assay was performed by pre-incubating affinity-purified anti-GST::GmZF-HD1 antibody and nuclear extracts together, and the samples were then processed as for EMSA. Control experiments were performed by incubating the same amount of IgG fraction of preimmune antiserum in the binding reaction mixtures.
Transient expression assay with Arabidopsis protoplasts
A tetramer of the A2 region in the GmCaM4 promoter was fused to a minimal −46 CaMV 35S promoter-GUS reporter gene (A2 region-m35S-GUS) (27). To construct the effector plasmid, full-length GmZF-HD1 cDNA was inserted into a plant expression vector (pHBT95) containing the 35SC4PPDK promoter and nos terminator (28). Isolation of Arabidopsis mesophyll protoplasts and polyethylene glycol-mediated DNA transfection were performed as previously described (29). In each transformation, an Arabidopsis protoplast suspension (5 × 106 per ml) was transfected with 10 μg of reporter construct alone or together with 15 μg of the effector construct or a vector DNA control (pHBT95). Transfected protoplasts were incubated in W5 solution for 16 h in the dark. Glucuronidase (GUS) expression was detected fluorometrically with the substrate 4-methyl umbelliferyl glucuronide, as described (27). LUC assays were performed using the Promega luciferase assay system according to the manufacturer's instructions. To normalize the transfection efficiency, the CaMV 35S promoter-LUC control vector pJD 300 was cotransfected in each experiment as described (30).
RESULTS
Identification of a binding site for pathogen-activated nuclear proteins in the GmCaM4 promoter
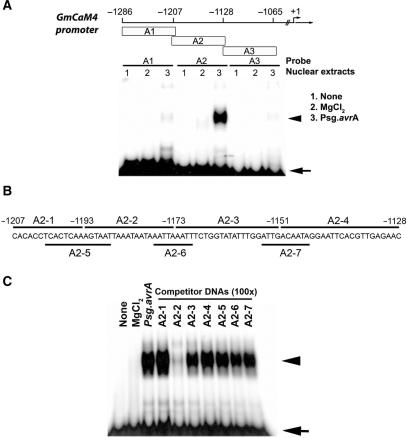
Based on DNA binding assays (electrophoretic mobility shift assay, or EMSA) and transient expression assays in Arabidopsis protoplasts with GmCaM4 deletion constructs, we previously showed that two regions of the GmCaM4 promoter are involved in pathogen-mediated gene regulation (13). One is a GT-1 cis-acting element within the −858 to −728 region of the GmCaM4 promoter. However, another region upstream of the GT-1 cis-acting element, from −1286 to −1065, remained to be further characterized. In this study, we undertook to identify core cis-acting elements upstream of the GT-1 cis-acting element that specifically interact with nuclear extracts from pathogen-treated cells by dividing this 220-nt region into three overlapping fragments (A1, A2 and A3), which were used as probes for EMSA (Figure 1). The EMSA data showed that a stable binding complex formed only between the A2 probe (−1207 to −1128) and nuclear extracts from pathogen-treated cells (Figure 1A). To first prove that the complex originated from the interaction of the A2 probe with proteins but not with contaminants in the extracts, we also performed EMSA using the A2 region as a probe with extracts heated for 5 min at 65°C, or treated with RNase A or proteinase K. We found that the binding complex was heat-labile and sensitive to proteinase K but not RNase A, confirming that it was a DNA–protein complex (data not shown).
Figure 1.
Identification of a pathogen-responsive protein-binding site in the GmCaM-4 promoter. (A) Schematic diagram of DNA fragments derived from the GmCaM-4 promoter used for gel mobility shift assay (EMSA) and their patterns of binding with nuclear extracts prepared from W82 soybean suspension culture cells treated with MgCl2 (lanes 2, MgCl2) or Pseudomonas syringae pv glycinea carrying avrA (Psg. avrA) (lanes 3, Psg. avrA). None (lanes 1) represents free DNA probe. The positions of fragments upstream of the GmCaM4 transcriptional start site are indicated. (B) Oligonucleotides (A2-1–A2-7) corresponding to the GmCaM-4 promoter A2 region that were used as competitors. (C) Competitive gel mobility shift assay. The DNA binding reaction was performed by pre-incubating unlabeled competitors (A2-1–A2-7) and then adding the 32P-labeled A-2 fragment as a probe. A 100-fold molar excess of competitor DNA was added to each reaction mixture. Free (arrow) and protein-complexed (arrowhead) probes were separated on an 8% polyacrylamide gel and visualized by autoradiography.
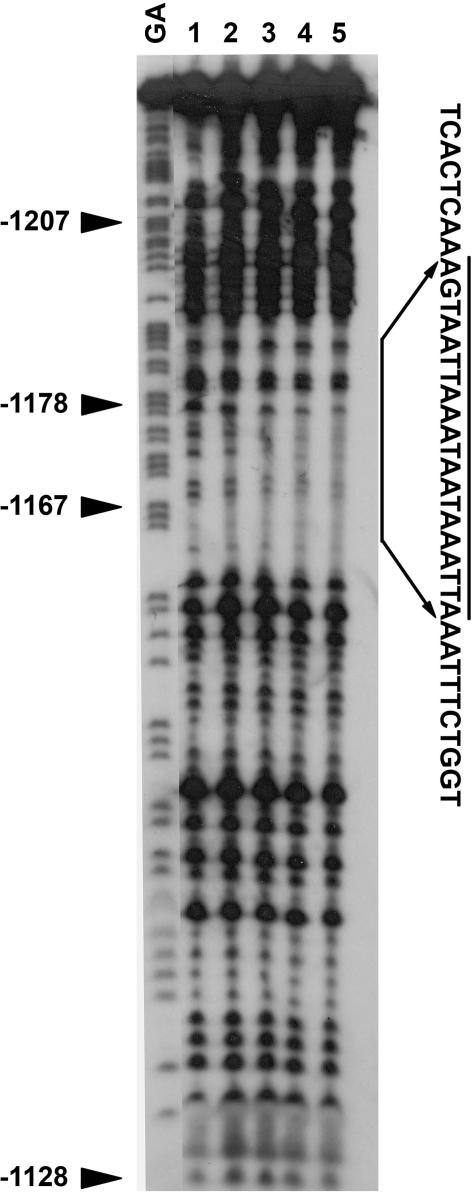
We then designed seven overlapping double-stranded oligonucleotides (A2–1 to −7) to further map the binding site and performed competition assays using the A2 region as a probe (Figure 1B). Remarkably, a 100-fold molar excess of the unlabeled A2–2 oligonucleotide completely inhibited the binding of nuclear proteins to the A2 region, but an excess of any of the other six oligonucleotides did not have this effect (Figure 1C). We then employed a general and powerful tool for identifying transcription factor-binding sites in promoter sequences, the DNase I footprinting assay. We used the A2 region as a probe with various concentrations of nuclear extracts from pathogen-treated cells. Similar to what we found with the competition assay (Figure 1C), a region covering ∼20-nt (AGTAATTAAATAATAAATTA) and almost identical to the A2–2 sequence was protected from DNase I digestion by nuclear extracts (Figure 2). These results suggest that a cis-acting element within the A2–2 region of the GmCaM4 promoter interacts with pathogen-activated nuclear protein(s). The 20-nt sequence identified by the DNase I foot-printing assay, AGTAATTAAATAATAAATTA, contains two core consensus sequences (underlined) that have been identified as motifs bound by homeodomain proteins. As shown in the competition assay (Figure 1C), neither the A2–5 nor the A2–6 fragment, which contain a partial or full core sequence, respectively, competitively inhibited the binding of nuclear proteins to the A2 probe, suggesting that the full length of the A2–2 fragment is required for binding nuclear proteins. To determine the overall sequence sufficient for binding nuclear proteins, we tested fragments of varying length that include the A2–2 sequence at the 5′ end. The result of this experiment indicated that an additional 10 bp of sequence flanking the A2–2 fragment was necessary for the maximal binding of pathogen-induced nuclear proteins (data not shown). Therefore, we used this 30-bp sequence for further experiments.
Figure 2.
DNase I footprint analysis of the A2 region of the GmCaM-4 promoter bound by pathogen-responsive proteins. End-labeled fragments were digested with DNase I after incubation with 0, 5, 10, 20 or 25 μg nuclear proteins from Psg. avrA-treated W82 soybean cells. The resulting DNA fragments were purified and electrophoresed on a 6% non-denaturing polyacrylamide sequencing gel. GA, G + A sequencing ladder (non-coding strand); lane 1, no nuclear protein; lanes 2–5, 5, 10, 20 and 25 μg nuclear protein, respectively. The DNase I-protected region is indicated by a solid bar and the sequence is shown.
Isolation of cDNAs encoding DNA binding proteins that recognize the A2 fragment of the GmCaM4 promoter
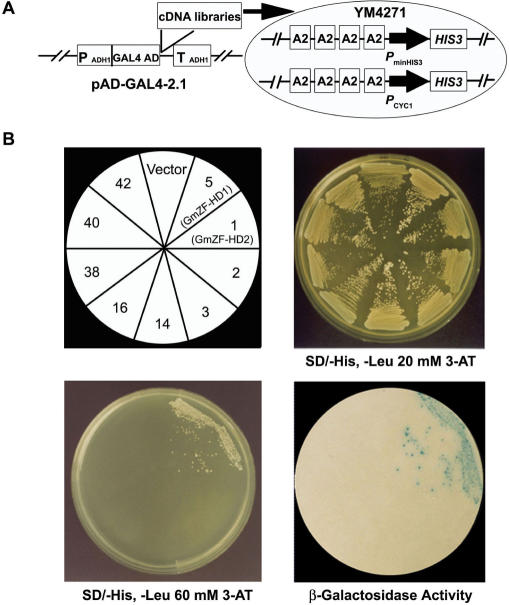
We used the yeast one-hybrid screening system (31) to isolate cDNAs encoding DNA binding proteins that interact with the 80-nt A2 region in a sequence-specific manner. As a dual reporter system, we first constructed a yeast strain (YM4271) carrying integrated copies of HIS3 and lacZ with four tandem repeats of the 80-nt A2 fragment of the GmCaM4 promoter (Figure 3A). In the resulting strain, the HIS3 gene is transcribed at a basal level, permitting growth on minimal medium lacking histidine but not in the presence of 20 mM 3-aminotriazole (3-AT), a competitive inhibitor of the HIS3 gene product. YM4271 was then transformed with soybean cDNA libraries by the lithium acetate/polyethylene glycerol method, and a ∼1.5 × 107 transformants were screened. Forty-two positive clones were selected on minimal medium lacking histidine and containing 20 mM 3-AT. Plasmids were recovered and electroporated into an E. coli host for propagation. The cDNAs from the isolated plasmids were subjected to restriction and sequencing analysis, which allowed the 42 cDNA clones to be classified into 16 distinct groups (Table 1).
Figure 3.
Isolation of cDNA clones encoding proteins that bind to the A2 region of the GmCaM4 promoter. (A) Strategy for cDNA isolation by selection in yeast. Soybean cDNA libraries constructed in the yeast expression vector pAD-GAL4-2.1 were transformed into YM4271 cells carrying the dual reporter genes HIS3 and lacZ under the control of four tandem repeats of the A2 region of the GmCaM-4 promoter. The yeast expression cDNA libraries encode an N-terminal fusion of the GAL4 activation domain (AD) to soybean proteins. PminHIS3 indicates the minimal promoter of the HIS3 gene, and PminCYC1 indicates the CYC1 minimal promoter. PADH1 indicates the alcohol dehydrogenase 1 (ADH1) promoter, and TADH1 indicates the ADH1 terminator. (B) Specific interactions of GmZF-HD1 and −2 with the A2 region of the GmCaM-4 promoter. Yeast transformants harboring the indicated clones (upper left) were grown in the presence of 20 mM (upper right) or 60 mM (lower left) 3-AT. Nine of the 16 positive clones are shown in this figure. Numbers indicate representative clones as seen in Table 1. The β-galactosidase activity of yeast colonies grown in YPD medium was determined by a filter-lift assay (lower right).
Table 1.
General characteristics of the cDNA clones isolated in this study
| Group | Clone | Insert Size (kb) | No. of Clones Obtained | Results of BLAST Search |
|---|---|---|---|---|
| 1 | 5 | 0.77 | 22 | GmZF-HD1 |
| 2 | 1 | 0.66 | 6 | GmZF-HD2 |
| 3 | 2 | 1.3 | 1 | Poly(A) binding protein |
| 4 | 3 | 1.2 | 1 | Storage protein |
| 5 | 9 | 0.8 | 1 | FerredoxinIII |
| 6 | 10 | 0.3 | 1 | Yeast pheromone receptor |
| 7 | 11 | 1.1 | 1 | Ankyrin-like protein |
| 8 | 13 | 0.5 | 1 | Glutamate decarboxylase |
| 9 | 14 | 0.8 | 1 | Yippee-like protein |
| 10 | 16 | 1.3 | 1 | Heatshock protein |
| 11 | 18 | 1.25 | 1 | Glycoprotein |
| 12 | 25 | 1.8 | 1 | Aspartate aminotransferase |
| 13 | 27 | 1.3 | 1 | Oligouridylate binding protein |
| 14 | 38 | 1.35 | 1 | Ribosomal protein |
| 15 | 40 | 1.4 | 1 | Peroxidase |
| 16 | 42 | 0.6 | 1 | Oxygen evolving enhancer |
Forty-two positive clones were isolated by yeast one-hybrid screening and divided into 16 distinct cDNA groups.
To select true positive cDNAs that encode transcriptional regulators among the 16 representative cDNA clones, we examined their lacZ activity by a filter-replica method using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) as a substrate. Only two cDNA clones (clones 5 and 1) later named GmZF-HD1 and −2 (for Glycine max zinc finger-homeodomain protein1 and -2, respectively) formed blue colonies on filter papers containing X-gal (Figure 3B). With the selected 16 cDNA clones, we also performed a cell growth assay on selection medium without histidine but with a higher concentration of 3-AT (60 mM). The data again showed that only clones 5 (GmZF-HD1) and 1 (GmZF-HD2) survived on medium containing 60 mM 3-AT. These results confirmed that the GmZF-HD1 and −2 proteins specifically and strongly bound to the A2 region of the GmCaM4 promoter and activated transcription of the dual reporter genes in yeast, suggesting that they were good candidates as transcriptional regulators of pathogen-induced GmCaM4 gene expression.
Sequence analysis of GmZF-HD1 and −2 cDNAs
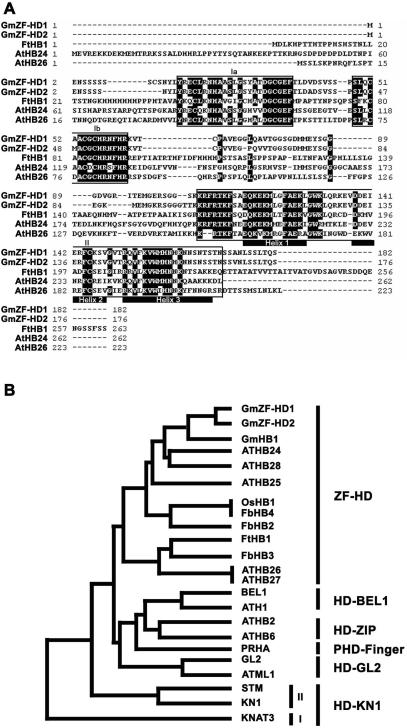
To examine the structures of the GmZF-HD1 and −2 cDNA clones, we sequenced the two cDNAs, ∼770-nt and 660-nt, respectively, which revealed that both were full-length. The GmZF-HD1 cDNA encodes an open reading frame of 182 amino acids, which specifies a putative protein with a predicted molecular mass of about 20 kDa. The GmZF-HD2 cDNA encodes an open reading frame of 176 amino acids, which specifies a putative protein with a predicted molecular mass of about 19.4 kDa (Figure 4A). We submitted the nucleotide sequences of the two cDNAs to the GenBank/EMBL/DDBJ database (accession numbers AY695729 and AY695730 for the GmZF-HD1 cDNA and GmZF-HD2 cDNA, respectively). We then searched databases for sequences homologous to those of the GmZF-HD1 and −2 proteins. We found that each GmZF-HD protein has two highly conserved putative zinc finger domains in the N-terminus and a DNA binding homeodomain in the C-terminus, which is characteristic of homologs in the plant ZF-HD family, including AtHB24 and −26 of the C3 plant Arabidopsis thaliana and FtHB1 of the C4 plant Flaveria trinervia (Figure 4A). The ZF-HD class of homeodomain proteins may be involved in the photosynthesis-related mesophyll-specific gene expression of phosphoenolpyruvate carboxylase in C4 species of the genus Flaveria (32). More recently, it was proposed that ZF-HD transcriptional regulators play overlapping regulatory roles in Arabidopsis floral development (33). However, there is no prior evidence that ZF-HD homeodomain proteins are involved in pathogen signaling and plant defense mechanisms.
Figure 4.
Amino acid sequence alignment and phylogenetic analysis of GmZF-HD1 and −2. (A) Alignment of the GmZF-HD1 and −2 amino acid sequences with the Flaveria trinervia (FtHB1) and Arabidopsis thaliana (AtHB24 and AtHB26) homeodomain proteins. The highly conserved domains Ia, Ib and II are boxed, identical residues are indicated with white letters in black boxes, dashes indicate the absence of residues, and the trihelix-DNA binding regions of the homeodomain are underlined. (B) Phylogenetic tree of plant homeodomain proteins. The GenBank accession numbers for the following proteins are given in parenthesis: GmHB1 (AW760234), ATHB24 (AC006439), ATHB28 (AL049862.1), ATHB25 (AB011479), OsHB1 (AC007858), FbHB4 (Y18581), FbHB2 (Y18579), FtHB1 (Y18577), FbHB3 (Y18580), ATHB26 (AB011483), ATHB27 (AB007647), BEL1 (U39944), ATH1 (X80126), ATHB2 (X68145), ATHB6 (X67034), PRHA (L21991), GL2 (L32873), ATML1 (U37589), STM (U32344), KN1 (X61308), KNAT3 (X92392).
Interestingly, PRHA in the PHD finger sub-family has been reported to be involved in the transcriptional regulation of the Arabidopsis pr2 gene (20). A phylogenetic tree constructed by comparing deduced amino acid sequences indicates that GmZF-HD1 and −2 are more closely related to FtHB1 than to PRHA (Figure 4B). Therefore, these observations imply that the GmZF-HD1 and −2 proteins represent a new class of ZF-HD proteins that is involved in pathogen signaling in soybean and that the characteristic zinc finger domains play a role in protein regulation.
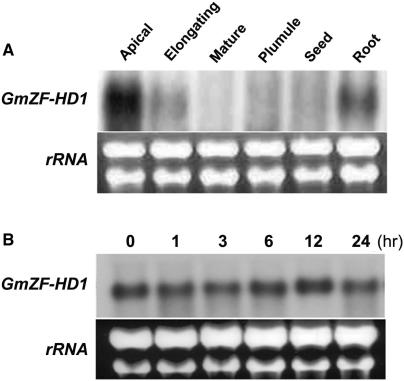
Expression pattern of the GmZF-HD1 gene
The copy number of the GmZF-HD1 gene in the soybean genome was estimated by Southern blot analysis. Simple patterns of hybridization were observed under high-stringency conditions, suggesting that GmZF-HD1 is present in single- or low-copy number in the soybean genome (data not shown). The tissue-specific expression pattern of GmCaM4 were previously investigated in soybean plants by northern analysis (11) and by expression of a promoter-β-glucuronidase (GUS) reporter in transgenic Arabidopsis plants (13). These studies showed that GmCaM4 is primarily expressed in the apical hook and elongating hypocotyls in both soybean and transgenic Arabidopsis. To determine the profile of GmZF-HD1 expression, we performed northern analysis with tissue-specific RNAs. The GmZF-HD1 transcript was highly abundant in apical hooks and roots, but much less abundant in elongating hypocotyls. It was almost undetectable in mature hypocotyls, plumules, and seeds (Figure 5A). These results are very similar to the expression patterns of GmCaM4 in soybean seedlings, suggesting that the GmZF-HD1 protein may be involved in regulating the expression of GmCaM4. It was previously reported that expression of the GmCaM4 gene is dramatically induced by pathogen treatment (14). Therefore, we investigated the expression of GmZF-HD1 in response to pathogen treatment. Although GmZF-HD1 was highly expressed in soybean suspension cells under normal conditions, it was not significantly induced by pathogen treatment (Figure 5B). This result suggests that the activity of GmZF-HD1 in the induction of GmCaM4 gene expression in response to pathogen may be controlled at post-transcriptional instead of transcriptional levels.
Figure 5.
Expression pattern of the GmZF-HD1 mRNA in soybean. (A) Northern blot analysis of GmZF-HD1 gene expression in various soybean tissues. (B) Time-course of expression of the GmZF-HD1 transcript after pathogen (Psg. avrA) treatment in soybean suspension cells. Total RNA (20 μg) prepared from apical hypocotyls (Apical), elongating hypocotyls (Elongating), mature hypocotyls (Mature), plumule, seed, root and soybean suspension cells treated with pathogen (Psg. avrA) were separated on a 1.5% formaldehyde/agarose gel, transferred onto a nylon membrane and hybridized with a 32P-labeled GmZF-HD1 cDNA probe. To verify equal loading of total RNA, rRNAs were visualized by staining with ethidium bromide.
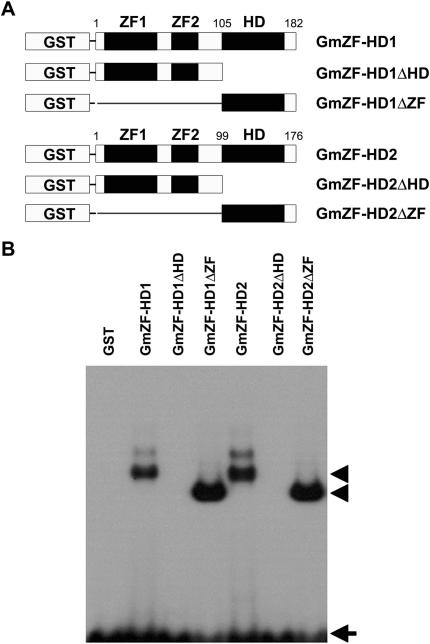
The C-terminal homeodomain, but not the N-terminal zinc finger domains, binds to the 30-nt GmCaM4 promoter sequence
To evaluate whether GmZF-HD1 and -2 bind to the 30-nt sequence containing the two repeats of a conserved ATTA homeodomain binding site in the GmCaM4 promoter, we produced recombinant full-length GmZF-HD1 and −2 proteins fused to GST in E. coli. The two full-length recombinant proteins were similarly capable of binding to the 30-nt sequence in the gel shift assay (Figure 6B). Then, to test which domain (ZF or HD) is involved in DNA binding, we performed EMSA with C-terminal HD deletion derivatives (ΔHD) and N-terminal ZF deletion derivatives (ΔZF) of each protein (Figure 6A). As shown in Figure 6B, EMSA performed using the 30-nt sequence as a probe with the purified deletion mutants showed that the N-terminal ZF deletion mutants (i.e. those containing only the homeodomain) strongly bound to the 30-nt sequence, but that the C-terminal HD deletion mutants did not (i.e. those containing only the zinc finger domains) (Figure 6B). We then performed a competition assay and found that DNA–protein complex formation was significantly reduced by an up to 200-fold molar excess of unlabeled 30-nt competitor (data not shown). Since the binding affinity to the 30-nt sequence of proteins containing only the homeodomain (GmZF-HD1ΔZF and −2ΔZF) was much stronger than that of the full-length proteins (GmZF-HD1 and −2), and since GmZF-HD1ΔZF and GmZF-HD2ΔZF bound with similar affinity to this sequence, GmZF-HD1ΔZF was chosen for further EMSA experiments to characterize its interaction with this region of the GmCaM4 promoter.
Figure 6.
The DNA-binding abilities of GmZF-HD1 and −2. (A) Schematic representation of GmZF-HD1 and −2 GST fusion constructs. Numbers indicate the positions of boundaries in fusion constructs. Amino acid positions of domain boundaries are indicated. (B) Interaction of the 30-nt sequence containing the A2-2 region of the GmCaM-4 promoter with recombinant GmZF-HD1 and −2 proteins. The 32P-labeled 30-nt oligonucleotide was incubated in the presence of recombinant GmZF-HD1 or −2 protein or GST alone. All lanes contain 10 μg of each bacterial extract. Free (arrow) and protein-complexed (arrowhead) probes were separated on an 8% polyacrylamide gel and visualized by autoradiography.
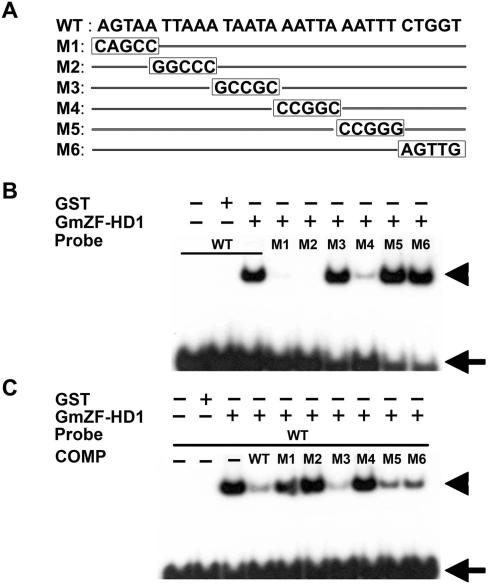
Identification of GmZF-HD1 binding sites in the GmCaM4 promoter
To more precisely map the binding site of the GmZF-HD1 protein, we designed six mutated versions of the 30-nt sequence (M1 to M6), each with 5-nt mutations as outlined in Figure 7A. The ability of the recombinant GmZF-HD1(ΔZF) fusion protein to bind wild-type or mutated 30-nt sequences was examined using EMSA and competition assays. As shown in Figure 7B, the GmZF-HD1(ΔZF) protein strongly bound to the M3, M5 and M6 sequences and weakly bound to M4, but not to M1 or M2. To confirm this result, we performed a competition assay using each mutated DNA sequence as a competitor. A 200-fold molar excess of M3, M5 or M6 almost completely inhibited binding to the 30-nt sequence, similar to the inhibition conferred by the wild-type sequence. On the other hand, M1, M2 and M4 as competitors did not affect the formation of DNA–protein complexes, which were similar to those observed in the DNA binding assay (Figure 7C). These results strongly suggest that GmZF-HD1 binds in a sequence-specific manner to the GmCaM4 promoter and that the 20-nt sequence AGTAATTAAANNNNNAATTA containing two ATTA repeats may be a putative core cis-acting element in this promoter.
Figure 7.
Specific interaction of the GmZF-HD1 protein with 30-nt oligonucleotides derived from the GmCaM4 promoter. (A) Nucleotide sequences of a 30-nt fragment of the GmCaM-4 promoter (WT) and mutated fragments (M1 to M6) used as probes or competitors. (B) DNA-binding ability of recombinant GmZF-HD1 with 30-nt DNA (WT) and mutant fragments (M1 to M6). The radioactive probes were incubated in the presence of recombinant GmZF-HD1 or GST alone. (C) Competitive gel mobility shift assay using 30-nt DNA (WT) and mutated DNA fragments (M1 to M6) as competitors. The DNA binding reaction was performed with recombinant GST-fused GmZF-HD1 protein and 32P-labeled 30-nt DNA fragment (WT) in the absence or presence of unlabeled competitors. Each competitor DNA (200-fold molar excess) was added to a reaction mixture. Free (arrow) and protein-complexed (arrowhead) probes were separated on an 8% polyacrylamide gel and visualized by autoradiography.
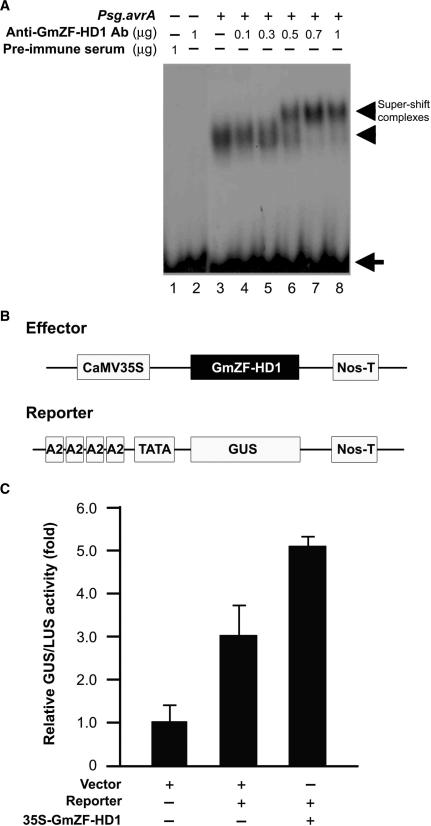
GmZF-HD1 actively functions as a transcriptional regulator involved in pathogen-induced gene expression in vivo
To confirm that GmZF-HD1 is an active component of the protein-DNA complex observed in EMSA experiments with the 30-nt sequence of the GmCaM4 promoter and nuclear extracts from pathogen-treated soybean suspension cells, we raised anti-GmZF-HD1 polyclonal antibodies against the affinity-purified GST::GmZF-HD1 fusion protein. We used these antibodies for supershift assays together with nuclear extracts from pathogen-treated cells (Figure 8A). The specificity of the antiserum was confirmed by western analysis with recombinant GST::GmZF-HD1 proteins and total and nuclear soybean proteins. The antiserum showed no cross-reactivity to any other proteins derived from total bacterial cell extracts or to any other soybean proteins (data not shown). Incubation of 0.5–1 μg of antiserum, nuclear extracts and the 30-nt probe in the gel shift reaction clearly resulted in a supershift of a DNA–protein complex (Figure 8A, lanes 6–8), which was not observed in the presence of preimmune serum (Figure 8A, lane 1) or in the absence of antiserum (Figure 8A, lane 3). These observations strongly confirm that the GmZF-HD1 protein is a component of the nuclear protein–DNA complex formed with the 30-nt sequence of the GmCaM4 promoter and that this complex forms when cells are stimulated by pathogens, but not under basal conditions.
Figure 8.
Direct interaction with and activation of the GmCaM4 promoter by the GmZF-HD1 protein. (A) Gel mobility super-shift assay using an anti-GmZF-HD1 antibody. Various concentrations of polyclonal antibodies raised against the GST::GmZF-HD1 protein were incubated with the protein–DNA complex formed with the 32P-labeled 30-nt fragment of GmCaM-4 promoter and nuclear extracts from pathogen-treated soybean suspension culture cells. Lane 1 contains preimmune serum, lane 2 contains 1 μg affinity-purified GmZF-HD1 antibody, and lane 3 contains nuclear extracts alone. Nuclear extracts from pathogen-treated cells were preincubated with 0.1 μg (lane 4), 0.3 μg (lane 5), 0.5 μg (lane 6), 0.7 ug (lane 7) or 1 μg (lane 8) affinity-purified GmZF-HD1 antibody before adding the 32P-labeled 30-nt fragment of GmCaM-4 promoter as a probe. Free (arrow) and protein-complexed (arrowhead) probes were separated on an 8% polyacrylamide gel and visualized by autoradiography. (B) Schematic representation of the effector and reporter constructs used in transient expression assays in Arabidopsis protoplasts. In the effector construct, GmZF-HD1 cDNA was inserted between the CaMV 35S promoter and a Nos terminator (Nos-T). In the reporter construct, four tandem repeats of the A2 region containing the 30-nt fragment of the GmCaM-4 promoter were subcloned upstream of a minimal TATA promoter-GUS construct. (C) Effect of the GmZF-HD1 protein on GmCaM-4 promoter activity in Arabidopsis protoplasts. The empty vector without GmZF-HD1 and the A2 region of the GmCaM-4 promoter were used as controls. For normalization of transformation efficiency, the CaMV 35S promoter-luciferase (LUC) plasmid was co-transfected in each experiment. The data are presented as mean ± SE of results from three independent samples. The numbers show the relative increase in expression compared to the value obtained with the vector control.
After demonstrating that GmZF-HD1 is a component of this pathogen-induced complex, we then examined whether the protein activates downstream transcription in vivo in a manner dependent on the 30-nt sequence. To do this, we used Arabidopsis leaf mesophyll protoplasts harboring a GUS reporter plasmid, which was constructed by fusion of the GUS gene with four tandem repeats of the A2 region of the GmCaM4 promoter cloned upstream of the cauliflower mosaic virus (CaMV) 35S minimal promoter in pDel 151-8 (34). We also constructed an effector plasmid by inserting the full-length GmZF-HD1 cDNA sequence after the CaMV 35S promoter (Figure 8B). As shown in Figure 8C, cells transfected with the reporter construct alone exhibited an increase in GUS activity of about 2.9-fold, compared to control cells transfected with the vector, indicating that endogenous Arabidopsis proteins may recognize the promoter sequence in the reporter construct. Co-expression with the effector protein GmZF-HD1 increased GUS activity about 5.1-fold. These results support the conclusion that the GmZF-HD1 protein actively functions as a transcriptional regulator and that it is involved in pathogen-induced GmCaM4 gene expression.
DISCUSSION
It is important to understand how plants recognize and transduce stresses such as pathogen infection and high environmental salinity to activate transcription of key cellular-response mediators such as calcium binding proteins, or CaMs. We previously showed that GmCaM4 gene expression induced by environmental stresses such as pathogen and salinity is partially mediated by the binding of a GT-1-like transcription factor to the GT-1 cis-acting element (GAAAAA) in the −858 to −728 region of the GmCaM4 promoter (13). In addition, we reported that the −1286 to −1065 region upstream of the GT-1 cis-acting element is also involved in pathogen-stimulated GmCaM4 gene regulation (13). In this report, we identified repeats of a conserved homeodomain-binding site, ATTA, in the GmCaM4 promoter that are relevant to pathogen responses. To isolate transcription factors that bind the cis-acting element, we screened soybean cDNA libraries using a yeast one-hybrid system and cloned two cDNAs, GmZF-HD1 and −2, encoding homeodomain DNA binding proteins that specifically interact with the A2 region (80 bp) of the GmCaM4 promoter (Figure 3). A previous yeast one-hybrid screening performed in plants had a success rate of 87.8% (36 true/41 total clones) (31). In this study, we obtained a success rate of 66.7% (28 true/42 total clones), indicating that our screen was effective for isolating new proteins that bind to the GmCaM4 promoter.
The homeodomain is one of the most important DNA-binding motifs in diverse organisms such as humans, Drosophila, Caenorhabditis elegans and plants, and it has been widely studied as a model system for protein–DNA interactions (35). The first crystal structure of the homeodomain–DNA complex was revealed in 1990 by Kissinger et al. (36), and subsequent structures of homeodomain–DNA complexes have been found to be very similar (37–40). Our data also show that the homeodomain but not the zinc finger domain is involved in sequence-specific DNA binding (Figure 6B). Most homeodomains bind to ATTA as a core consensus sequence (40).
To precisely map the GmZF-HD1 and −2 binding site in the GmCaM4 promoter, we performed EMSA using bacterially expressed recombinant GmZF-HD proteins (Figure 6). Recombinant GmZF-HD1 and −2 proteins specifically bound to the 30-nt sequence of the GmCaM4 promoter (Figure 6B), which is consistent with the results obtained with nuclear extracts from pathogen-treated cells (Figures 1A, 1C and Figure 2). A supershift assay using an anti-GmZF-HD1 antibody confirmed that GmZF-HD1 is indeed a component of nuclear extracts that form DNA–protein complexes with the 30-nt sequence of the GmCaM4 promoter (Figure 8A). More detailed EMSA and competition assays with a series of mutant oligonucleotides revealed that two repeats of the conserved ATTA homeodomain binding site in the GmCaM4 promoter are required for full binding activity with the GmZF-HD1 and −2 proteins (Figures 7A, B and C). The functional role of GmZF-HD1 and −2 in GmCaM4 gene regulation in vivo was also confirmed by a transient expression assay performed with Arabidopsis protoplasts (Figure 8C). The Arabidopsis zinc finger homeodomain protein ATHB33 was recently reported to bind to a 20-bp sequence (AGTGTCTTGTAATTAAAA), which contains an ATTA core sequence (underlined) that is similar to that of other homeodomain proteins in plants (39). In contrast, we showed here that both of the ATTA core sequences in the 30-bp sequence are required for full binding of the GmZF-HD1 protein (Figure 7). These data suggest that GmZF-HD1 proteins may recognize distinct promoter motifs that other plant homeodomain proteins do not. These distinctions may play pivotal roles in the regulation of different target promoters.
It is not yet clear how the GmZF-HD1 and −2 proteins are regulated after pathogen stimulation. To better understand the mechanism of this response, the localization of GmZF-HD1 and −2 before and after pathogen exposure must be determined. Members of the ZF-HD family have two highly conserved N-terminal zinc finger domains and a C-terminal homeodomain (32). Interestingly, Windhövel et al. (32) demonstrated that the two conserved N-terminal motifs constitute a novel dimerization domain that is involved in homo- or heterodimer formation. Mutagenesis of the second zinc finger domain showed that highly conserved cysteine residues are essential for protein–protein interaction (32). In the case of GmZF-HD1 and −2, their binding to ATTA repeats in the GmCaM4 promoter may not require dimerization (either homo- or heterodimerization) since a mutant deleted for the N-terminal ZF (i.e. containing only the homeodomain) still strongly bound to the 30-nt sequence containing this site. This result is perhaps unexpected because dimerization through the zinc finger could stabilize binding. Here we speculate that two monomers with N-terminal zinc finger domain truncation may have easier access to the closely spaced binding sites because of reduced steric hindrance.
We propose a model in which GmZF-HD1 translocation to the nucleus is important for regulating its transcription of the CaM4 promoter. This proposal is based on three observations: (i) there is no significant increase in GmZF-HD1 transcript level upon pathogen treatment (Figure 5B); (ii) the GmZF-HD proteins are present in nuclear extracts from pathogen-treated cells but not in extracts from untreated cells (Figure 1A); and (iii) post-translational modifications do not seem to play a major role in the binding of GmZF-HD to the GmCaM4 promoter because recombinant GmZF-HD1 and −2 expressed in bacteria, in which proteins are not post-translationally modified, exhibit DNA binding activity (Figures 6 and 7). Therefore, a possible scenario is that in response to pathogen attack, an unknown inhibitory protein present in the cytosol no longer prevents the GmZF-HD1 and −2 transcription factors from entering the nucleus where they can interact with the GmCaM4 promoter. GmZF-HD1 and −2 are small proteins (20 and 19.4 kDa, respectively) so in principle they could easily diffuse into the nucleus even without a nuclear localization signal sequence. Nuclear proteins extracted from unstimulated cells did not bind to the conserved homeodomain-binding site in the GmCaM4 promoter, suggesting that GmZF-HD1 and −2 are not present in the nucleus under basal conditions. The best example of this model would be the activation of the NF-κB transcription factor in animal cells exposed to bacterial pathogens. NF-κB is released from the inhibition of IκB in the cytosol and is then translocated into the nucleus, where it regulates the expression of immune response genes (41). This suggestion is consistent with our data, but alternative models can be proposed. For example, post-translational modifications may be important for optimal GmZF-HD activity but not for DNA binding. There may also be an increase in protein expression upon pathogen infection although transcriptional up-regulation does not occur (as judged by northern analysis).
At present, it is not known whether discrete cis-acting elements respond to different developmental and environmental cues or whether single or overlapping DNA motifs bind multiple regulatory factors, for example, as postulated for the parsley phenylalanine ammonia lyase1 (pal1) gene (42) and the bean pal2 gene (43). We observed that the expression patterns of the GmZF-HD1 and GmCaM4 genes were very similar soybean seedling tissues (Figure 5A), consistent with a role for the GmZF-HD1 protein in binding and controlling the activity of the GmCaM4 promoter. Their overlapping patterns of gene expression are also important for the transcriptional activation of GmCaM4 in response to the transduction of external pathogen-specific signals into the nucleus.
To summarize, we have shown that a new class of plant homeodomain proteins, GmZF-HD1 and GmZF-HD2, specifically bind to a 30-nt A/T-rich DNA sequence that contains two repeats of the conserved homeodomain binding site ATTA in the GmCaM4 promoter following exposure to plant pathogens. Thus our results strongly support regulation of the GmCaM4 gene by the GmZF-HD transcription factors as a significant component of the plant defense-signaling pathway.
ACKNOWLEDGEMENTS
This work was supported by grants from the Plant Diversity Research Center (#PF06303-01) of the 21st Century Frontier Research Program and the Environmental Biotechnology National Core Research Center (#R15-2003-012-02003-0) funded by the Ministry of Science and Technology in the Republic of Korea. S.M.L. was supported by a scholarship from the BK21 program of the Ministry of Education and Human Resources. The sequences reported in this article have been deposited in the GenBank database (accession numbers AY695729 for GmZF-HD1 cDNA and AY695730 for GmZF-HD2 cDNA). Funding to pay the Open Access publication charges for this article was provided by the Environmental Biotechnology National Core Research Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gilroy S, Bethke PC, Jones RL. Calcium homeostasis in plants. J. Cell Sci. 1993;106:453–461. doi: 10.1242/jcs.106.2.453. [DOI] [PubMed] [Google Scholar]
- 2.Knight MR, Campbell AK, Smith SM, Trewavas AJ. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature. 1991;352:524–526. doi: 10.1038/352524a0. [DOI] [PubMed] [Google Scholar]
- 3.Dixon RA, Harrison MJ, Lamb CJ. Early events in the activation of plant defense responses. Annu. Rev. Phytopathol. 1994;32:479–501. [Google Scholar]
- 4.Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;24:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 5.Sacks WR, Ferreira P, Hahlbrock K, Jabs T, Nürnberger T, Renelt A, Scheel D. In: Advances in Molecular Genetics of Plant-Microbe Interactions. Nester EW, Verma DPS, editors. Vol. 2. Dordrecht, The Netherlands: Kluwer; 1993. pp. 485–495. [Google Scholar]
- 6.Sanders D, Brownlee C, Harper JF. Communicating with calcium. Plant Cell. 1999;11:691–706. doi: 10.1105/tpc.11.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy AS. Calcium: silver bullet in signaling. Plant Sci. 2001;160:381–404. doi: 10.1016/s0168-9452(00)00386-1. [DOI] [PubMed] [Google Scholar]
- 8.Snedden WA, Fromm H. Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 2001;151:35–66. doi: 10.1046/j.1469-8137.2001.00154.x. [DOI] [PubMed] [Google Scholar]
- 9.Hoeflich KP, Ikura M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 2002;108:739–742. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 10.Yang T, Poovaiah BW. Calcium/calmodulin-mediated signal network in plants. Trends plant Sci. 2003;8:505–512. doi: 10.1016/j.tplants.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Lee SH, Kim JC, Lee MS, Heo WD, Seo HY, Yoon HW, Hong JC, Lee SY, Bahk JD, et al. Identification of a novel divergent calmodulin isoform from soybean which has differential ability to activate calmodulin-dependent enzymes. J. Biol. Chem. 1995;270:21806–21812. doi: 10.1074/jbc.270.37.21806. [DOI] [PubMed] [Google Scholar]
- 12.Yamakawa H, Mitsuhara I, Ito N, Seo S, Kamada H, Ohashi Y. Transcriptionally and post-transcriptionally regulated response of 13 calmodulin genes to tobacco mosaic virus-induced cell death and wounding in tobacco plant. Eur. J. Biochem. 2001;268:3916–3929. doi: 10.1046/j.1432-1327.2001.02301.x. [DOI] [PubMed] [Google Scholar]
- 13.Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, et al. Pathogen- and NaCl-induced expression of the GmCaM4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004;135:21806–21812. doi: 10.1104/pp.104.041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heo WD, Lee SH, Kim MC, Kim JC, Chung WS, Chun HJ, Lee KJ, Park CY, Park HC, et al. Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl Acad. Sci. USA. 1999;96:766–771. doi: 10.1073/pnas.96.2.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo JH, Park CY, Kim JC, Heo WD, Cheong MS, Park HC, Kim MC, Moon BC, Choi MS, et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J Biol Chem. 2005;280:3697–3706. doi: 10.1074/jbc.M408237200. [DOI] [PubMed] [Google Scholar]
- 16.Williams RW. Plant homeobox genes: many functions stem from a common motif. Bioessays. 1998;20:280–282. doi: 10.1002/(SICI)1521-1878(199804)20:4<280::AID-BIES2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Langdale JA. Cellular differentiation in the leaf. Curr. Opin. Cell Biol. 1998;10:734–738. doi: 10.1016/s0955-0674(98)80115-4. [DOI] [PubMed] [Google Scholar]
- 18.Hudson A. Axioms and axes in leaf formation? Curr. Opin. Plant Biol. 1999;2:56–60. doi: 10.1016/s1369-5266(99)80011-9. [DOI] [PubMed] [Google Scholar]
- 19.Muehlbauer GJ, Fowler JE, Girard L, Tyers R, Harper L, Freeling M. Ectopic expression of the maize homeobox gene Liguleless3 alters cell fates in the leaf. Plant Physiol. 1999;119:651–662. doi: 10.1104/pp.119.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korfhage U, Trezzini GF, Meier I, Hahlbrock K, Somssich IE. Plant homeodomain protein involved in transcriptional regulation of a pathogen defense-related gene. Plant Cell. 1994;6:695–708. doi: 10.1105/tpc.6.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajasekhar VK, Lamb C, Dixon RA. Early events in the signal pathway for the oxidative burst in soybean cells exposed to avirulent pseudomonas syringae pv glycinea. Plant Physiol. 1999;120:1137–1146. doi: 10.1104/pp.120.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagao RT, Goekjian VH, Hong JC, Key JL. Identification of protein-binding DNA sequences in an auxin-regulated gene of soybean. Plant Mol. Biol. 1993;21:1147–1162. doi: 10.1007/BF00023610. [DOI] [PubMed] [Google Scholar]
- 23.Hong JC, Cheong YH, Nagao RT, Bahk JD, Key JL, Cho MJ. Isolation of two soybean G-box binding factors which interact with a G-box sequence of an auxin-responsive gene. Plant J. 1995;8:199–211. doi: 10.1046/j.1365-313x.1995.08020199.x. [DOI] [PubMed] [Google Scholar]
- 24.Galas DJ, Schmitz A. DNase footprinting: a simple method for detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 26.Park HC, Kang YH, Chun HJ, Koo JC, Cheong YH, Kim CY, Kim MC, Chung WS, Kim JC, et al. Characterization of a stamen-specific cDNA encoding a novel plant defensin in Chinese cabbage. Plant Mol. Biol. 2002;50:59–69. doi: 10.1023/a:1016005231852. [DOI] [PubMed] [Google Scholar]
- 27.Jefferson A, Kavanagh TA, Bevan MW. GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheen J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science. 1996;274:1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- 29.Abel S, Theologis A. Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J. 1994;5:421–427. doi: 10.1111/j.1365-313x.1994.00421.x. [DOI] [PubMed] [Google Scholar]
- 30.Luehrsen KR, de Wet JR, Walbot V. Transient expression analysis in plants using firefly luciferase reporter gene. Methods Enzymol. 1992;216:397–414. doi: 10.1016/0076-6879(92)16037-k. [DOI] [PubMed] [Google Scholar]
- 31.Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Windhövel A, Hein I, Dabrowa R, Stockhaus J. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol. Biol. 2001;45:201–214. doi: 10.1023/a:1006450005648. [DOI] [PubMed] [Google Scholar]
- 33.Tan QK-G, Irish VF. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 2006;140:1095–1108. doi: 10.1104/pp.105.070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Martienssen R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- 35.Veraksa A, Del Campo M, McGinnis W. Developmental patterning genes and their conserved functions: from model organisms to humans. Mol. Genet. Metab. 2000;69:85–100. doi: 10.1006/mgme.2000.2963. [DOI] [PubMed] [Google Scholar]
- 36.Kissinger CR, Liu B, Martin-Bianco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 37.Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991;30:11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- 38.Kornberg TB. Understanding the homeodomain. J. Biol. Chem. 1993;268:26813–26816. [PubMed] [Google Scholar]
- 39.Gehring WJ, Affolter M, Bürglin T. Homeodomain proteins. Annu. Rev. Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 40.Wolberger C. Homeodomain interactions. Curr. Opin. Struct. Biol. 1996;6:62–68. doi: 10.1016/s0959-440x(96)80096-0. [DOI] [PubMed] [Google Scholar]
- 41.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2124. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 42.Lois R, Dietrich A, Hahlbrock K, Schulz W. A phenylalanine ammonia-lyase gene from parsley: Structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 1989;8:1641–1648. doi: 10.1002/j.1460-2075.1989.tb03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leyva A, Liang X, Pintor-Toro JA, Dixon RA, Lamb CJ. Cis-element combinations determine phenylalanine ammoni-lyase gene tissue-specific expression patterns. Plant Cell. 1992;4:263–271. doi: 10.1105/tpc.4.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]