Abstract
Mitoxantrone is an anti-cancer agent used in the treatment of breast and prostate cancers. It is classified as a topoisomerase II poison, however can also be activated by formaldehyde to generate drug–DNA adducts. Despite identification of this novel form of mitoxantrone–DNA interaction, excessively high, biologically irrelevant drug concentrations are necessary to generate adducts. A search for mitoxantrone analogues that could potentially undergo this reaction with DNA more efficiently identified Pixantrone as an ideal candidate. An in vitro crosslinking assay demonstrated that Pixantrone is efficiently activated by formaldehyde to generate covalent drug–DNA adducts capable of stabilizing double-stranded DNA in denaturing conditions. Pixantrone–DNA adduct formation is both concentration and time dependent and the reaction exhibits an absolute requirement for formaldehyde. In a direct comparison with mitoxantrone–DNA adduct formation, Pixantrone exhibited a 10- to 100-fold greater propensity to generate adducts at equimolar formaldehyde and drug concentrations. Pixantrone–DNA adducts are thermally and temporally labile, yet they exhibit a greater thermal midpoint temperature and an extended half-life at 37°C when compared to mitoxantrone–DNA adducts. Unlike mitoxantrone, this enhanced stability, coupled with a greater propensity to form covalent drug–DNA adducts, may endow formaldehyde-activated Pixantrone with the attributes required for Pixantrone–DNA adducts to be biologically active.
INTRODUCTION
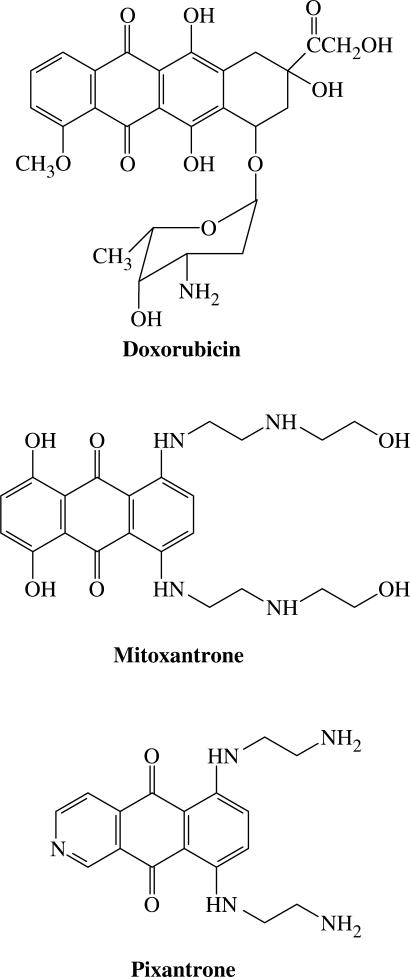
The anthracycline doxorubicin (Figure 1) is among the most versatile chemotherapeutic agents currently used in the clinic (1,2). The proven clinical utility of doxorubicin, a DNA-directed drug, has been tempered by dose-limiting cardiotoxicity, and this prompted a search for analogues with comparable therapeutic efficacy yet lacking the characteristic cardiotoxicity (3–5).
Figure 1.
Chemical structures of doxorubicin, mitoxantrone and Pixantrone.
The anthracenedione class of compounds were identified as good drug candidates designed to satisfy these criteria. The anthracenediones, most notably mitoxantrone (Novatrone™) and its 5,8-dehydroxy analogue ametantrone, are simplified anthracycline analogues, which retain the planar ring structure characteristic of anthracyclines that permits intercalation between base pairs of DNA (6,7) (Figure 1). The biochemical mechanism by which mitoxantrone exerts its cytotoxic effects is likely to be multifaceted, however its role as a topoisomerase II poison and subsequent induction of cytotoxic double-strand DNA breaks has been well established (8–10).
Despite an improved clinical tolerability of mitoxantrone chemotherapy, it still exerts a range of toxic side-effects including myelosuppression and cardiotoxicity (11–13). This cardiotoxicity may be attributed to the 5,8-dihydroxy substituents of mitoxantrone since mice treated with this drug exhibited a delayed mortality (14) yet those treated with ametantrone did not.
A second-generation group of anthracenediones were prepared in an effort to develop compounds endowed with better therapeutic efficacy and reduced side-effects. These compounds retained the anthraquinone nucleus of mitoxantrone, however, the 5,8-substituents implicated in its cardiotoxicity were removed and nitrogen atoms introduced into the chromophore. Krapcho et al. (15) rationalized that these nitrogen atoms may provide basic sites or improved hydrogen bonding, therefore providing the analogues with a potentially greater affinity for DNA and topoisomerase II.
A series of these novel anthracenediones were prepared and included compounds bearing either one (mono-aza) or two (di-aza) nitrogen atoms within the chromophore (15–17). Interestingly, an in vitro and in vivo screen of these compounds for anti-tumour activity revealed that only mono-aza analogues comprising the nitrogen atom at position 2 of the chromophore demonstrated significant anti-cancer efficacy (15–18). Within this select group of compounds, BBR 2778 (6,9-bis[(2-aminoethyl)amino]benzo[g]isoquinoline-5,10-dione dimaleate) emerged as the most promising drug candidate. BBR 2778 (subsequently named Pixantrone™; Figure 1) demonstrated superior anti-leukemic activity in mice over a wide, well-tolerated range of doses when compared with mitoxantrone (15). Further preclinical studies in mice showed that Pixantrone has a wide spectrum of anti-tumour activity, and a marked efficacy against haematological malignancies, particularly lymphomas and leukemias (19). Histopathological evaluation of the heart tissue in these studies revealed that Pixantrone induced no detectable delayed cardiotoxicity (19). These key findings prompted the entry of Pixantrone into clinical trials for further development. The drug is progressing through these trials with encouraging results as a single agent and in combination regimens, and is currently in Phase-III studies in the treatment of indolent and aggressive non-Hodgkin's lymphoma (20).
Like mitoxantrone, the mechanism of action of Pixantrone is not fully understood but likely to be multimodal. Pixantrone interacts with DNA with modest affinity via intercalation (21–23). The drug can function as a topoisomerase II poison by stabilizing the normally transiently bound protein–DNA complex (21,22,24), giving rise to double-strand DNA breaks. However, these breaks do not correlate directly with the potency of Pixantrone as a cytotoxic compound (21,22). This suggests that Pixantrone may be operating via an additional, currently unknown, mechanism of action.
Although mitoxantrone functions as a topoisomerase II poison via its ability to intercalate within DNA, a novel form of mitoxantrone–DNA interaction has been identified. Mitoxantrone can be readily oxidatively metabolized to generate reactive species that bind covalently to DNA (25–27). A common theme amongst these studies was that each of the oxidative systems utilized hydrogen peroxide which can generate in vitro formaldehyde by oxidation of substrates present in the system (28). Subsequent studies using cell-free systems revealed that formaldehyde alone, rather than oxidative metabolism, was sufficient for activation of mitoxantrone and subsequent covalent binding of the drug to DNA (29).
Presently, it is believed that these DNA adducts are linked via a ‘secondary’ amino function of a single side-chain of mitoxantrone (29–32). Although mitoxantrone and Pixantrone share close structural similarity, Pixantrone bears a ‘primary’ amino group in each of its side-chains and is therefore more susceptible to formaldehyde activation and consequently has a greater potential to form DNA adducts. The present study explored the potential of Pixantrone to bind covalently to DNA through pre-activation by formaldehyde.
MATERIALS AND METHODS
Materials
Pixantrone was provided by Cell Therapeutics Europe, Bresso, Italy. Mitoxantrone dihydrochloride and formamide were purchased from Sigma Chemical Co., St. Louis, MO, USA. Formaldehyde solution (40% v/v) was obtained from BDH. The plasmid pCC1 containing the lac UV5 promoter was constructed by Carleen Cullinane (Peter MacCallum Cancer Centre, Melbourne, VIC, Australia). A Maxi Plasmid Purification Kit was purchased from Qiagen, Valencia, CA, USA. Ultra-pure dNTPs, [α32P] dATP (3000 Ci/mmol), [α32P] dCTP (3000 Ci/mmol) and ProbeQuant G-50 micro-columns were purchased from GE Healthcare, Piscataway, NJ, USA. The restriction enzyme HindIII was purchased from Promega, Madison, WI, USA and calf thymus DNA was from Worthington Biochemical Corporation, Lakewood, NJ, USA. Klenow fragment from Escherichia coli DNA polymerase I and BSA were both from New England Biolabs, Beverly, MA, USA. Tris-saturated phenol was obtained from Invitrogen, Carlsbad, CA, USA and glycogen was from Roche Molecular Biochemicals, Nutley, NJ, USA. The remaining chemicals and reagents were of analytical grade. Distilled water passed through a four stage Milli-Q purification system was used to prepare all solutions.
Drugs
Pixantrone and mitoxantrone stock solutions (stored at −20°C) were prepared by dissolving each in Milli-Q water to an approximate concentration of 2 mM. The precise concentrations of each drug were determined spectrophotometrically using ε = 19 200 M−1 cm−1 at 608 nm and ε1%1 cm = 296 at 641 nm for mitoxantrone and Pixantrone, respectively. Formaldehyde solutions were freshly prepared on the day of each experiment.
DNA source
Escherichia coli HB101 cells containing the plasmid pCC1 were grown overnight in selective LB broth containing 50 μg/ml ampicillin. The plasmid was subsequently isolated using a Qiagen Maxi Plasmid Purification Kit. The plasmid was linearized by restriction digestion with the sticky-end generating enzyme HindIII. The 5′-overhang of the fragment was filled in using the Klenow fragment of DNA polymerase I in the presence of either [α32P] dATP or [α32P] dCTP. Unincorporated label was removed from the labelled fragment by passing the reaction mixture through a G-50 ProbeQuant chromatography column. The eluted fragment was subsequently subjected to phenol/chloroform extraction, ethanol precipitated and then resuspended in 1× TE (10 mM Tris, 1 mM EDTA, pH 8.0). The final DNA concentration was adjusted to 400 μMbp by the addition of sonicated calf thymus DNA.
In vitro crosslinking assay
Covalent drug–DNA adducts were formed in a reaction mixture typically consisting of the following: end-labelled DNA (25 μMbp) was reacted with Pixantrone or mitoxantrone and formaldehyde in phosphate-buffered saline (pH 7.0) at 37°C. Intercalated drug (but not covalently bound drug) was removed from DNA by extraction with Tris-saturated phenol twice followed by a single chloroform extraction. DNA was then precipitated with ethanol and sodium acetate in the presence of glycogen as an inert carrier. Samples were subsequently resuspended in 10 μl 1× TE buffer and denatured in two volumes of loading dye (90% formamide, 10 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanol) at 52°C for 5 min.
Stability studies
DNA adduct stability studies were performed as described above, however an additional extraction procedure was employed following incubation of drug–DNA adducts for defined time periods. Samples were extracted once with phenol, once with chloroform and then ethanol precipitated to remove any non-covalently bound drug resulting from dissociation as a consequence of the thermal lability of the adducts.
Electrophoresis, phosphorimaging and quantitation
Samples were subjected to electrophoresis through 0.8% agarose gels overnight at 30–40 V in 1× TAE buffer. Gels were dried under vacuum in a Bio-Rad Model 583 gel drier and subsequently exposed to a phosphor screen overnight. Phosphorimaging analysis of each dried gel was performed using a Molecular Dynamics Model 400B PhosphorImager and the bands quantitated using ImageQuant software.
RESULTS
The basis of the in vitro crosslinking assay is that DNA-stabilizing drug adducts will prevent the complete denaturation of double-stranded DNA (ds DNA) upon exposure to denaturing conditions (33). When samples are subjected to electrophoresis, ds DNA will migrate more slowly compared to rapidly moving single-stranded DNA. Stabilization of ds DNA is a functional consequence of the formation of some types of drug–DNA adducts. Consequently, the percentage of ds DNA provides a direct measure of drug–DNA adduct formation. Each of the following experiments included controls to confirm that DNA reacted in the absence of drug was efficiently denatured, indicating that the denaturation conditions were appropriate.
Pixantrone and formaldehyde concentration-dependence of drug–DNA adduct formation
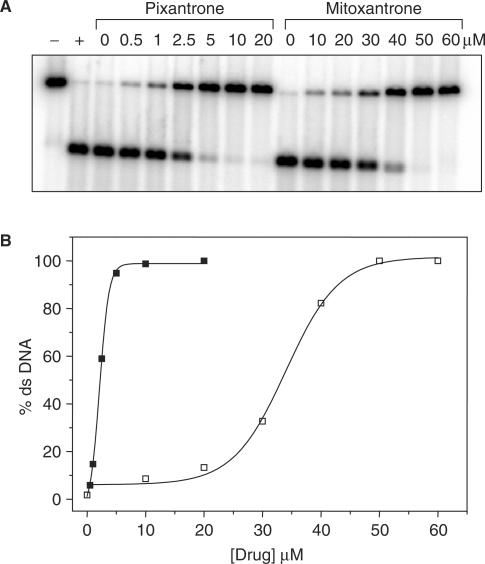
Initially, it was necessary to determine if Pixantrone binds covalently to DNA in the presence of formaldehyde, and this was confirmed using a simple drug concentration-dependence assay. Samples were prepared by incubating end-labelled DNA (25 μMbp) with varying concentrations of Pixantrone in the presence of 2 mM formaldehyde and phosphate-buffered saline (pH 7.0). The reaction was performed at 37°C and allowed to proceed overnight. Following a cleanup procedure to remove non-covalently bound drug, samples were denatured at 52°C in 60% formamide for 5 min, and resolved electrophoretically. The phosphorimage in Figure 2A reveals that Pixantrone-reacted samples stabilize a large percentage of ds DNA even at relatively low drug concentrations. Quantitation of these bands (Figure 2B) showed that 50% ds DNA stabilization was achieved by ∼2.5 μM Pixantrone. By comparison, mitoxantrone required ∼10-fold more drug to generate a similar level of ds DNA stabilization. Using the Poisson distribution to calculate adduct levels (34), this percentage of ds DNA stabilization reflects adduct levels of ∼0.7 adducts per fragment (or 2.1 adducts per 10 kb). Results were presented as percentage ds DNA stabilization rather than adducts per fragment because the Poisson distribution is not meaningful for values approaching 100% stabilized DNA. With regards to the range of adduct levels that can be calculated accurately, it should be noted that 20% ds DNA stabilization reflects 0.2 adducts per fragment while 80% ds DNA stabilization reflects 1.6 adducts per fragment (35).
Figure 2.
Pixantrone binds covalently to DNA in the presence of formaldehyde and stabilizes ds DNA in denaturing conditions. (A) DNA samples were reacted with 2 mM formaldehyde and either 0–20 μM Pixantrone or 0–60 μM mitoxantrone overnight. Each sample was routinely subjected to phenol/chloroform extraction, ethanol precipitated, denatured and subsequently electrophoresed through an agarose gel. Control ss DNA (+) was generated by subjecting unreacted DNA to thermal denaturation while the corresponding ds DNA control (−) was not thermally denatured. (B) The relative amount of ds DNA represented in (A) was quantitated and is expressed as a function of Pixantrone (solid squares) or mitoxantrone (open squares) concentration.
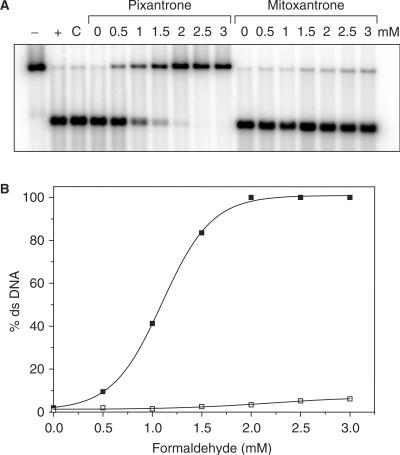
To confirm that the Pixantrone–DNA interaction required formaldehyde, a formaldehyde concentration-dependence study was performed. Samples were reacted as outlined above with increasing concentrations of formaldehyde, and processed as described previously. The phosphorimage in Figure 3A demonstrates a clear formaldehyde-dependence for Pixantrone–DNA adduct formation, which is reflected by the increasing stabilization of ds DNA with increasing formaldehyde concentration. Importantly, no DNA adducts were formed in the absence of formaldehyde, indicating a critical role for formaldehyde in the activation of Pixantrone to form drug–DNA adducts. Quantitation of this gel (Figure 3B) reveals that stabilization of ds DNA by Pixantrone–DNA adducts is maximal at 2 mM formaldehyde. Mitoxantrone also requires formaldehyde for reaction with DNA but is ∼100-fold less efficient in this reaction at equivalent concentrations.
Figure 3.
Formaldehyde is an absolute requirement for the formation of Pixantrone–DNA adducts. (A) Samples were reacted with increasing concentrations of formaldehyde (0–3 mM as indicated) and either 15 μM Pixantrone or 15 μM mitoxantrone overnight. Each sample was routinely processed and analysed as described previously. C is a control incubated in the absence of drug. (B) The relative amount of ds DNA represented in (A) was quantitated and is expressed as a function of formaldehyde concentration (Pixantrone, solid squares; mitoxantrone, open squares).
Time dependence of Pixantrone–DNA adduct formation
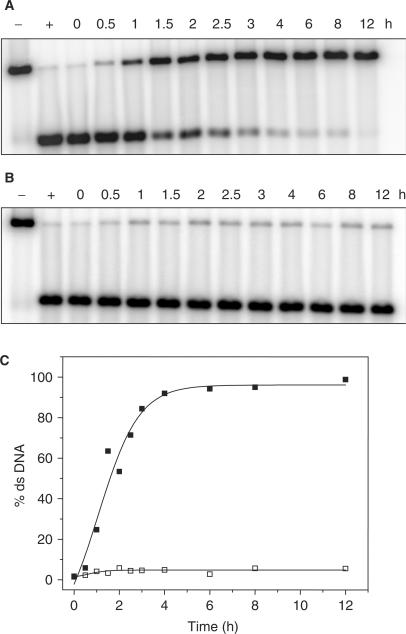
The time dependence of Pixantrone–DNA adduct formation was then investigated (Figure 4). Samples were reacted with formaldehyde as outlined above with either 10 μM Pixantrone or mitoxantrone. Samples were incubated at 37°C for defined time periods and frozen at −20°C until all samples were obtained. Figure 4A demonstrates that Pixantrone–DNA adduct formation is a time-dependent process, reaching a steady state within 4 h (Figure 4C). Mitoxantrone did not form adducts at an equimolar concentration of drug, regardless of the length of time of incubation (Figure 4B).
Figure 4.
Time-dependence of Pixantrone–DNA adduct formation. Samples were reacted with 2 mM formaldehyde and either (A) 10 μM Pixantrone or (B) 10 μM mitoxantrone for increasing time periods up to 12 h as indicated. Samples were then typically processed and analysed as described in the Materials and Methods section. (C) The relative amount of ds DNA represented in (A) and (B) was quantitated and is expressed as a function of time (Pixantrone, solid squares; mitoxantrone, open squares).
pH dependence of Pixantrone–DNA adduct formation
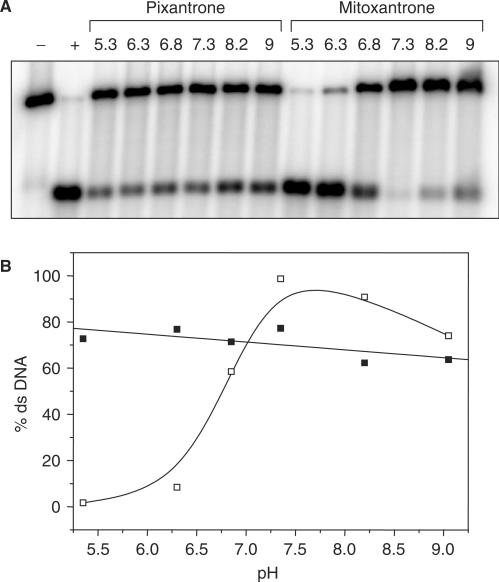
The pH dependence of formation of Pixantrone and mitoxantrone–DNA adducts was also examined by incubating end-labelled DNA with Pixantrone or mitoxantrone and formaldehyde in PBS adjusted to various pHs ranging from 5 to 9 (Figure 5A). Quantitation of these bands revealed that Pixantrone–DNA adduct formation was independent of pH in the range examined (Figure 5B). In contrast, mitoxantrone–DNA adduct formation was highly sensitive to extremes of pH and favoured neutral pH for optimal adduct formation (Figure 5B).
Figure 5.
pH-dependence of Pixantrone–DNA adduct formation. (A) Each DNA sample was incubated with 2 mM formaldehyde and either 10 μM Pixantrone or 50 μM mitoxantrone in phosphate-buffered saline adjusted to various pHs ranging from 5.3 to 9 for 4 h. Each sample was routinely processed and analysed as described in the Materials and Methods section. (B) The relative amount of ds DNA represented in (A) was quantitated and is expressed as a function of pH (Pixantrone, solid squares; mitoxantrone, open squares).
Thermal stability of Pixantrone–DNA adducts
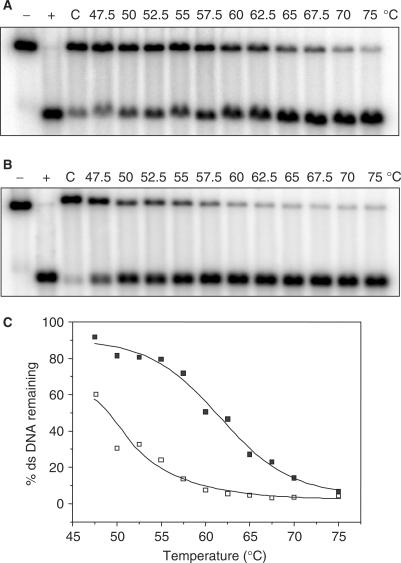
Given the thermal lability of mitoxantrone–DNA adducts (29), it was important to probe the potential thermal instability of Pixantrone–DNA adducts. DNA was reacted with formaldehyde and Pixantrone to generate partially ds DNA samples (∼70–80%) that were subsequently incubated at various temperatures in a non-denaturing (1× TE, pH 8.0) environment. Figure 6A reveals that, like mitoxantrone, Pixantrone–DNA adducts are lost with increasing temperature, indicating thermal lability of the drug–DNA adducts. Quantitation of the bands in Figure 6A and B yielded a melting curve (Figure 6C). The Pixantrone curve revealed that 50% of DNA adducts are lost at 62.5°C. In contrast, the same percentage of mitoxantrone–DNA adducts were lost at the much lower temperature of 49°C (Figure 6C), suggesting that Pixantrone adducts exhibit greater thermal stability than the mitoxantrone equivalent.
Figure 6.
Pixantrone–DNA adducts are thermally labile. DNA samples were reacted with 2 mM formaldehyde and either (A) 15 μM Pixantrone or (B) 50 μM mitoxantrone for 4 h and then subjected to a standard phenol/chloroform extraction and ethanol precipitation. Following resuspension in TE, samples were exposed to various temperatures ranging from 47.5 to 75°C for 5 min as indicated. C is a drug-treated control that remained on ice for this 5 min incubation. DNA was subsequently cleaned via a second round of phenol/chloroform extraction and the samples prepared for electrophoresis as usual. (C) The relative amount of ds DNA represented in (A) and (B) was quantitated and is expressed as a function of temperature (Pixantrone, solid squares; mitoxantrone, open squares).
Temporal stability of Pixantrone–DNA adducts at 37°C
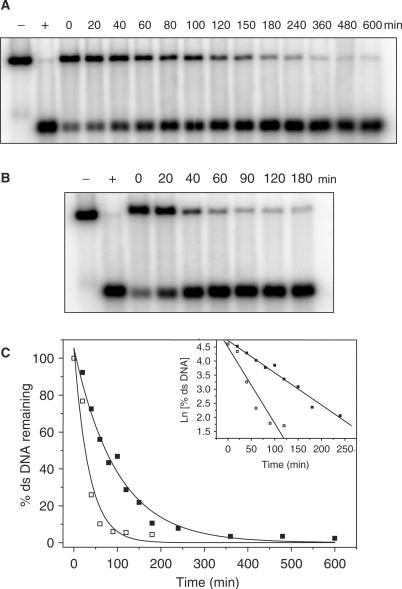
Next, it was important to investigate the stability of Pixantrone–DNA adducts at a physiologically relevant temperature. Following reaction of DNA with formaldehyde and Pixantrone to generate a sub-maximal level of ds DNA, samples were incubated at 37°C for defined time periods up to 10 h. Figure 7A and B demonstrates that both Pixantrone and mitoxantrone adducts are lost with time, indicating that the drug–DNA adducts are labile at 37°C. Pixantrone–DNA adducts exhibited a half-life of 80 min at 37°C, considerably more stable than mitoxantrone–DNA adducts with a half-life of 25 min. The loss of these adducts was a single first-order decay process, as indicated by a linear plot of ln(ds DNA) with time (inset, Figure 7C).
Figure 7.
Pixantrone–DNA adducts are temporally labile at 37°C. DNA samples were reacted with 2 mM formaldehyde and either (A) 15 μM Pixantrone or (B) 50 μM mitoxantrone for 4 h and then subjected to a standard phenol/chloroform extraction and ethanol precipitation. Following resuspension in TE, samples were incubated at 37°C for defined time periods up to 10 h as indicated. DNA was subsequently cleaned via a second round of phenol/chloroform extraction and the samples prepared for electrophoresis as normal. (C) The relative amount of ds DNA represented in (A) and (B) was quantitated and is expressed as a function of time (Pixantrone, solid squares; mitoxantrone, open squares). The inset shows the first-order decay of adducts.
DISCUSSION
A relatively small but growing body of evidence has recently implicated formaldehyde in the activation of mitoxantrone, which can subsequently bind covalently to DNA (29–31). The strong structural similarity between Pixantrone and mitoxantrone prompted an investigation into the possible activation of Pixantrone by formaldehyde to form similar covalent drug–DNA adducts.
Formaldehyde-activated Pixantrone forms covalent drug–DNA adducts
The drug concentration-dependence curve (Figure 2) indicates that both Pixantrone and formaldehyde combine to generate a product capable of stabilizing ds DNA, which is detected via the in vitro crosslinking assay. Indeed, neither agent alone was sufficient to stabilize duplex DNA. By analogy with formaldehyde-activated mitoxantrone, the identity of this duplex-stabilizing product is the covalent formaldehyde-mediated Pixantrone–DNA adduct.
The generation of these adducts is also strongly dependent upon the level of formaldehyde (Figure 3), indicating a crucial role for formaldehyde in the formation of Pixantrone–DNA adducts. Presumably, formaldehyde undergoes nucleophilic attack by either amine within the side-chains of Pixantrone to form a highly reactive Schiff base intermediate. This reactive intermediate provides at least one potential binding site to DNA and enables the subsequent formation of a Pixantrone–DNA adduct. Extending the analogy even further, it is likely that the nature of the covalent linkage forged between Pixantrone and DNA is a methylene bridge provided by formaldehyde.
An additional feature of both concentration-dependence curves is that Pixantrone demonstrates a far greater propensity to form covalent drug–DNA adducts than mitoxantrone (Figures 2 and 3). While Pixantrone possesses a primary amino group in each side-chain, mitoxantrone contains a secondary amine function in the corresponding positions. The primary amino groups of Pixantrone may be favourable for adduct formation for several reasons. First, the primary amino group is expected to be more reactive because it is more nucleophilic and more accessible (i.e. less steric restriction) than a secondary amino group. Second, since the primary amino group is expected to be substantially more reactive, this should provide a greater concentration of reactive Pixantrone–Schiff base precursor. Finally, the Pixantrone–Schiff base may be relatively more stable, thus ensuring a sustained time for reaction with its molecular target, DNA. It is not clear which of the many steps involved in adduct formation contribute to the observed pH dependence, but it is likely that it is due primarily to the secondary amino being more basic than the primary amino group, making it less nucleophilic and less likely to form an imine with formaldehyde at lower pHs.
Pixantrone–DNA adducts are thermally and temporally labile
The results also demonstrate that Pixantrone–DNA adducts are lost with increasing temperature (Figure 6) and time (Figure 7), indicating that the adduct is intrinsically unstable. The lability of this drug–DNA interaction suggests that Pixantrone–DNA adducts do not form conventional stable covalent crosslinks. In this case, both DNA strands are covalently coupled together by the drug, generating a genuine cross-link, which is characteristically much more stable. Rather, Pixantrone–DNA adducts function as ‘virtual’ crosslinks, which are responsible for stabilizing ds DNA in the in vitro crosslinking assay. The Pixantrone adduct is covalently bound via a methylene bridge (provided by formaldehyde) to a single DNA strand. This monoadduct is presumably stabilized by strong hydrogen bonding to the opposite DNA strand with the drug chromophore intercalated between neighbouring base pairs.
Cellular implications for Pixantrone–DNA adducts
Formaldehyde can be derived from a variety of sources in biological systems. It occurs naturally at low levels as a consequence of normal cellular metabolism (36) and is elevated in some cancers, including haematological malignancies (37). Indeed, Pixantrone has shown promising anti-cancer activity against some of these tumours (20,38,39) and it may be that elevated formaldehyde levels naturally predispose these cancers to Pixantrone cytotoxicity. A sensitive technique for detecting endogenous levels of formaldehyde involves a complex preconcentration-chemical ionization mass spectrometry technique being utilized (40). Intracellular levels of formaldehyde in cancer cells in vitro were found to range from 1.5 to 4 μM using this technique, and this generally correlated well with doxorubicin cytotoxicity. Although these levels are well below the formaldehyde concentrations utilized to form Pixantrone adducts in the current study, it would be interesting to study a broad range of cell lines for formaldehyde levels and Pixantrone cytotoxicity to assess a possible correlation. To address the question of whether Pixantrone forms adducts in cells in the absence of externally supplied formaldehyde, [14C] labelled Pixantrone was utilized. Preliminary results employing liquid scintillation analysis of DNA isolated from [14C] Pixantrone-treated cells indicate that adduct formation only occurs in the presence of formaldehyde-releasing prodrugs. However due to a lack of sensitivity using this technique, more reliable data for cells treated with Pixantrone as a single agent need to be generated using accelerator mass spectrometry as described (41).
Formaldehyde can be introduced at high levels into biological systems by formaldehyde-releasing prodrugs. AN-9 (pivaloyloxymethyl butyrate; Pivanex™) is amongst the most promising drug candidates of this class. Originally designed as a histone deacetylase inhibitor, AN-9 undergoes cellular hydrolysis by esterases to release butyric acid, pivalic acid and formaldehyde (42). Cutts et al. (35) reported that the combination of AN-9 and the anthracycline doxorubicin dramatically enhanced the formation of doxorubicin–DNA adducts in human cancer cells. This study attributed the generation of doxorubicin–DNA adducts to formaldehyde release via cellular hydrolysis of AN-9 and this provided a molecular rationale for the synergistic response observed between doxorubicin and AN-9.
Mitoxantrone can also form covalent DNA adducts in cells when administered in combination with AN-9, however excessively high (i.e. ⩾20 μM) drug levels are required (32). This suggests that it is unlikely that the induction of mitoxantrone–DNA adducts contributes significantly to the biological activity of this drug.
A highlight of the present study is that formaldehyde-activated Pixantrone reacts far more efficiently with DNA in vitro to generate covalent drug–DNA adducts than mitoxantrone. The enhanced efficiency of this reaction can potentially be harnessed in a cellular environment by combining Pixantrone with AN-9 to generate Pixantrone–DNA adducts in cells. Indeed, efficient formation of Pixantrone–DNA adducts was achieved with drug concentrations ranging from 0.5 to 20 μM Pixantrone (Figure 2A), levels that typically correspond with IC50 values reported for Pixantrone in experimental cancer cell lines (15,19,21,22).
A second key feature of this study is that Pixantrone–DNA adducts also exhibit greater stability than the corresponding mitoxantrone species. This may also be important in a cellular context since an extended Pixantrone–DNA adduct half-life may maximize DNA damage and enable an enhanced disruption of critical cellular processes such as DNA replication and transcription. Moreover, these adducts have the potential to elicit a potent apoptotic response, as recently demonstrated for doxorubicin–DNA adducts that were mediated by AN-9 (43). Studies utilizing growth inhibition and clonogenic assays are currently in progress to establish whether Pixantrone exhibits a similar favourable interaction with AN-9. Preliminary results (Mansour,O.C. et al., unpublished data) using tumour cells in culture indicate that Pixantrone and AN-9 have a greater than additive effect in growth inhibition and cell death assays, while the mitoxantrone/AN-9 interaction is additive, indicating that formaldehyde-activated Pixantrone indeed possesses superior biological activity.
ACKNOWLEDGEMENT
This work was carried out with support from the Australian Research Council. Funding to pay the Open Access publication charges for this article was provided by the Australian Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Creighton TE. Encyclopedia of Molecular Biology. New York, Chichester: Wiley; 1999. [Google Scholar]
- 2.Hellman S, DeVita VT, Rosenberg SA. Cancer: Principles & Practice of Oncology. 6th. Philadelphia, London: Lippincott-Raven; 2001. [Google Scholar]
- 3.Murdock KC, Child RG, Fabio PF, Angier RB, Wallace RE, Durr FE, Citarella RV. Antitumor agents. 1. 1,4-Bis[(aminoalkyl)amino]-9,10-anthracenediones. J. Med. Chem. 1979;22:1024–1030. doi: 10.1021/jm00195a002. [DOI] [PubMed] [Google Scholar]
- 4.Wallace RE, Murdock KC, Angier RB, Durr FE. Activity of a novel anthracenedione, 1,4-dihydroxy-5,8-bis(((2-[(2-hydroxyethyl)amino]ethyl)amino])-9,10-anthracenedione dihydrochloride, against experimental tumors in mice. Cancer Res. 1979;39:1570–1574. [PubMed] [Google Scholar]
- 5.Zee-Cheng RK, Cheng CC. Antineoplastic agents. Structure-activity relationship study of bis(substituted aminoalkylamino)anthraquinones. J. Med. Chem. 1978;21:291–294. doi: 10.1021/jm00201a012. [DOI] [PubMed] [Google Scholar]
- 6.Durr FE. In: Anthracycline and Anthracenedione-based Anticancer Agents. Lown JW, editor. Amsterdam, The Netherlands: Elsevier; 1988. pp. 163–200. [Google Scholar]
- 7.Pratt WB, Ruddon RW, Ensminger WD, Maybaum J. The Anticancer Drugs. 2nd. New York, USA: Oxford University Press; 1994. [Google Scholar]
- 8.Smith PJ, Morgan SA, Fox ME, Watson JV. Mitoxantrone-DNA binding and the induction of topoisomerase II associated DNA damage in multi-drug resistant small cell lung cancer cells. Biochem. Pharmacol. 1990;40:2069–2078. doi: 10.1016/0006-2952(90)90237-f. [DOI] [PubMed] [Google Scholar]
- 9.Davies SM, Robson CN, Davies SL, Hickson ID. Nuclear topoisomerase II levels correlate with the sensitivity of mammalian cells to intercalating agents and epipodophyllotoxins. J. Biol. Chem. 1988;263:17724–17729. [PubMed] [Google Scholar]
- 10.Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 11.Cornbleet MA, Stuart-Harris RC, Smith IE, Coleman RE, Rubens RD, McDonald M, Mouridsen HT, Rainer H, van Oosterom AT, et al. Mitoxantrone for the treatment of advanced breast cancer: single-agent therapy in previously untreated patients. Eur. J. Cancer Clin. Oncol. 1984;20:1141–1146. doi: 10.1016/0277-5379(84)90122-6. [DOI] [PubMed] [Google Scholar]
- 12.Faulds D, Balfour JA, Chrisp P, Langtry HD. Mitoxantrone. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in the chemotherapy of cancer. Drugs. 1991;41:400–449. doi: 10.2165/00003495-199141030-00007. [DOI] [PubMed] [Google Scholar]
- 13.Pratt WB, Pratt WB. The Anticancer Drugs. 2nd. New York, USA: Oxford University Press; 1994. [Google Scholar]
- 14.Corbett TH, Griswold DP, Jr, Roberts BJ, Schabel M., Jr Absence of delayed lethality in mice treated with aclacinomycin A. Cancer Chemother. Pharmacol. 1981;6:161–168. doi: 10.1007/BF00262337. [DOI] [PubMed] [Google Scholar]
- 15.Krapcho AP, Petry ME, Getahun Z, Landi JJ, Jr, Stallman J, Polsenberg JF, Gallagher CE, Maresch MJ, Hacker MP, et al. 6,9-Bis[(aminoalkyl)amino]benzo[g]isoquinoline-5,10-diones. A novel class of chromophore-modified antitumor anthracene-9,10-diones: synthesis and antitumor evaluations. J. Med. Chem. 1994;37:828–837. doi: 10.1021/jm00032a018. [DOI] [PubMed] [Google Scholar]
- 16.Krapcho AP, Landi JJ, Jr, Hacker MP, McCormack JJ. Synthesis and antineoplastic evaluations of 5,8-bis[(aminoalkyl)amino]-1-azaanthracene-9,10-diones. J. Med. Chem. 1985;28:1124–1126. doi: 10.1021/jm00146a029. [DOI] [PubMed] [Google Scholar]
- 17.Krapcho AP, Maresch MJ, Helgason AL, Rosner KE, Hacker MP, Spinelli S, Menta E, Oliva A. The synthesis of 6,9-bis-[(aminoalkyl)amino] substituted benzo[g]quinoxaline-, benzo[g]quinazoline- and benzo[g]phthalazine-5,10-diones via regiospecific displacements. J. Heterocyclic. Chem. 1993;30:1597–1606. [Google Scholar]
- 18.Krapcho AP, Maresch MJ, Hacker MP, Menta E, Oliva A, Giuliani FC, Spinelli S. Aza and diaza bioisosteric anthracene-9,10-diones as antitumor agents. Acta Biochim. Pol. 1995;42:427–432. [PubMed] [Google Scholar]
- 19.Beggiolin G, Crippa L, Menta E, Manzotti C, Cavalletti E, Pezzoni G, Torriani D, Randisi E, Cavagnoli R, et al. Bbr 2778, an aza-anthracenedione endowed with preclinical anticancer activity and lack of delayed cardiotoxicity. Tumori. 2001;87:407–416. doi: 10.1177/030089160108700611. [DOI] [PubMed] [Google Scholar]
- 20.Borchmann P, Reiser M. Pixantrone (Novuspharma) IDrugs. 2003;6:486–490. [PubMed] [Google Scholar]
- 21.De Isabella P, Palumbo M, Sissi C, Capranico G, Carenini N, Menta E, Oliva A, Spinelli S, Krapcho AP, et al. Topoisomerase II DNA cleavage stimulation, DNA binding activity, cytotoxicity, and physico-chemical properties of 2-aza- and 2-aza-oxide-anthracenedione derivatives. Mol. Pharmacol. 1995;48:30–38. [PubMed] [Google Scholar]
- 22.Hazlehurst LA, Krapcho AP, Hacker MP. Comparison of aza-anthracenedione-induced DNA damage and cytotoxicity in experimental tumor cells. Biochem. Pharmacol. 1995;50:1087–1094. doi: 10.1016/0006-2952(95)00246-v. [DOI] [PubMed] [Google Scholar]
- 23.Hazlehurst LA, Krapcho AP, Hacker MP. Correlation of DNA reactivity and cytotoxicity of a new class of anticancer agents: aza-anthracenediones. Cancer Lett. 1995;91:115–124. doi: 10.1016/0304-3835(95)91035-5. [DOI] [PubMed] [Google Scholar]
- 24.Zwelling LA, Mayes J, Altschuler E, Satitpunwaycha P, Tritton TR, Hacker MP. Activity of two novel anthracene-9,10-diones against human leukemia cells containing intercalator-sensitive or -resistant forms of topoisomerase II. Biochem. Pharmacol. 1993;46:265–271. doi: 10.1016/0006-2952(93)90413-q. [DOI] [PubMed] [Google Scholar]
- 25.Reszka K, Hartley JA, Kolodziejczyk P, Lown JW. Interaction of the peroxidase-derived metabolite of mitoxantrone with nucleic acids. Evidence for covalent binding of 14C-labeled drug. Biochem. Pharmacol. 1989;38:4253–4260. doi: 10.1016/0006-2952(89)90523-6. [DOI] [PubMed] [Google Scholar]
- 26.Panousis C, Kettle AJ, Phillips DR. Oxidative metabolism of mitoxantrone by the human neutrophil enzyme myeloperoxidase. Biochem. Pharmacol. 1994;48:2223–2230. doi: 10.1016/0006-2952(94)00429-3. [DOI] [PubMed] [Google Scholar]
- 27.Panousis C, Kettle AJ, Phillips DR. Myeloperoxidase oxidizes mitoxantrone to metabolites which bind covalently to DNA and RNA. Anticancer Drug Des. 1995;10:593–605. [PubMed] [Google Scholar]
- 28.Taatjes DJ, Gaudiano G, Resing K, Koch TH. Alkylation of DNA by the anthracycline, antitumor drugs adriamycin and daunomycin. J. Med. Chem. 1996;39:4135–4138. doi: 10.1021/jm960519z. [DOI] [PubMed] [Google Scholar]
- 29.Parker BS, Cullinane C, Phillips DR. Formation of DNA adducts by formaldehyde-activated mitoxantrone. Nucleic Acids Res. 1999;27:2918–2923. doi: 10.1093/nar/27.14.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker BS, Cutts SM, Cullinane C, Phillips DR. Formaldehyde activation of mitoxantrone yields CpG and CpA specific DNA adducts. Nucleic Acids Res. 2000;28:982–990. doi: 10.1093/nar/28.4.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker BS, Cutts SM, Phillips DR. Cytosine methylation enhances mitoxantrone-DNA adduct formation at CpG dinucleotides. J. Biol. Chem. 2001;276:15953–15960. doi: 10.1074/jbc.M009216200. [DOI] [PubMed] [Google Scholar]
- 32.Parker BS, Rephaeli A, Nudelman A, Phillips DR, Cutts SM. Formation of mitoxantrone adducts in human tumor cells: potentiation by AN-9 and DNA methylation. Oncol. Res. 2004;14:279–290. doi: 10.3727/096504003773994815. [DOI] [PubMed] [Google Scholar]
- 33.Luce RA, Sigurdsson ST, Hopkins PB. Quantification of formaldehyde-mediated covalent adducts of adriamycin with DNA. Biochemistry. 1999;38:8682–8690. doi: 10.1021/bi990553q. [DOI] [PubMed] [Google Scholar]
- 34.Vos JM. In: DNA Repair: A Laboratory Manual of Research Procedures. Friedberg EC, Hanawalt PC, editors. Vol. 3. NY: Marcel Dekker; 1988. pp. 367–398. [Google Scholar]
- 35.Cutts SM, Rephaeli A, Nudelman A, Hmelnitsky I, Phillips DR. Molecular basis for the synergistic interaction of adriamycin with the formaldehyde-releasing prodrug pivaloyloxymethyl butyrate (AN-9) Cancer Res. 2001;61:8194–8202. [PubMed] [Google Scholar]
- 36.Iborra FJ, Renau-Piqueras J, Portoles M, Boleda MD, Guerri C, Pares X. Immunocytochemical and biochemical demonstration of formaldhyde dehydrogenase (class III alcohol dehydrogenase) in the nucleus. J. Histochem. Cytochem. 1992;40:1865–1878. doi: 10.1177/40.12.1453005. [DOI] [PubMed] [Google Scholar]
- 37.Thorndike J, Beck WS. Production of formaldehyde from N5-methyltetrahydrofolate by normal and leukemic leukocytes. Cancer Res. 1977;37:1125–1132. [PubMed] [Google Scholar]
- 38.Borchmann P, Morschhauser F, Parry A, Schnell R, Harousseau JL, Gisselbrecht C, Rudolph C, Wilhelm M, Gunther H, et al. Phase-II study of the new aza-anthracenedione, BBR 2778, in patients with relapsed aggressive non-Hodgkin's lymphomas. Haematologica. 2003;88:888–894. [PubMed] [Google Scholar]
- 39.Borchmann P, Schnell R, Knippertz R, Staak JO, Camboni GM, Bernareggi A, Hubel K, Staib P, Schulz A, et al. Phase I study of BBR 2778, a new aza-anthracenedione, in advanced or refractory non-Hodgkin's lymphoma. Ann. Oncol. 2001;12:661–667. doi: 10.1023/a:1011139016294. [DOI] [PubMed] [Google Scholar]
- 40.Kato S, Burke PJ, Koch TH, Bierbaum VM. Formaldehyde in human cancer cells: detection by preconcentration-chemical ionization mass spectrometry. Anal. Chem. 2001;73:2992–2997. doi: 10.1021/ac001498q. [DOI] [PubMed] [Google Scholar]
- 41.Hah SS, Stivers KM, de Vere White RW, Henderson PT. Kinetics of carboplatin-DNA binding in genomic DNA and bladder cancer cells as determined by accelerator mass spectrometry. Chem. Res. Toxicol. 2006;19:622–626. doi: 10.1021/tx060058c. [DOI] [PubMed] [Google Scholar]
- 42.Rephaeli A, Rabizadeh E, Aviram A, Shaklai M, Ruse M, Nudelman A. Derivatives of butyric acid as potential anti-neoplastic agents. Int. J. Cancer. 1991;49:66–72. doi: 10.1002/ijc.2910490113. [DOI] [PubMed] [Google Scholar]
- 43.Swift LP, Rephaeli A, Nudelman A, Phillips DR, Cutts SM. Doxorubicin-DNA adducts induce a non-topoisomerase II-mediated form of cell death. Cancer Res. 2006;66:4863–4871. doi: 10.1158/0008-5472.CAN-05-3410. [DOI] [PubMed] [Google Scholar]