Abstract
Fanconi anemia (FA) is a chromosomal instability disorder in which DNA-damage processing defects are reported for translesion synthesis (TLS), non-homologous end joining (NHEJ) and homologous recombination (HR; both increased and decreased). To reconcile these diverse findings, we compared spontaneous mutagenesis in FA and HR mutants of hamster CHO cells. In the fancg mutant we find a reduced mutation rate accompanied by an increased proportion of deletions within the hprt gene. Moreover, in fancg cells gene amplification at the CAD and dhfr loci is elevated, another manifestation of inappropriate processing of damage during DNA replication. In contrast, the rad51d HR mutant has a greatly elevated rate of hprt mutations, >85% of which are deletions. Our analysis supports the concept that HR faithfully restores broken replication forks, whereas the FA pathway acts more globally to ensure chromosome stability by promoting efficient end joining of replication-derived breaks, as well as TLS and HR.
INTRODUCTION
Fanconi anemia (FA) is a genetic disease characterized by diverse congenital abnormalities, early predisposition to cancer and progressive bone marrow failure due to defective hematopoiesis (1,2). Patients have mutations in one of at least 12 genes: FANCA, B, C, D1 (also known as BRCA2), D2, E, F, G, I, J (BRIP1/BACH1), L, M (Hef), and N (PALB2). Many of the FANC proteins (FANCA/B/C/E/F/G/L/M/FAAP24/FAAP100) form a nuclear ‘core complex’ (3–7), the integrity of which is essential for the monoubiquitination of FANCD2 in response to DNA damage, including that from mitomycin C crosslinking or oxidative lesions from ionizing radiation (IR). During the cell cycle, monoubiquitinated FANCD2 appears during S phase and co-localizes at sites of putative double-strand breaks (DSBs) with nuclear foci of BRCA1 and Rad51(8), two key proteins in the DSB repair pathway of homologous recombination repair (HRR). HRR uses the sister chromatid, when it is available, as a template for error-free repair of DSBs caused by insults such as ionizing radiation, as well as for restarting broken replication forks during DNA synthesis (9,10). These processes are facilitated by the Rad51 recombinase, which requires mediator proteins including BRCA2 and five Rad51 paralogs (XRCC2, XRCC3, RAD51B, RAD51C, RAD51D) (11). The identification of FANCD1 as BRCA2 (12), and the physical associations between other FANC and HRR proteins such as XRCC3 (13), also suggest a role for the FANC ‘pathway’ in preventing or repairing broken replication forks, and highlight a potential link between the FA proteins and the better defined HRR pathway.
Cells from FA patients typically show increased spontaneous chromatid breaks and gaps (14), and consistently show high sensitivity for cell killing and chromosomal aberrations in response to DNA crosslinking agents (2,15). Treatment of FA cells with low doses of mitomycin C produces excessive chromosomal interchanges due to misrepair of chromatid breaks (16,17) (arising in S or G2 phases) by an end-joining repair pathway, such as DNA-PK-dependent (18,19) or PARP1-dependent (20,21) non-homologous end joining (NHEJ). Such cellular phenotypes are reminiscent of cells defective in HRR, including the well-studied rodent cell lines deficient in the Rad51 paralogs (11,22,23), which show high levels of spontaneous chromosomal aberrations and high sensitivity to crosslinking agents, suggesting an overlapping role for the FANC and HRR proteins in maintaining genome integrity. HRR capacity in FA cells, as indicated by synthetic reporter genes, is reported to be decreased (24–26), increased (27,28) or unaltered (29). These approaches have not helped assess the functional overlap between the FA and HRR pathways.
The role of the FA pathway in DSB repair through DNA end joining mechanisms also has not been established. FA cells generally do not exhibit phenotypes associated with gross NHEJ deficiency, such as high IR sensitivity. It is not understood why various NHEJ assays in FA cells provide conflicting results. For example, studies based on chromosomally integrated reporter constructs containing a I–Sce-I restriction site and PCR analysis found no reduction in NHEJ activity in FA cells from three complementation groups (A, D2 and G) compared to gene-complemented control cells (26). However, an intact FA pathway was required for the end-joining repair of DSBs in plasmid-based assays (both in vitro and in vivo) and for survival of cells after electroporation with restriction enzymes was reported (30–32).
Although chromosomal rearrangements such as those associated with FA and HRR deficiencies are known to play a role in carcinogenesis, single-gene mutation and amplification, are not well characterized in FA and HHR mutant cells. Using an isogenic CHO rad51d knockout mutant, we recently showed that HRR deficiency causes a substantial increase (∼12-fold) in the rate of spontaneous mutagenesis in the X-linked hprt (hypoxanthine phosphoribosyltransferase) gene, and in the rate of amplification at two loci (dhfr, ∼10-fold; CAD, ∼4-fold) (23). This mutant phenotype suggests a major role for HRR in rescuing broken DNA replication forks (23). Mutagenesis studies in human FA cells have given seemingly conflicting results (33). FA lymphoblasts had a ‘reduced’ rate of mutagenesis in the hprt gene in response to treatments with monofunctional and bifunctional psoralens, whereas the spontaneous mutant frequencies were markedly ‘increased’ in FA patients at two autosomal loci: glycophorin A (GPA) in erythrocytes (34,35) and PIG-A in lymphoblasts (36). In chicken DT40 cells a requirement for a FANC protein (FANCC) to promote translesion synthesis (TLS) during crosslink repair was reported (37). Thus, the precise role of the FA proteins in mutagenesis remains unclear, and may indeed involve multiple cellular mechanisms of maintaining genomic integrity.
In this study, we use a model mutagenesis system of isogenic CHO fancg (38) and rad51d (23) knockout mutants to understand how the FA pathway influences spontaneous mutagenesis and to distinguish the roles of the FA and HRR pathways in mutation control. CHO cells have been widely used to perform highly quantitative mutagenesis studies at the hprt locus (39) and, using the dhfr locus to analyze gene amplification, to assay a specific type of carcinogenic mutagenesis (40). We find both a ‘reduced’ rate of occurrence of viable hprt mutants and ‘increased’ rates of gene amplification in fancg cells. In addition, we compare the spectra of hprt mutations in the fancg and rad51d mutant lines with those of their gene-complemented control cells. These comparisons of mutation rate and spectrum in this model genetic system reveal fundamental differences between the contributions of the FA and HRR pathways in preventing mutagenesis. Our findings suggest that the FA pathway may deal with spontaneous DNA damage by promoting efficient DNA end joining as well as TLS and HRR.
MATERIALS AND METHODS
Cell culture
Cells lines used were the CHO parental AA8 cells (41), the fancg knockout line (KO40), the hamster Fancg-complemented KO40 cells (40BP6) (38), the rad51d knockout cell line (51D1) and the hamster Rad51d-complemented 51D1 cells (51D1.3) (23). Cells were grown in monolayer or suspension culture in αMEM supplemented with 10% fetal bovine serum and antibiotics (41).
Mutation and gene amplification rates
Mutation rate was determined by fluctuation analysis (42). For hprt mutants, replica cultures were seeded with 500 cells and grown in suspension to 1–2 × 106 cells/replica, plated and incubated under 6S-Gua selection (41). Hprt, dhfr and CAD mutation rates were calculated using the Poisson P0 term (42), the maximum likelihood method (43) and the method of the mean (44). To recover cells having amplified dhfr or CAD genes, selection was done in 300 nM methotrexate or in 360 µM N-(phosphonacetyl)-L-aspartate (PALA) and 1 µM dipyridimole, respectively. Verification of cad gene amplification was done using real-time, quantitative PCR analysis. Equal numbers of cells from 10 PALA resistant colonies, picked in fluctuation tests, were pooled and genomic DNA isolated. The comparative threshold cycle (CT) method was used to quantify relative gene copy number between the CAD loci in DNA of PALA-resistant cell pools and in DNA isolated from stock populations. CT values, defined as the cycle number at which fluorescence of the reporter dye becomes higher than the background level, were determined for the target (CAD) and an internal reference (APE1) in each sample. The relative gene copy number of the CAD locus of the PALA resistant (PALA-R) clones versus the stock cells was calculated as 2−ΔΔCT, where ΔΔCT = ΔCTPALA-R -ΔCTStock and each ΔCT = CTCAD − CTAPE1. PCR reactions for both primer sets and all DNA samples were performed in triplicate with the DyNAmo™ SYBR® Green qPCR enzyme kit (Finnzymes). PCR and fluorescence detection was performed by the DNA Engine Opticon (MJ Research). Fluorescence and sample comparison was done with Opticon MONITOR analysis software.
Hprt mutation spectrum analysis
After nine days incubation, independent 6S-Gua-resistant clones were isolated for hprt mutation analysis in a three-step process: (i) Gene disrupting mutations of hprt were determined by RT–PCR of the hprt gene transcript and sequencing. After outgrowth of the 6S-Gua resistant clones, RNA was isolated using the RNeasy Mini Kit (Qiagen, Inc. Valencia, CA, USA), and cDNA was made by RT–PCR using SuperScript III First-Strand Synthesis System for RT–PCR (Invitrogen Corp. Carlsbad, CA, USA), and subsequent amplification using the forward primer 5′ TTCCTCCTCACACCGCTCTT, located 47 bp upstream of the hprt start codon (exon 1), and reverse primer 5′ TGCAGATTCAACTTGAACTCTC, located 3 bp downstream of the hprt termination codon (exon 9). PCR-amplified cDNAs were sent for sequencing (Elim Biopharmaceuticals, Inc. Hayward, CA, USA), using the forward and reverse primers noted above. (ii) Genomic DNA was isolated (QiaAmp DNA Blood Mini Kit, Qiagen, Inc.) from all clones without RT–PCR products and tested for the presence of exons 1 and 9 (RT–PCR primer sites) by PCR amplification with the primers: exon 1 forward: 5′ CCTCACCGCTTTCTCGTGCC (3 bp from 5′ end of exon); exon 1 reverse 5′ CACGACGCTGGGGCTGCGGG (last bp of 3′ end of exon 1); exon 9 forward 5′ GTGAAACTGGGAAAGCCAAA (17 bp from 5′ end of exon 9); exon 9 reverse 5′ TGAAAGAATCCAAGTGGGAAA (56 bp from 3′ end of exon 9. (iii) Genomic DNA was isolated for all clones missing exons from the sequence of the RT–PCR product, and tested for the presence of the exons by PCR. Primer pairs used to verify of the presence of the following exons were: Exons 2 and 3, forward: 5′ TGATGAACCAGGCTATGACC, located in exon2, and reverse: 5′ AATCCAGCAGGTCAGCAAAG located in exon 3; Exon 4, forward: 5′ TGATCAGTCAACAGGGGACA, located at the 5′ end of exon 4, and reverse: 5′ TTGAGAGATCATCCCCACCA, located at the 3′end of exon 4; Exons 6, 7 and 8, forward: 5′ CAATGCAAACTCTGCTTTCC, located at the 5′ end of exon 6, an additional confirmation forward primer 5′ CTGGTGAAAAGGACCTCTCG, located at the 5′ end of exon7, and reverse: 5′ TCATTATAG T CAAGGGCATATCCA, located at the 3′ end of exon 8. All primers were synthesized by (Qiagen Inc. Valencia, CA, USA).
RESULTS
Reduced occurrence of spontaneous hprt mutants in fancg cells
Given the finding of abnormally low frequencies of viable hprt mutants in human FA lymphoblasts treated with psoralens (45,46), we wished to determine whether the spontaneous mutation rate (the calculated probability that a mutation arises in a cell's division cycle) is also altered in fancg CHO cells and the relevance of any observed changes to chromosome instability. Although spontaneous frequencies (frequency = fraction of cell population that is mutant) of viable hprt mutants were reported to be either normal or elevated in FA cells (34,46,47) (perhaps reflecting variation in culture history), mutation rates have not been reported. For rate measurements, small, hprt-mutant-free replicate cultures were expanded to ∼106 cells, counted and then selected in 6S-Gua medium, which is toxic to cells having functional hprt. Cells with mutations in the hprt locus formed visible, countable colonies, from which mutation rates were calculated by classical Luria–Delbrück fluctuation analysis.
The mutation rate for viable mutants was reduced by >67% in fancg (KO40) cells compared to both the parental AA8 and the Fancg-complemented cells (40BP6), based on the average of the three statistical methods of calculation (Table 1). This reduction is statistically significant [P < 0.05 for mutation rates calculated by each method (42–44)]. Thus, the reduction in mutation rate in fancg cells suggests that most of the mutational events that would lead to viable hprt mutants in wild-type cells are lethal in fancg cells due to conversion to large deletions or rearrangements. The reduced recovery of fancg cells is not explained by reduced plating efficiency since the plating efficiency of KO40 is 84% versus 90% for AA8 and 40BP6 (38). Although the high concentration of 6S-Gua used for selection of hprt mutants (2 µg/ml) far exceeds the levels at which cell survival assays are done, we tested the possibility that increased sensitivity of KO40 cells to 6S-Gua (38) might affect the outcome of the mutation rate analyses by measuring mutant frequencies at both 2 µg/ml (the standard concentration) and 0.4 µg/ml 6S-Gua (the equitoxic dose relative to AA8 cells at 2 µg/ml). There was little difference in the frequency between the two doses (2.2 × 10−5 versus 2.7 × 10−5, respectively), which implies hprt mutant recovery is unrelated to Fancg status.
Table 1.
Rate of occurrence of viable hprt mutants in fancg (KO40) cells versus parental (AA8) and Fancg-complemented cells (40BP6)
| Cell line | Mutations per cell per generation (units × 10−7) calculated by method of: | ||
|---|---|---|---|
| P0 | Maximum likelihood | Mean | |
| AA8 (6)a | 1.0 ± 0.1b | 1.5 ± 0.3 | 7 ± 1 |
| KO40 (5) | <0.4 ± 0.1c | <0.5 ± 0.1 | <2 ± 0.3 |
| 40BP6 (5) | 0.8 ± 0.1 | 1.3 ± 0.2 | 4 ± 1 |
aThe number in parenthesis is the number of times the experiment was performed; each experiment had 12 replicate dishes. bSEM. cSince no 6S-Gua-resistant colonies were recovered in two of the five KO40 experiments (indicated by ‘<’), these are conservative estimates of the rates. These values are significantly different (P < 0.05) from the AA8 value using a t-test.
Thus, we infer that although the observed rate of occurrence of viable mutants is reduced in fancg cells, the true rate of gene disruption mutagenesis may be the same, or even increased, as in the HRR-deficient CHO cells, but with a high proportion of the events falling into a lethal class of mutation and remaining undetected.
Increased proportion of deletions in spontaneous hprt mutants of fancg cells
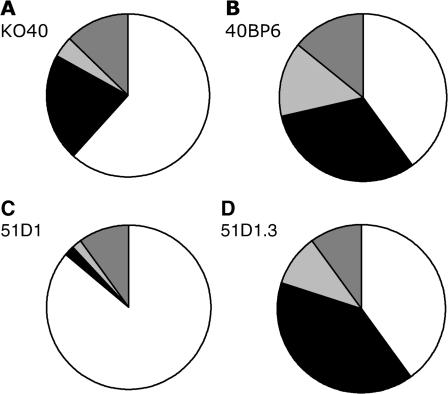
Since the spontaneous rate of forming viable hprt mutants was decreased in the fancg CHO cells, we wished to determine whether the spectrum of the recovered mutants could provide insight into the particular classes of mutation that were being converted into lethal events. Mutations were assigned to four classes (base substitution, deletion, insertion, splicing) based on analysis of mRNA and, in many cases, genomic DNA. ‘Splicing’ mutations are those with alterations of mRNA whose causation was not identified by analysis of genomic DNA. For example, in clones showing loss of one or more exons in the RT–PCR product sequence, genomic-DNA PCR amplification of the missing exon(s) was used to distinguish deletions of the exons from other splicing errors, referred to as exon skips. Another form of spicing error, leading to exon duplications in the mRNA, was also detected among some clones. It was reported that splicing alterations, which make up 12% of hprt mutations in a human database, are primarily base substitution mutations (48). However, since we are unable to classify the types of mutations leading to most exon skips and duplications, we consider them as a separate class of events referred to generically as ‘splice mutants’ in our analysis. Consistent with previous studies in FA lymphoblasts compared with non-FA cells in response to psoralens (45,46), we found an almost statistically significant (P = 0.052) increase in the proportion of hprt mutants that were deletions in the fancg cells (KO40; 21% base substitutions, 62% deletions, 2% insertions and 15% splice mutants; n = 47, Figure 1A) when compared to the Fancg-complemented control cells (40BP6; 31% base substitutions, 40% deletions, 11% insertions and 17% splice mutants; n = 35, Figure1B). This trend towards deletions was significant when compared to both complemented cell lines combined (P = 0.02). Deletions in both KO40 and 40BP6 cell lines ranged in size from a few base pairs up to potentially the entire locus (as implied by no amplification of exon 1 and exon 9). Insertion mutations ranged in size from 1 to 4 bp, of which all were duplications of local sequence and did not occur in nucleotide repeats (Supplementary Table 1).
Figure 1.
Proportion of base substitution (black), deletion (white), insertion (light gray), and splicing mutants (exon skips, duplications, and 17-bp deletions; dark gray) hprt mutations among clones of fancg KO40 cells (A), Fancg gene-corrected BP6 cells (B), rad51d 51D1 (C), and Rad51d gene-corrected 51D1.3 cells (D).
Excessive spontaneous hprt deletions in HRR-defective rad51d cells
Although our previous study discovered a high rate of hprt mutagenesis in CHO rad51d cells (∼12–fold elevated parental and gene-complemented cells) (23), the spectrum was not determined. We find that the rad51d-associated HRR deficiency leads to a large increase in deletions (86%, n = 50, Figure1C) and vast reduction in the proportion of base substitutions and insertions (2% each), with the remaining 10% of mutations being splice mutants, all exon skips. This proportion of deletions is significantly (P < 0.001) different from the proportion in the Rad51d-complemented control cells (51D1.3), which have a mutant spectrum with 36% deletions, 40% base substitutions, 7% insertions and 25% splice mutants (n = 40, Figure 1D), similar to that of the 40BP6 cells. As seen in the fancg cells, rad51d hprt deletions ranged in size from 1 bp to deletion of the entire locus. The description of each mutant is provided in Supplementary Table 1. For comparison, the hprt mutation spectrum in brca2 V79 hamster cells (defective in the one known gene common to the FA and HRR pathways) also showed a spectrum shift toward more deletions (49) while showing only a moderate increase in spontaneous mutation rate (∼4–fold) (50).
Elevated gene amplification rates in fancg cells
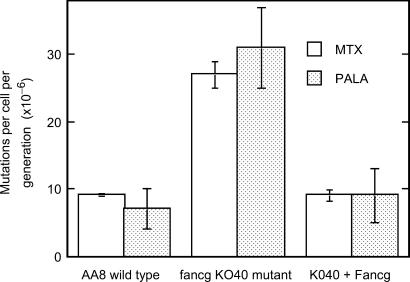
Our inference that the fancg mutation results in aberrantly repaired DSBs during DNA replication suggested that the process of gene amplification, which is typically associated with conditions that cause inappropriate rejoining of DSBs (51,52), might be elevated in fancg cells, as seen previously in rad51d cells (23). To test this prediction, we measured gene amplification using the extensively studied dhfr locus (where an increased gene-copy number confers methotrexate resistance) and the CAD (carbamyl-P-synthetase, aspartate transcarbamylase, dihydro-orotase) locus where amplification confers PALA resistance. We find that rates of spontaneous gene amplification are substantially increased (3- to 4-fold) at both loci in fancg cells (Figure 2), a change that is statistically significant for both loci in comparison to wild-type AA8 cells (P < 0.01 and P < 0.05 for the dhfr and CAD loci, respectively) and Fancg-corrected 40BP6 cells (P < 0.001 and P < 0.05 for the dhfr and CAD loci, respectively). It has been shown previously that all PALA-resistant AA8 clones have detectable amplification at the CAD locus (51). We also verified amplification by measuring its ‘extent’ at the CAD locus using quantitative PCR of genomic DNA from pools of 10 independent PALA-resistant clones. As expected, there was an increase in the relative number of CAD gene copies with respect to an internal-standard reference gene (APE1 locus) when compared to the DNA from cells in the respective stock cultures. In wild-type cells, the fold increase normalized to APE1 was 2.3 ± 0.8 (SEM), while in fancg cells we found a 2.8-fold increase. Taken together, we infer that the extent of gene amplification was the same in those wild-type and fancg cells that were PALA resistant. Overall, the increased rate of mutagenesis in the form of gene amplification in fancg is consistent with increased aberrant repair of spontaneous DSBs, most likely during S phase. Interestingly, these results are similar to those seen in the rad51d CHO cells, which show even greater amplification at both reporter loci (23).
Figure 2.
Rate of amplification mutations at dhfr (methotrexate resistance) and CAD (PALA resistance) loci in fancg and control cells. Each methotrexate experiment was done three or four times with 20 replica cultures, and each PALA experiment was done twice with 20 replicates.
DISCUSSION
Implications of changes in spontaneous hprt mutation rate and spectrum in mutant lines
In the rad51d cells, the greatly increased yield of hprt mutants suggests a prominent role for this pathway in preventing frequent mutagenic events from occurring during normal DNA replication in the face of spontaneous (oxidative) lesions (23). Indeed, HRR was shown to act on collapsed replication forks caused by endogenous DNA single-strand breaks (53). In our study we found that the spectrum of mutations from rad51d cells reveals that the mutagenic events prevented by intact HRR are deletions, likely caused by efficient DNA end-joining mechanisms that repair DSBs that persist when broken replication forks are not restarted.
In the fancg CHO cells, we see a major reduction in the yield of four classes of spontaneous hprt mutants compared with wild-type. The reduced yield of both base substitution and deletion/insertion events in the fancg cells points to a role for Fancg and the FA proteins in promoting TLS at replication-blocking lesions (the source of base substitution mutations), as well as in coordinating HRR to restart broken replication forks and NHEJ to restore broken-fork-associated DSBs. The latter two processes are necessary events for assuring either conservative repair or recoverable hprt deletion mutants, respectively. There was a tendency toward more deletions among spontaneous mutations of the CHO fancg cells. It is interesting to note that the spectrum of spontaneous mutations in hprt in FA lymphoblasts and T-lymphocytes is also shifted toward more frequent deletions versus base substitutions (45–47). In the fancg cells a high proportion of replication fork breaks, which in normal or rad51d cells often result in recoverable hprt mutants, must result in lethality to account for the reduced yield of viable mutants. The most likely source of this reduction is the failure to rejoin the breaks due to both impaired HRR and end joining, or by erroneous rejoining causing multigenic, lethal deletion or translocation (as depicted by the model in Figure 3 and discussed subsequently).
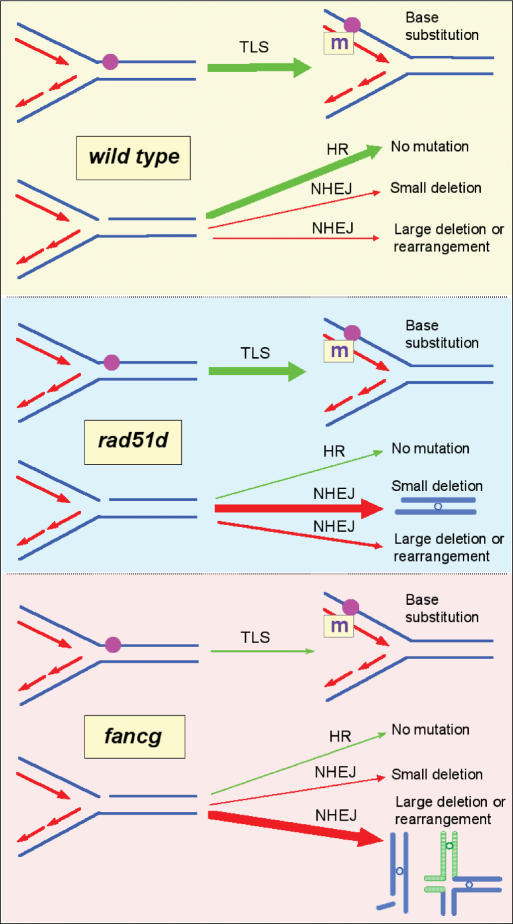
Figure 3.
Model of spontaneous hprt mutational outcome in wild-type, rad51d and fancg mutant cells during replication. Each panel represents one of the three genotypes: wild type, rad51d and fancg. On the right in each panel are shown the potential mutational outcomes due to a replication fork encountering a polymerase-blocking lesion (upper fork) or a single-strand break or gap such as a repair intermediate (lower fork). Arrows represent the options for resolving each situation, leading to the viable or lethal mutagenic events. Upper panel: In wild-type cells, there are two major outcomes (large green arrows): translesion synthesis (TLS) bypasses fork blocking lesions (sometimes leading to base substitution, ‘m’), and homologous recombination (HR) restarts broken replication forks and prevents mutations. Middle panel: The HR-defective rad51d cells, which are presumed to have normal TLS, inefficiently restart broken forks in an error-free manner, but retain efficient NHEJ activity, as evidenced by a high frequency and proportion of small-sized (non-lethal) deletions (large red arrow). NHEJ acts on replication-associated DSBs that arise when replication forks break and free-ends persist as replication continues. Lower panel: In the fancg cells, the reduced absolute number of base substitutions and small deletions, along with the increased rate of gene amplification, suggest diminished TLS and HR, as well as the loss of NHEJ activity that produces deletions that are recoverable in the hprt mutagenesis assay (as in rad51d cells). The predominant events in fancg cells appear to be inviable deletions or rearrangements (large red arrow).
Relevance of the FA pathway of gene amplification
Gene amplification is a type of mutation often associated with tumors and is elevated in cultured tumor cells versus non-tumorigenic cells (54,55). Many studies have shown the importance of DSBs in gene amplification although the mechanisms remain incompletely understood (55–58). Treatments with IR or H2O2, which cause lesions that include DSBs, enhance gene amplification (52). CHO cells defective in NHEJ due to a DNA-PKcs mutation have greatly elevated (20- to 150-fold) amplification (51), while the rad51d cells show 4- to 10-fold increases (23).
Our findings of 3- to 4-fold increased gene amplification in the fancg cells support the idea that DSBs arising during DNA replication are aberrantly repaired. The breakage–fusion-bridge mechanism of amplification is a popular model (59), in which the amplification process may be initiated by a DSB that arises during replication and persists until being removed by end joining between sister chromatids. During the next anaphase, an asymmetrical mechanical break in the dicentric ‘bridge’ chromosome can then result in duplication of the target gene in one daughter cell. This process can be repeated in subsequent mitoses.
Model of spontaneous mutational outcomes in wild-type and FA and HRR mutant cells
It is noteworthy that fancg CHO cells are hypersensitive to killing by a variety of mutagens besides crosslinking agents, i.e. γ-rays, methyl methanesulfonate, methylnitrosourea, ethylnitrosourea and 6S-Gua (38). This finding implies that loss of Fancg and, consequently, Fancd2 monoubiquitination, causes a defect in dealing with a much broader class of DNA damage than simply inter-strand crosslinking, e.g. oxidative and alkylation lesions commonly caused by normal cellular metabolism. Induced mutagenesis data also support a role for FA proteins in promoting replication past a variety of DNA lesions, as the fancg cells have decreased recovery of hprt mutants after exposure to various DNA damaging agents, including UV-C, γ-rays and ethylnitrosourea, relative to the parental control cells (33). The unusually high sensitivity of fancg cells (38) (and FA cells generally) to crosslinking agents may be explained by the unique, dual requirement for TLS and HRR to bring about repair of broken replication forks resulting from cross-link processing.
In summary, our gene amplification and hprt mutation studies emphasize both similarities and marked differences in phenotype between the FA and HRR mutants in an isogenic mammalian system. Our data combined with that in the literature support a model in which FANCD2 monoubiquitination acts upstream of TLS, HRR and NHEJ by supporting all three processes during S phase in response to endogenous and exogenous DNA damage (Figure 3). Although endogenous inter-strand crosslinks may contribute to the FA phenotype, our induced mutagenesis and survival studies argue that the FANC pathway is more globally important for diverse lesions (33). We conclude that pathway coordination by FA proteins supports a ‘Fire Captain’ model (33), in which they act to limit the severity of mutagenesis by promoting efficient TLS, HRR and NHEJ.
Supplementary Material
ACKOWLEDGEMENTS
The authors would like to thank Vicki Kopf, Lynn Carr and Tricia Allen for technical assistance and Maureen Hoatlin for extensive comments on the manuscript. They also thank the Drug Synthesis and Chemistry Branch, Division of Cancer Treatment, National Cancer Institute, for providing PALA. This work was performed under the auspices of the US Department of Energy by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48. The DOE Low-Dose Program and NCI/NIH grants CA89405 and CA112566 funded this work. Funding to pay the Open Access publication charges for this article was provided by NIH grant CA112566.
Conflict of interest statement. None declared.
REFERENCES
- 1.Alter BP. Fanconi's anemia and malignancies. Am. J. Hematol. 1996;53:99–110. doi: 10.1002/(SICI)1096-8652(199610)53:2<99::AID-AJH7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 2.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 3.Thompson LH. Unraveling the Fanconi anemia–DNA repair connection. Nat. Genet. 2005;37:921–922. doi: 10.1038/ng0905-921. [DOI] [PubMed] [Google Scholar]
- 4.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi Anemia (Cross)linked to DNA Repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 6.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, Yan Z, Xue Y, Oostra AB, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 9.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 10.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 13.Hussain S, Wilson JB, Blom E, Thompson LH, Sung P, Gordon SM, Kupfer GM, Joenje H, Mathew CG, et al. Tetratricopeptide-motif-mediated interaction of FANCG with recombination proteins XRCC3 and BRCA2. DNA Repair. 2006;5:629–640. doi: 10.1016/j.dnarep.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 14.German J. The chromosome-breakage syndromes: rare disorders that provide models for studying somatic mutation. In: Spatz L, Bloom AD, Paul NW, editors. Detection of Cancer Predisposition: laboratory Approaches, March of Dimes Birth Defects Foundation. NY: White Plains; 1990. pp. 85–111. [PubMed] [Google Scholar]
- 15.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 16.Newell AE, Akkari YM, Torimaru Y, Rosenthal A, Reifsteck CA, Cox B, Grompe M, Olson SB. Interstrand crosslink-induced radials form between non-homologous chromosomes, but are absent in sex chromosomes. DNA Repair. 2004;3:535–542. doi: 10.1016/j.dnarep.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Thompson LH, Hinz JM, Yamada NA, Jones NJ. How Fanconi anemia proteins promote the four Rs: Replication, recombination, repair, and recovery. Environ. Mol. Mutagen. 2005;45:128–142. doi: 10.1002/em.20109. [DOI] [PubMed] [Google Scholar]
- 18.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair. 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 21.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 22.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat. Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 23.Hinz JM, Tebbs RS, Wilson PF, Nham PB, Salazar EP, Nagasawa H, Urbin SS, Bedford JS, Thompson LH. Repression of mutagenesis by Rad51D-mediated homologous recombination. Nucleic Acids Res. 2006;34:1358–1368. doi: 10.1093/nar/gkl020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto K, Ishiai M, Matsushita N, Arakawa H, Lamerdin JE, Buerstedde JM, Tanimoto M, Harada M, Thompson LH, et al. Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells. Mol. Cell. Biol. 2003;23:5421–5430. doi: 10.1128/MCB.23.15.5421-5430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, Namikoshi K, Kimura M, Matsushita N, Arakawa H, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol. Cell. Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl Acad. Sci. USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thyagarajan B, Campbell C. Elevated homologous recombination activity in fanconi anemia fibroblasts. J. Biol. Chem. 1997;272:23328–23333. doi: 10.1074/jbc.272.37.23328. [DOI] [PubMed] [Google Scholar]
- 28.Donahue SL, Lundberg R, Saplis R, Campbell C. Deficient regulation of DNA double-strand break repair in Fanconi anemia fibroblasts. J. Biol. Chem. 2003;278:29487–29495. doi: 10.1074/jbc.M213251200. [DOI] [PubMed] [Google Scholar]
- 29.Ohashi A, Zdzienicka MZ, Chen J, Couch FJ. FANCD2 functions independently of BRCA2 and RAD51 associated homologous recombination in response to DNA damage. J. Biol. Chem. 2005;280:14877–14883. doi: 10.1074/jbc.M414669200. [DOI] [PubMed] [Google Scholar]
- 30.Lundberg R, Mavinakere M, Campbell C. Deficient DNA end joining activity in extracts from fanconi anemia fibroblasts. J. Biol. Chem. 2001;276:9543–9549. doi: 10.1074/jbc.M008634200. [DOI] [PubMed] [Google Scholar]
- 31.Donahue SL, Campbell C. A DNA double strand break repair defect in Fanconi anemia fibroblasts. J. Biol. Chem. 2002;277:46243–46247. doi: 10.1074/jbc.M207937200. [DOI] [PubMed] [Google Scholar]
- 32.Donahue SL, Campbell C. A Rad50-dependent pathway of DNA repair is deficient in Fanconi anemia fibroblasts. Nucleic Acids Res. 2004;32:3248–3257. doi: 10.1093/nar/gkh649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinz JM, Nham PB, Salazar EP, Wilson PF, Thompson LH. The Fanconi anemia pathway limits the severity of mutagenesis. DNA Repair. 2006;34:1358–1368. doi: 10.1016/j.dnarep.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 34.Sala-Trepat M, Boyse J, Richard P, Papadopoulo D, Moustacchi E. Frequencies of HPRT- lymphocytes and glycophorin A variants erythrocytes in Fanconi anemia patients, their parents and control donors. Mutat. Res. 1993;289:115–126. doi: 10.1016/0027-5107(93)90137-5. [DOI] [PubMed] [Google Scholar]
- 35.Evdokimova VN, McLoughlin RK, Wenger SL, Grant SG. Use of the glycophorin A somatic mutation assay for rapid, unambiguous identification of Fanconi anemia homozygotes regardless of GPA genotype. Am. J. Med. Genet. A. 2005;135:59–65. doi: 10.1002/ajmg.a.30687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Araten DJ, Golde DW, Zhang RH, Thaler HT, Gargiulo L, Notaro R, Luzzatto L. A quantitative measurement of the human somatic mutation rate. Cancer Res. 2005;65:8111–8117. doi: 10.1158/0008-5472.CAN-04-1198. [DOI] [PubMed] [Google Scholar]
- 37.Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Tebbs RS, Hinz JM, Yamada NA, Wilson JB, Salazar EP, Thomas CB, Jones IM, Jones NJ, Thompson LH. New insights into the Fanconi anemia pathway from an isogenic FancG hamster CHO mutant. DNA Repair. 2005;4:11–22. doi: 10.1016/j.dnarep.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 39.O’Neill JP, Hsie AW. Chemical mutagenesis of mammalian cells can be quantified. Nature. 1977;269:815–817. doi: 10.1038/269815a0. [DOI] [PubMed] [Google Scholar]
- 40.Milbrandt JD, Heintz NH, White WC, Rothman SM, Hamlin JL. Methotrexate-resistant Chinese hamster ovary cells have amplified a 135-kilobase-pair region that includes the dihydrofolate reductase gene. Proc. Natl Acad. Sci. USA. 1981;78:6043–6047. doi: 10.1073/pnas.78.10.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson LH, Fong S, Brookman K. Validation of conditions for efficient detection of HPRT and APRT mutations in suspension-cultured Chinese hamster cells. Mutat. Res. 1980;74:21–36. doi: 10.1016/0165-1161(80)90188-0. [DOI] [PubMed] [Google Scholar]
- 42.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng Q. Update on estimation of mutation rates using data from fluctuation experiments. Genetics. 2005;171:861–864. doi: 10.1534/genetics.104.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capizzi RL, Jameson JW. A table for the estimation of the spontaneous mutation rate of cells in culture. Mutat. Res. 1973;17:147–148. doi: 10.1016/0027-5107(73)90265-0. [DOI] [PubMed] [Google Scholar]
- 45.Papadopoulo D, Porfirio B, Moustacchi E. Mutagenic response of Fanconi's anemia cells from a defined complementation group after treatment with photoactivated bifunctional psoralens. Cancer Res. 1990;50:3289–3294. [PubMed] [Google Scholar]
- 46.Papadopoulo D, Guillouf C, Mohrenweiser H, Moustacchi E. Hypomutability in Fanconi anemia cells is associated with increased deletion frequency at the HPRT locus. Proc. Natl Acad. Sci. USA. 1990;87:8383–8387. doi: 10.1073/pnas.87.21.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laquerbe A, Sala-Trepat M, Vives C, Escarceller M, Papadopoulo D. Molecular spectra of HPRT deletion mutations in circulating T-lymphocytes in Fanconi anemia patients. Mutat. Res. 1999;431:341–350. doi: 10.1016/s0027-5107(99)00177-3. [DOI] [PubMed] [Google Scholar]
- 48.O'Neill JP, Rogan PK, Cariello N, Nicklas JA. Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum. Mutat. Res. 1998;411:179–214. doi: 10.1016/s1383-5742(98)00013-1. [DOI] [PubMed] [Google Scholar]
- 49.Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, Swaminathan S, van Buul PP, Errami A, et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol. 2002;22:669–679. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraakman-Van Der Zwet M, Wiegant WW, Zdzienicka MZ. Brca2 (XRCC11) deficiency results in enhanced mutagenesis. Mutagenesis. 2003;18:521–525. doi: 10.1093/mutage/geg032. [DOI] [PubMed] [Google Scholar]
- 51.Mondello C, Rebuzzini P, Dolzan M, Edmonson S, Taccioli GE, Giulotto E. Increased gene amplification in immortal rodent cells deficient for the DNA-dependent protein kinase catalytic subunit. Cancer Res. 2001;61:4520–4525. [PubMed] [Google Scholar]
- 52.Mondello C, Guasconi V, Giulotto E, Nuzzo F. Gamma-ray and hydrogen peroxide induction of gene amplification in hamster cells deficient in DNA double strand break repair. DNA Repair. 2002;1:483–493. doi: 10.1016/s1568-7864(02)00035-6. [DOI] [PubMed] [Google Scholar]
- 53.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tlsty TD, Margolin BH, Lum K. Differences in the rates of gene amplification in nontumorigenic and tumorigenic cell lines as measured by Luria-Delbruck fluctuation analysis. Proc. Natl Acad. Sci. USA. 1989;86:9441–9445. doi: 10.1073/pnas.86.23.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 56.Poupon MF, Smith KA, Chernova OB, Gilbert C, Stark GR. Inefficient growth arrest in response to dNTP starvation stimulates gene amplification through bridge-breakage-fusion cycles. Mol. Biol. Cell. 1996;7:345–354. doi: 10.1091/mbc.7.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paulson TG, Almasan A, Brody LL, Wahl GM. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol. Cell. Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singer MJ, Mesner LD, Friedman CL, Trask BJ, Hamlin JL. Amplification of the human dihydrofolate reductase gene via double minutes is initiated by chromosome breaks. Proc. Natl Acad. Sci. USA. 2000;97:7921–7926. doi: 10.1073/pnas.130194897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp. Cell Res. 2005;302:233–243. doi: 10.1016/j.yexcr.2004.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.