Abstract
LINE-1 elements (L1s) are a family of highly successful retrotransposons comprising ∼17% of the human genome, the majority of which have inserted through an endonuclease-dependent mechanism termed target-primed reverse transcription. Recent in vitro analyses suggest that in the absence of non-homologous end joining proteins, L1 elements may utilize an alternative, endonuclease-independent pathway for insertion. However, it remains unknown whether this pathway operates in vivo or in cell lines where all DNA repair mechanisms are functional. Here, we have analyzed the human genome to demonstrate that this alternative pathway for L1 insertion has been active in recent human evolution and characterized 21 loci where L1 elements have integrated without signs of endonuclease-related activity. The structural features of these loci suggest a role for this process in DNA double-strand break repair. We show that endonuclease-independent L1 insertions are structurally distinguishable from classical L1 insertion loci, and that they are associated with inter-chromosomal translocations and deletions of target genomic DNA.
INTRODUCTION
Long interspersed element-1 (LINE-1 or L1) is a ubiquitous retrotransposon family in the human genome, with ∼520 000 insertions comprising ∼17% of total genomic sequence (1,2). A full-length L1 element is ∼6-kb long and contains two open reading frames (ORF1 and ORF2) (3). While ORF1 encodes an RNA-binding protein with nucleic acid chaperone activity (4), ORF2 encodes for endonuclease (EN) and reverse transcriptase (RT) activities (5,6), and both ORFs are required for L1 retrotransposition (7,8). In addition to insertional mutagenesis (9–11), L1 elements have also been associated with exon shuffling, creation of deletions through unequal homologous recombination and intra-chromosomal and inter-chromosomal translocation of genomic sequence (12–14). As such, the dynamic nature of L1 elements makes them important agents of genomic rearrangement (15,16).
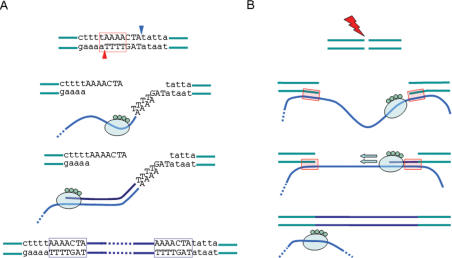
The currently accepted model for genomic integration of L1 elements is termed target-site primed reverse transcription (TPRT) (17,18) (Figure 1). During TPRT, the L1 EN cleaves one strand of the target DNA at a motif approaching the consensus 5′-TTTT/A-3′ (where “/”denotes the cleavage site), producing a free 3′-hydroxyl (5,19). Next, the L1 RNA anneals to the nick site using its 3′ poly (A) tail, and the L1 RT initiates reverse transcription using the L1 RNA as a template. Cleavage of the second DNA strand by the L1 EN usually occurs 7–20 base pairs downstream of the initial nicking site, creating staggered breaks in the target DNA that are later filled in to form direct repeats flanking the newly inserted element (termed target site duplications or TSDs) (20). Integration of the newly synthesized cDNA and completion of second-strand synthesis are the remaining steps in the TPRT model; however, the order in which they occur and their exact mechanism remain unclear (21). Apart from the presence of TSDs, other structural hallmarks of TPRT-mediated L1 insertion include frequent 5′ truncations (or truncation/inversions) and intact 3′ ends with variable-length A-rich tails (20).
Figure 1.
Comparison of TPRT and NCLI L1 insertions. (A) Classical TPRT-mediated L1 insertion in the human genome. First-strand cleavage by the L1 EN (red arrowhead) at the 5′-TTTT/A-3′ consensus (red dotted box) allows L1 mRNA (blue line) to anneal to genomic DNA using its poly(A) tail. RT activity of L1 ORF2 (green oval) synthesizes L1 cDNA (purple line) using L1 mRNA as template and 3′ OH from nicked genomic DNA as primer. Second-strand cleavage (blue arrowhead) occurs 7–20 bp downstream from first-strand cleavage site, creating staggered nicks which are later filled in to form TSDs (blue dotted boxes). Attachment of the L1 cDNA and synthesis of the second strand complete the insertion process. TSD sequences for this diagram are from a 637-bp human L1 element located at chr1:65036188–65036824. (B) Schematic representation of an NCLI event. Following creation of a genomic double-strand break (red thunderbolt), free-floating L1 mRNA (blue line) attaches to newly separated ends using small stretches of complementary bases. Once gap is bridged, it may be filled in by DNA synthesis by either the L1 RT, cellular repair polymerases or both. L1 insertion thus created lacks structural features of TPRT-mediated insertion.
In recent years, increasing evidence from cell culture retrotransposition assays suggests that in addition to TPRT-mediated insertion, a second, less-characterized L1 integration pathway may exist that is independent of L1-encoded endonuclease (18,22). However, with a few isolated exceptions (23–25), the majority of endonuclease-independent (ENi) L1 insertions have been recovered in cell lines lacking one or more components of the cellular non-homologous end joining (NHEJ) mechanism, a principal form of DNA double-strand break (DSB) repair (26). Consequently, whether ENi L1 insertion occurs at detectable frequencies when normal DNA repair pathways are functional has been the subject of continued debate (3, 22, 27–29). Additionally, existing analyses of human genomic L1 elements (20,30), by focusing solely on TPRT-mediated insertions, have left this question unanswered in a systematic fashion.
In this study, we have utilized computational analyses of the draft sequence of the human genome to recover L1 elements that utilized this alternative pathway of integration (which we term non-classical L1 insertion or NCLI). We report 21 loci where L1 elements appear to have inserted without any hallmarks of endonuclease activity. In each case, we verified the ancestral (i.e. no L1 insertion) state of the loci by re-sequencing the orthologous positions in the common chimpanzee and rhesus macaque genomes. Overall, our results suggest that NCLI has been active in recent human evolution, and that it provides an alternative ‘non-selfish’ pathway for L1 integration in the human genome. Interestingly, we find that NCLI loci are clustered in gene-rich regions of the genome, in contrast to the distribution of the more common TPRT-mediated L1 insertions. Based on the unique structural features of NCLI-mediated L1 elements, we suggest that this process may be capable of repairing genomic lesions and that it may confer a slight selective advantage to what may be the otherwise deleterious nature of the L1 family. We conclude that non-LTR retrotransposons may have a previously unrecognized role in maintaining human genomic integrity.
MATERIALS AND METHODS
Computational screening for putative ENi L1 insertions
To identify NCLI loci in the publicly available human genome, we first downloaded the file chromOut.zip from the UCSC Genome Bioinformatics website (http://hgdownload.cse.ucsc.edu/downloads.html#human). This archive contains output files from the RepeatMasker (RM) software package (http://www.repeatmasker.org/) run at the −s (sensitive) setting on individual human chromosomes. For this project, the archived files corresponded to RM output from the May 2004 freeze of the human genome (hg17). Next, using our own script, we extracted all L1 insertions from each chromosome. To find elements missing the segment of the 3′ UTR normally used during TPRT-mediated insertion, we developed a set of computer programs that scanned the comprehensive list of L1 elements to find all elements truncated beyond 20 bases from the 3′ end. We chose the 20 bp truncation limit for two specific reasons. Firstly, from aligning six previously published consensus sequences of relatively young L1 elements, we found the shortest length of the poly(A) tail to be 13 bp. Secondly, we added a 7 bp window to the 13-bp poly(A) tail to account for the possibility of small internal deletions near the 3′ end of the L1 insertions that would mimic the appearance of a 3′-truncated insertion. As RM assigns a size of 6155 bp to full-length L1 elements from subfamilies L1Hs and L1PA2, our initial output files thus contained sets of L1 insertions ending at position 6135 or lower. To verify the effectiveness of this strategy, for each chromosome, we manually inspected sets of 50 loci on either side of this truncation limit. The sets of L1 elements with 3′ truncations <20 bp did not return any loci matching all of these three criteria; absence of TSDs of any length, absence of a poly(A) tail and significant deviation from the consensus L1 EN cleavage site. Thus, these L1 elements most likely integrated into the genome through traditional TPRT-mediated insertion. As such, after visual inspection of the computational output, all loci that we selected for further experimental verification came from the set of insertions with 3′ truncations 20 bp or longer. To further narrow our list to relatively young L1 insertions, we discarded all elements >2% diverged from their respective consensus sequences according to the RM algorithm. We rejected all L1 insertions that had TSDs of any length, even if they bordered a 3′ truncated element. Our RM output parsing software accounted for L1 elements fragmented by small insertions/deletions and for truncated/inverted L1 insertions, both of which commonly occur during the TPRT process and are sometimes annotated by RM as separate insertions. All the computer programs are available from the authors upon request.
Manual inspection of sequence and verification of ancestral (pre-insertion) status
To confirm the ancestral (i.e. no insertion) stage for computationally recovered NCLI loci, we extracted 10 000 bp of flanking sequence on either side of the L1 element. First, we ran each extracted segment (L1 insertion plus flanking sequence) through RM to verify that the potential NCLI candidates were not fragments of 3′ intact L1 elements separated by large blocks of intervening non-L1 sequence. We then used the BLAT software package (http://www.genome.ucsc.edu/cgi-bin/hgBlat) to construct triple alignments of the human, chimpanzee and rhesus macaque genomes at each locus. Next, we manually inspected each alignment to verify that the 5′ and 3′ ends of each putative human NCLI event corresponded to either gaps or extra, non-L1 sequence in the ancestral sequence (the presence of non-L1 sequence indicated a deletion in the ancestral genome whose boundaries exactly matched the human L1 insertion). In addition, to further confirm the endonuclease-independent nature of putative NCLI loci, we analyzed them for divergence from the TTTT/A L1-EN cleavage site consensus, based on an earlier analysis of EN site preferences (22). This left us with a final data set of 21 potential NCLI loci that fit all four of the following criteria: 3′ truncation, absence of TSDs, absence of a poly(A) tail and significant divergence from the L1-EN consensus.
PCR amplification and DNA sequence analysis of NCLI loci
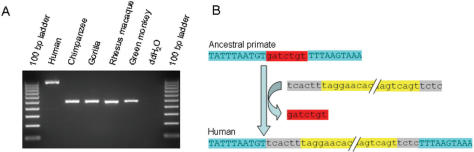
To experimentally confirm that these 21 loci represented truncated L1 insertions rather than deletions of the 3′ UTR, we designed oligonucleotide primers in the non-repetitive sequence flanking the L1 elements and amplified them by PCR on a panel of five primate species (Figure 2), including Homo sapiens (HeLa; cell line ATCC CCL-2), Pan troglodytes (common chimpanzee; cell line AG06939B), Gorilla gorilla (Western lowland gorilla; cell line AG05251), Macaca mulatta (Rhesus macaque; cell line NG07098) and Chlorocebus aethiops (Green monkey; cell line ATCC CCL70). PCR amplification of NCLI loci was performed in 25 µl reactions using 10–50 ng genomic DNA, 200 nM of each oligonucleotide primer, 200 µM dNTPs in 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–HCl (pH 8.4) and 2.5 units Taq DNA polymerase. The conditions for the PCR were an initial denaturation step of 94°C for 4 min, followed by 32 cycles of 1 min of denaturation at 94°C, 1 min of annealing at optimal annealing temperature and 1 min of extension at 72°C, followed by a final extension step at 72°C for 10 min. For loci with large insertions or deletions (>2 kb), we used Ex Taq™ polymerase (TaKaRa) and carried out PCR in 50 µl reactions following the manufacturer's suggested protocol. PCR amplicons were separated on 1% agarose gels, stained with ethidium bromide and visualized using UV fluorescence. Detailed information for each locus including primer sequences, annealing temperature and PCR product sizes is available from the ‘Publications’ section of the Batzer laboratory website (http://batzerlab.lsu.edu).
Figure 2.
Analysis of NCLI elements. (A) Gel chromatograph of PCR products from a phylogenetic analysis of a human genome-specific NCLI locus (NCLI34). DNA template used in each lane is shown at top. (B) Schematic diagram of NCLI locus (NCLI53) showing L1 insertion (yellow box) associated with 7-bp deletion of target DNA (red box). Matching flanking sequence is shown as light blue boxes. Grey boxes indicate small segments of non-L1 ‘filler’ DNA at either end of the L1 insertion.
Repetitive DNA may correspond to sites of genome assembly errors; therefore we re-sequenced all loci from the chimpanzee and rhesus macaque genomes to confirm that the computationally recovered pre-insertion sequence was accurate. Individual PCR products were purified from gels using Wizard® gel purification kits (Promega). Amplicons <1.3 kb were cloned into vectors using TOPO-TA Cloning® kits (Invitrogen) and three colonies were randomly selected and sequenced in both directions using M13 forward and reverse primers to verify that the PCR product matched the computationally recovered sequence. For PCR products larger than 1.3 kb, gel-purified PCR products were sequenced directly using the respective primers to verify that sequence boundaries matched the computational predictions. All sequencing was performed by the chain termination method (31) on an Applied Biosystems ABI3130XL automated DNA sequencer. Analysis of all of the re-sequenced loci showed that the sequences were exact matches to those in the draft genome sequence assemblies.
RESULTS
A whole-genome scan for non-classical L1 insertions
To analyze the human genome sequence for potential NCLI loci, we combined computational and experimental approaches. First, using RM, we computationally extracted young L1 insertions lacking structural hallmarks of TPRT-mediated ‘classical’ retrotransposition (see ‘Materials and Methods’ section). Next, we constructed triple alignments of the human, chimpanzee and rhesus macaque genomes at these loci to reconstruct the ancestral (i.e., pre-L1 insertion) state and manually inspected the structure of each locus to detect signs of non-TPRT mediated insertion. Finally, we used PCR and re-sequencing to experimentally verify the sequence architecture for both the post-insertion and pre-insertion states of the loci (Figure 2). Specifically, the loci included in this analysis after experimental confirmation of the computational output possessed all four of the following characteristics: 3′ truncation beyond 25 bp (i.e., 5 bp more than the minimum truncation level set during computational screening) relative to the L1HS_3′end consensus from the RepeatMaskerLib.embl repetitive element library, downloadable from: http://www.girinst.org/repbase/index.html) (32), absence of TSDs of any length, absence of a poly(A) tail and significant deviation from the consensus L1 EN cleavage site. Structural features of the NCLI loci that were extracted using this approach closely mimic ENi L1 insertions reported in earlier cell-culture analyses (18,22,28), further consolidating our hypothesis that they represent products of a similar insertion mechanism in the human genome. We found a total of 21 NCLI loci in the May 2004 freeze of the human genome (hg17)(Table 1), of which we were able to recover the pre-insertion site of seven loci from the chimpanzee genome assembly (panTro2; March 2006 freeze) (33) and 14 loci from the rhesus macaque genome assembly (rheMac2; January 2006 freeze) (34). As we were only interested in NCLI loci for which we could verify the pre-insertion sequence, we discarded all L1 insertions that were shared between these three genomes and thus represented older ancestral L1 elements. The L1 elements at NCLI loci ranged between 34 and 4410 bp in length, with a total of 12 018 bp L1 DNA (along with 1365 bp of non-L1 sequence) being captured between the matching 5′ and 3′ ends of the pre-insertion and post-insertion states. In addition, 18 of 21 NCLI loci were associated with deletions of target site DNA, ranging between 5 bp and 14 534 bp and totaling 31 009 bp.
Table 1.
Human NCLI loci and insertion site characteristics
| Locus | Coordinates | L1 bp_ins | non-L1 bp_ins | bp_del | L1 seq 5′ or 3′? | AT% ±200 bp | AT% ±20 Kb | Lineage | Intragenic? |
|---|---|---|---|---|---|---|---|---|---|
| NCLI1 | chr3:196416805–196421321 | 4410 | 107 | 109 | 3′ | 59.5 | 49.04 | H | C3ORF1 |
| NCLI3 | chr4:67544153–67545039 | 589 | 298 | 1574 | 3′ | 63 | 62.41 | H | – |
| NCLI9 | chr17:36395952–36396018 | 67 | 0 | 0 | Both | 60.5 | 60.86 | HC | KRT40 |
| NCLI11 | chr19:15679181–15680403 | 1223 | 0 | 2867 | Both | 67.5 | 59.16 | H | – |
| NCLI23 | chr2:29588579–29590824 | 2246 | 0 | 17 | Both | 65 | 56.7 | HC | ALK |
| NCLI32 | chr4:112069027–112069153 | 122 | 5 | 23 | 3′ | 60.5 | 63.86 | HC | – |
| NCLI33 | chr4:60239707–60239936 | 108 | 122 | 2485 | 3′ | 65.5 | 67.18 | HC | – |
| NCLI34 | chr4:87186203–87186706 | 483 | 21 | 30 | 5′ | 70.5 | 64 | H | MAPK10 |
| NCLI38 | chr5:51963332–51963788 | 441 | 16 | 1692 | 3′ | 53.5 | 61.77 | HC | – |
| NCLI40 | chr6:4414637–4415321 | 600 | 85 | 0 | Bone | 58 | 55.68 | HC | – |
| NCLI47 | chr9:108094757–108094921 | 160 | 5 | 8 | 5′ | 67.5 | 60.96 | HC | – |
| NCLI48 | chr10:60661882–60662013 | 34 | 98 | 5928 | 5′ | 67.5 | 66.94 | HC | PHYHIPL |
| NCLI51 | chr11:34668952–34669415 | 464 | 0 | 615 | Both | 58 | 63.31 | H | – |
| NCLI52 | chr12:59792048–59792392 | 336 | 9 | 46 | 3′ | 73 | 65.3 | HC | – |
| NCLI53 | chr12:14711194–14711264 | 61 | 10 | 7 | None | 67 | 60.16 | H | GUCY2C |
| NCLI55 | chr13:102553958–102554087 | 48 | 62 | 44 | None | 52 | 58.98 | HC | – |
| NCLI57 | chr13:80218694–80218899 | 202 | 4 | 5 | 5′ | 67.5 | 64.53 | HC | – |
| NCLI60 | chr16:35125561–35125651 | 86 | 0 | 0 | Both | 65.5 | 63.24 | H | – |
| NCLI61 | chr17:3071528–3071879 | 49 | 303 | 14534 | 5′ | 68 | 60.46 | HC | – |
| NCLI64 | chr22:45486099–45486153 | 35 | 0 | 1010 | Both | 67.5 | 51.07 | HC | CERK |
| NCLI65 | chr22:38619900–38620471 | 254 | 318 | 15 | None | 63.33 | 60.75 | HC | – |
| Total (bp) | 12018 | 1365 | 31009 | Average | 63.33 | 60.13 | |||
In the column for ‘Lineage’, H indicates a NCLI event specific to the human genome, while HC indicates an NCLI event shared between the human and chimpanzee genomes but absent from the rhesus macaque genome.
Our estimate of the total number of NCLI events is probably conservative, given that the RM algorithm we used to detect L1 elements, even at its −s (sensitive) setting, is unable to detect insertions smaller than 30 bp. Given that previous cell culture analyses of DSB repair by L1-mediated gene conversion have detected insertion tracts as small as 13 bp (35), it is quite possible that the number of recent human NCLI events is actually higher than our estimate. Further support for the existence of such ‘hyphen elements’ (24) in the genome comes from ongoing studies in our lab (Sen, S. K. et al., unpublished data), where we find that TPRT can produce severely 5′ truncated L1 and Alu insertions with a similar minimum size (∼28–30 bp). As such, it is possible that additional NCLI loci beyond the 21 analyzed here remain undetected in the human genome.
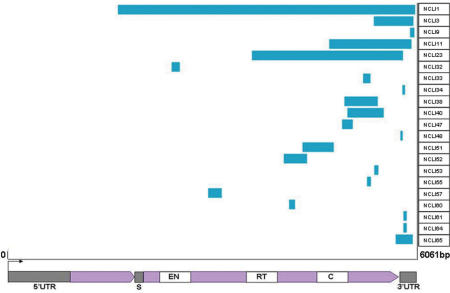
Alignment of L1 segments involved in NCLI events with the full-length consensus sequence of a human-specific L1 subfamily (L1Hs) revealed a tendency to cluster in the downstream half of the L1 consensus, with 18 out of 21 NCLI fragments having 5′ truncations 3000 bp or more in addition to their 3′ truncations (Figure 3; supplemental alignment 1, online). Previous analyses show that most TPRT-mediated genomic L1 insertions are severely 5′ truncated (20), which may reflect low processivity of the L1 RT or alternatively, host suppression of transcription (36). The analogous tendency of L1 fragments at NCLI loci to be confined within the downstream half of the element may either be due to the same reasons, or may be moderated by the dynamics of L1 ribonucleoprotein (RNP) positioning at the sites of DSBs (18).
Figure 3.
Schematic diagram of NCLI L1 element length. Length distribution of L1 segments at the 21 NCLI loci in this analysis along the sequence of a full-length L1 element (L1Hs) as shown by the blue bars. Location of different domains within the L1 element is shown in the lower panel. Of the non-coding regions (gray boxes) the 5′ UTR contains an internal RNA polII promoter, while a 63-bp spacer (S) separates the two ORFs (purple arrows). 40 kDa ORF1 has RNA-binding and nucleic acid chaperone activities, while 150 kDa ORF2 consists of an NH2-terminal endonuclease (EN) domain, a central reverse transcriptase (RT) domain, and a COOH-terminal zinc-knuckle like domain. The extreme 3′ end of the 3′UTR consists of a variable poly(A) tail, absent in all 21 NCLI-mediated insertions.
Random genomic deletions that remove the 3′ ends of classical TPRT-mediated L1 insertions (including the poly(A) tail and the downstream TSD) could mimic the sequence architecture of NCLI loci (23). However, by reconstructing the pre-insertion site of all loci (and verifying that the starting point of the 3′ flanking sequence remained unchanged before and after the L1 insertion), we effectively minimized the chances of including such events in our data, as it is unlikely that random deletions would repeatedly and precisely remove only L1 sequence, leaving the downstream sequence untouched. Also, for the 18 NCLI loci that were associated with target site deletions, this would require two independent, random deletion events to have taken place at exactly the same position in two separate primate species, which would have vanishingly small probability. The 3′ truncated L1 fragment at locus NCLI 40 was not associated with a deletion of target DNA and was followed by an adenosine-rich stretch, making it possible that an internal deletion had removed the 3′ UTR before the poly(A) tail. However, based on the absence of TSDs, high divergence from the L1-EN consensus and presence of non-L1 DNA at both 5′ and 3′ ends, we decided to include it in our analysis.
Analysis of insertion sites reveals divergence from L1-EN consensus
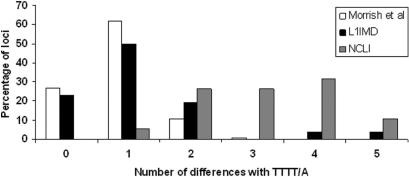
To find additional evidence supporting our hypothesis that NCLI events were created by an endonuclease-independent mechanism, we inspected all loci for deviations from the 5′-TTTT/A-3′ L1-EN consensus cleavage site. Histograms of divergence scores of NCLI events, compared to two other recent analyses of TPRT-mediated L1 insertions (Figure 4), revealed a marked shift in the maxima towards an increased number of differences. Statistical comparisons of the amounts of deviation from the consensus revealed a highly significant difference between the cleavage site preferences of NCLI loci versus a larger set of 282 recent TPRT-mediated L1 insertions (22) (unpaired t-test assuming unequal variances; P < 0.0001) (37), further bolstering our conclusion that breaks in the target DNA at NCLI loci were not products of L1-EN cleavage. Previous in vitro analyses have demonstrated that in addition to ‘preferred’ motifs for cleavage, a second set of ‘atypical’ motifs also exists which L1-EN can cleave at lower efficiencies during TPRT (19,22). However, none of the 21 loci involved insertions into any of these preferred or atypical motifs, further supporting our hypothesis that the NCLI mechanism is independent of L1-EN activity.
Figure 4.
L1 cleavage site analysis. Frequency spectra of deviations from the consensus L1 endonuclease cleavage site (5′-TTTT/A-3′). Two sets of TPRT-mediated insertions are represented along with NCLI events (gray bars): 282 L1_Ta subfamily elements identified in reference 22 (white bars) and 26 human genome-specific L1 insertion-mediated deletions (L1IMDs) identified in reference 10 (black bars).
An analysis of nucleotide composition in the 20 kb of genomic sequence flanking NCLI loci showed average AT content to be ∼ 60% (Table 1), which fits well with the global distribution of L1 elements in the human genome (1). Interestingly, even within these AT-rich surroundings, the 200 bp immediately surrounding the breakage sites within the ancestral genome (100 bp in either direction) showed a further increase in AT content (average of 63.3%). Given that AT-rich DNA is inherently unstable (38), this trend may reflect the possibility that such stretches in the local sequence architecture, being prone to mechanical or thermodynamic breakage, provide more frequent substrates for NCLI events than nearby GC-rich segments.
Structural characteristics of NCLI events
Structurally, NCLI loci closely resemble analogous insertions of non-LTR retrotransposons into pre-existing DSBs in cell culture models (22, 39–41), which supports our hypothesis that NCLI represents a DSB repair mechanism. Except for occasional 3′ transductions, which are a byproduct of the TPRT process (14), classical L1 insertions are rarely associated with insertions of non-L1 DNA. In contrast, 71% of NCLI events (15 out of 21) involved insertions of non-L1 DNA segments of lengths ranging from 4 to 312 bp along with the L1 DNA (we use the term ‘extra nucleotides’ to denote these segments). Extra nucleotides conjoined to the L1 element were at the 3′ and 5′ ends at eight and six NCLI loci, respectively, while at three loci such insertions flanked both sides of the L1 element. Closer examination of the extra nucleotides revealed some interesting clues about the possible mechanisms associated with NCLI events, which we discuss subsequently.
At two loci (NCLI1 and NCLI40), fragments of other cellular RNAs appeared to have been co-opted along with the L1 RNA during reverse transcription by the L1 reverse transcription. While chimeric L1-U6 snRNA insertions similar to NCLI1 have been previously described (42,43), an 18-bp fragment of GPD2 mRNA was present at the 5′ end of NCLI40, providing new evidence that the L1 RT can switch templates between L1 RNA and other cellular RNAs during the retrotransposition process. At one locus (NCLI3), an intact AluY element was present at the 5′ flank of the L1 insertion. While the AluY element may have been a later, TPRT-mediated insertion, the absence of TSDs and high divergence from the L1 EN consensus cleavage site suggest that this locus may also represent capture of a nearby Alu mRNA during NCLI or an instance of in vivo L1-Alu RNA recombination.
At two loci, BLAST searches using the extra nucleotides showed evidence for NCLI-mediated inter-chromosomal translocations. At NCLI65 (located on chr. 22), 267 of the 312 extra nucleotides at the 5′ flank of the L1 shared significant similarity with a 266-bp stretch on chr. 8 (94% identity; E = 2e−95). At the second locus (NCLI40, located on chr. 6), 24 out of 66 extra nucleotides at the 3′ end had a near-perfect match on chr. 2 (95% identity; E = 0.059). At a third locus (NCLI34), 11 of 21 extra nucleotides perfectly matched a segment of the AluJ consensus sequence. As this Alu subfamily has long been inactive in terms of retrotransposition, this may represent the use of an ancient insertion located elsewhere for SDSA-mediated DSB repair (44,45); alternatively, the homology could be purely due to chance. At locus NCLI48 (which was associated with a 5928-bp deletion in the ancestral genome), we found additional evidence for the SDSA repair pathway being a component of NCLI. Here, 98 bp extra nucleotide sequence at the 5′ end of the human L1 insertion had a highly significant match (96% identity; E = 4e−39) to a segment of equal length within the ancestral deletion referred to above. A viable mechanism explaining this structure involves local melting of the double helix within the segment deleted during the NCLI event to provide a transient single-stranded template for repair of the genomic lesion, conforming to the SDSA models described in the earlier studies referred to above. Extra nucleotide stretches at 12 of the 42 junctions (i.e. at either side of the 21 L1 fragments) either did not have statistically significant BLAST matches in the human genome, or were too small (<15 bp) to draw any definite conclusions. Two junctions (5′ end of NCLI33 and 3′ end of NCLI61) contained 122 bp and 303 bp insertions of AT-rich simple repeats, respectively, suggesting that the NCLI process may also contribute to the creation of new microsatellite loci in the human genome, in a manner similar to TPRT-mediated L1 insertion (46).
In contrast to previous computational analyses that estimate 19–25% of TPRT-mediated L1 insertions in the human genome to be 5′-truncated/inverted (20,47), only two of the 21 NCLI loci in our analysis showed internal rearrangements within the L1 segment. Interestingly, previous analyses of endonuclease-independent L1 insertions have not recovered any truncated/inverted structures as well (22). In view of these results, we suggest that linearly structured segments in the free-floating L1 mRNA are preferentially captured at the sites of DSBs. Strong support for this hypothesis comes from a previous analysis of ΦK174 DNA fragments transfected into enzymatically created DSBs in a thymidine kinase-deficient mouse cell line, where linear fragments were captured 9X more efficiently than supercoiled segments (39). Of the two NCLI loci that showed evidence for rearrangement within the L1, NCLI38 was a simple truncation/inversion structure most likely formed by twin priming (47). Locus NCLI34, where three consecutive L1 fragments formed a complex structure was more difficult to explain. However, the best BLAST match to the 377 bp highly diverged middle segment (98% identity; E = 0.0) was located downstream on the same chromosome. Thus, our model for this locus suggests an initial truncated/inverted NCLI event followed by a subsequent intra-chromosomal gene conversion which inserted the middle segment. Similar internal rearrangements in L1Hs elements have been documented by a previous analysis (48).
The total amount of deleted sequence between the pre-insertion and post-insertion states of the 21 NCLI events was 31 009 bp, more than twice the 13 383 bp of combined L1 and non-L1 sequence inserted at the same loci. Of the deleted sequence, almost 50% (14 534 bp) was associated with a single locus (NCLI61). For this locus, as for all others, we confirmed by both PCR and re-sequencing that the computationally detected deletion was authentic and matched the draft genome sequence.
Microhomology between ends of L1 inserts and flanking host DNA
Recent evidence suggests that microhomology between the L1 mRNA and single-stranded overhangs in the genomic DNA flanking the L1-EN cleavage site mediates 5′-end attachment during conventional TPRT, while the 3′ end of the mRNA anneals to the nicked DNA through its poly(A) tail (21,30). It is possible that a similar mechanism is used for attachment of the L1 RNA to the target DNA during the NCLI process as well. However, to support this assumption for NCLI loci, increased levels of microhomology would have to be present independently at the 5′ and 3′ ends of the L1 insertion rather than at the 5′ end alone. To detect such stretches of higher-than-random complementarity at the ends of a NCLI locus, wherever an exact junction was present between the L1 element and flanking pre-insertion host sequences, we located (i) the 5′ and 3′ extremities of the L1 insertion with respect to the L1Hs consensus sequence and; (ii) the starting points of 5′ and 3′-end flanking sequence (which we identified by aligning the pre-insertion and post-insertion states of the loci) (Figure 5A). Next, we isolated 6-bp stretches of sequence extending outwards from these points (i.e upstream of the 5′ end and downstream of the 3′ end) in both the L1Hs consensus and flanking sequence and aligned them to count the number of complementary bases (at loci where non-L1 DNA was present at one end of the L1 insertion, we only analyzed the other end). Given that microhomology-mediated single-strand annealing can resolve DSBs when the extent of complementarity is limited to even one match (49), the high numbers of complementary bases at the L1-genomic DNA junctions (particularly at the first two positions) noticed separately at both the 5′ and 3′ ends of NCLI loci (Figure 5B) strongly suggest that a similar mechanism is indeed likely to facilitate L1 mRNA binding during NCLI, and further consolidates our hypothesis that NCLI acts as a DSB repair mechanism.
Figure 5.
NCLI microhomology analysis. (A) Complementarity at the 5′ and 3′ ends of NCLI loci. Note that nucleotide positions are counted in opposite directions at the 5′ and 3′ ends, based on the first two nucleotides that would anneal during mRNA attachment. (B) Number of matches at each position and the corresponding P-values, that indicate the likelihood of obtaining the observed numbers of matches by chance alone. Bases are highlighted gray if they are complementary to the corresponding nucleotide on the L1 RNA. P-values were calculated based on a binomial probability distribution, where the chance of success (i.e. complementary pairing) at each position was 1/4 and the chance of failure was 3/4.
Genomic environment of NCLI events
To characterize the genomic context in which NCLI events occur, we scanned 2 Mb of sequence upstream and downstream of each locus for the presence of known or predicted human genes using NCBI MapViewer (http://www.ncbi.nlm.nih.gov/mapview/static/MVstart.html). Surprisingly, compared to the vast majority of L1s in the human genome which are located in gene-poor regions (1,20), NCLI events were concentrated in areas of relatively high gene density (one gene/83 kb), compared to both the global gene density in the human genome (one gene/150 kb)(50) and the average gene density in the vicinity of TPRT-mediated L1 insertions associated with human genomic deletions (one gene/200 kb) (10). In addition, 33% of NCLI loci (7 out of 21) were situated within the introns of known genes (Table 1), twice the figure of 13–17% for TPRT-mediated L1 insertions (20). Interestingly, when we analyzed the genomic sequences corresponding to the 19 NCLI-mediated deletions in the ancestral (i.e. chimpanzee or rhesus) genomes, we found that at one locus (NCLI61), a model rhesus gene (LOC721417) from the olfactory receptor family had been deleted during the L1 insertion process. Although the olfactory receptor gene family is one of the largest in primate genomes with ∼1000 members (51) and the deletion of a single gene is unlikely to create a significant difference in phenotype, this event further underscores the tendency of NCLI loci to be concentrated in gene-rich areas of the genome.
DISCUSSION
An alternative pathway for non-LTR retrotransposition in the human genome
In this analysis, we address one of the remaining questions in L1 element biology: does an alternative pathway exist for L1 retrotransposition in the human genome? (3,52) All through the late 1980s until the introduction in 1993 of the TPRT model for insertion of the R2Bm non-LTR retrotransposon in Bombyx mori (17), it was thought that L1 propagation occurred mainly through fortuitous insertion into DSBs (53,54). Subsequent research established beyond reasonable doubt that the L1 elements use a TPRT-like process as their predominant insertional mechanism (5,18,19) and the focus of L1 element biology has since shifted to resolving the unanswered questions of the TPRT model (21,55). Interestingly though, the hypothesis that an alternative, ENi mechanism acts concurrently with TPRT in the human genome, though often speculated upon (18,22), has never been fully investigated. Existing whole-genome analyses of L1 activity have focused solely on TPRT-mediated insertions, and while ENi L1 retrotransposition has earlier been detected in cell lines deficient for DNA repair proteins (22), the authors of these studies suggest that, in vivo (i.e. when cellular DNA repair mechanisms function normally), such insertions may not be present at detectable frequencies. Thus, the NCLI loci detected in our study represent the first whole-genome analysis of ENi L1 insertions in a phenotypically normal genetic background that is also subject to selection (i.e. an extant genome). Additionally, we find that the structures of NCLI events recovered in vivo closely mirror those previously found in vitro, reaffirming the validity of cell culture retrotransposition assays as surrogate models for analyzing retrotransposon biology and determining the impact that these elements have on the genome.
While it remains possible that further NCLI exist in the human genome that cannot be detected using our computational strategy, TPRT-mediated insertions will regardless be several orders of magnitude more frequent. This disparity in scale can be explained by the fundamentally different natures of the TPRT and NCLI mechanisms. From the retrotransposon point of view, TPRT is an ‘independent’ process, as L1 elements encode both the endonuclease and RT activities required for self-propagation through this mechanism. As such, TPRT-mediated insertion does not have to depend on pre-existing DSBs to provide integration sites. However, in contrast to the independent and organized nature of TPRT, structural features of NCLI loci suggest that it is a more random process, depending entirely on the presence of pre-existing DSBs to provide integration sites. Additionally, while only ∼2% of human-specific TPRT-mediated L1 insertions create deletions of target genomic DNA (10), the fact that 86% of NCLI loci (18 out of 21) are associated with genomic deletions would render it a rather inefficient mechanism, had L1 insertion been its sole function. Thus, it is possible that both these processes have co-existed over long periods of time, and while TPRT has doubtless been the primary mode of insertion, certain beneficial features of NCLI have probably contributed to its persistence despite the relative paucity of these events. The observation that at least seven NCLI events are restricted to the human lineage and absent from the chimpanzee and rhesus macaque genomes suggests that this process has been active in recent human genome evolution subsequent to the divergence of human and non-human primates.
Mechanistic aspects of NCLI suggest a role in DNA repair
The NCLI loci we analyzed may have been produced by three separate mechanisms: (i) capture of nearby L1 mRNAs at the site of DSBs and subsequent reverse transcription (Figure 1B); (ii) SDSA-mediated DSB repair in which the free-floating ends of a DSB transiently invade locally melted regions of neighboring double-stranded DNA to provide templates for transcription (44,45) and; (iii) conventional double-strand break-induced recombination (DSBR) (56). Since only three out of 21 NCLI loci (NCLI11, NCLI23 and NCLI51) involve insertions into pre-existing L1 elements, it is unlikely that conventional DSBR is a mechanism for NCLI, since the presence of sequence homology between the recombining strands is a prerequisite for this model. Of the other two pathways, while theoretically possible, we believe that SDSA is not a preferred mechanism for NCLI. Firstly, the SDSA pathway is highly efficient at minimizing loss of genomic DNA during the patching of DSBs (57), which contrasts with the ∼31 kb of genomic deletion detected at the NCLI loci in our analysis. Secondly, although L1 family insertions comprise ∼17% of the genome (1), subfamilies which have been active in recent human genome evolution comprise only a small fraction of this figure, while the vast majority of insertions belong to older, extinct subfamilies and have accumulated large numbers of mutations relative to the original consensus sequence (58–60). In this scenario, it is unlikely that the much smaller fraction of recent L1 insertions would be preferentially chosen as templates for SDSA-mediated repair at the 21 NCLI loci in our analysis, which invariably involve relatively young L1 elements with few internal mutations (i.e. <2% divergent by the RM algorithm; see ‘Materials and Methods’ section). However, at two loci (NCLI34 and NCLI48), we did find some evidence that SDSA may play a minor accessory role in the NCLI mechanism (see ‘Results’ section).
Consequently, our preferred model for NCLI is that L1 mRNAs occasionally act as genomic Band-Aids® by bridging pre-existing DSBs in the genome. Given that unrepaired DSBs are among the most lethal forms of DNA damage (26,61), it is not surprising that mammalian cells have evolved highly efficient repair pathways capable of patching DSBs with almost any DNA molecule available in the vicinity (39). Indeed, capture of mobile DNA (including DNA transposons and both LTR and non-LTR retrotransposons) at the site of genomic DNA lesions seems to be a recurring theme in eukaryotic cells (22, 39–41, 62). In addition, the exceptionally high levels of complementary bases at the L1-host DNA junctions at NCLI loci support our hypothesis of microhomology-mediated L1 mRNA capture between pre-existing DSB ends. A recent analysis shows that the L1-EN creates many more genomic DSBs than is required for its own retrotransposition (63), raising an interesting question: are some of these newly created breaks promptly filled in by NCLI? While we consider this to be a possibility, the NCLI loci analyzed in our study all show significant deviations from the L1-EN site, making it unlikely that any of them represent such an occurrence. However, NCLI could be considered a genomic ‘payoff’, through which L1 elements partially compensate for the excess of DSBs that they create. Additional studies of other non-autonomous L1-dependent retrotransposons such as Alu and SVA elements will provide further insight into the role these elements may play in NCLI.
Previous analyses have shown that cellular DNA repair proteins used in the NHEJ pathway mobilize to the sites of DSBs and compete with the DSBR repair machinery where both systems are available (64–66). Given that both NCLI and NHEJ are error-prone repair pathways associated with loss of genomic sequence, we consider it quite probable that the NHEJ machinery is co-opted at NCLI loci. Significantly, NHEJ proteins have previously been shown to co-fractionate with non-LTR retrotransposon cDNA intermediates, further supporting the hypothesis that they are involved in the genomic integration of mobile DNA (67,68).
CONCLUSION
In this study, we have demonstrated that NCLI has provided an alternative, endonuclease-independent pathway for L1 integration during human genome evolution, and highlighted its structural differences as compared to the more common and well-characterized TPRT-mediated mode of L1 insertion in the human genome. Based on the sequence architecture of NCLI loci, we propose that this mechanism has been a fortuitous mode for repair of genomic lesions. The distinct nature of the TPRT and NCLI processes suggests that they may have different genomic implications. TPRT-mediated L1 insertions in the human genome, apart from creating large numbers of DSBs, are associated with disruption of functional genes and may be prone to post-insertion ectopic recombination. On the contrary, both the genomic NCLI loci we have detected and similar insertions in previous cell-culture analyses show definite signs of being variants of DSB repair, and seven of the loci we have detected are located within protein-coding genes, breakage within which would otherwise have had direct consequences on the phenotype. Thus, it is interesting to speculate that this ‘non-selfish’ role of NCLI-mediated insertions in maintaining genomic integrity may result in a qualitative difference in the selective regimes acting on the TPRT and NCLI processes (69). Seven of the NCLI events we have recovered are specific to the human lineage. Assuming the total number of human lineage-specific L1 insertions to be ∼1300–1800 (10,70), NCLI thus occurs at the relatively low frequency of 0.5% in the human genome. However, extrapolating these numbers to the larger timescale of the primate radiation, the ∼520 000 L1 elements in primate genomes may thus include ∼2000–2800 NCLI events, making this process a significant factor in shaping the architecture of the genome. In our opinion, the finding that both L1 and Alu elements in the human genome are capable of acting as in vivo molecular Band-Aids® is significant, as it opens the possibility that active non-LTR retrotransposon families in primate genomes may have a role in maintaining genomic integrity that awaits further characterization.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank all members of the Batzer laboratory for their cooperation and assistance. They would also like to express their appreciation to Drs. J.Kim and J.R. Battista and two anonymous reviewers for their useful comments during preparation of the manuscript. They are especially grateful to J.A. Walker for logistical support and to Christopher Faulk for assistance with computational analyses. Mike McKenna, Pamela Bhattacharya and Alivia Dey provided generous help with statistical calculations. S.K.S. expresses his gratitude to Soma Chowdhury for her support during the course of this project. This research was supported by National Science Foundation grants BCS-0218338 (M.A.B.) and EPS-0346411 (M.A.B.); National Institutes of Health RO1GM59290 (M.A.B.), and the State of Louisiana Board of Regents Support Fund (M.A.B.). Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health and the National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Smit AF. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 3.Moran JV, Gilbert N. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, D.C.: ASM Press; 2002. pp. 836–869. [Google Scholar]
- 4.Martin SL. The ORF1 Protein Encoded by LINE-1: Structure and Function During L1 Retrotransposition. J. Biomed. Biotechnol. 2006;2006:45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 6.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 7.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 9.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 10.Han K, Sen SK, Wang J, Callinan PA, Lee J, Cordaux R, Liang P, Batzer MA. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 2005;33:4040–4052. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 12.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 13.Burwinkel B, Kilimann MW. Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J. Mol. Biol. 1998;277:513–517. doi: 10.1006/jmbi.1998.1641. [DOI] [PubMed] [Google Scholar]
- 14.Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat. Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 16.Kazazian HH., Jr L1 retrotransposons shape the mammalian genome. Science. 2000;289:1152–1153. doi: 10.1126/science.289.5482.1152. [DOI] [PubMed] [Google Scholar]
- 17.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 18.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 20.Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0052. research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zingler N, Willhoeft U, Brose HP, Schoder V, Jahns T, Hanschmann KM, Morrish TA, Lower J, Schumann GG. Analysis of 5' junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 2005;15:780–789. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 23.Mager DL, Henthorn PS, Smithies O. A Chinese G gamma + (A gamma delta beta)zero thalassemia deletion: comparison to other deletions in the human beta-globin gene cluster and sequence analysis of the breakpoints. Nucleic Acids Res. 1985;13:6559–6575. doi: 10.1093/nar/13.18.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Audrezet MP, Chen JM, Raguenes O, Chuzhanova N, Giteau K, Le Marechal C, Quere I, Cooper DN, Ferec C. Genomic rearrangements in the CFTR gene: extensive allelic heterogeneity and diverse mutational mechanisms. Hum. Mutat. 2004;23:343–357. doi: 10.1002/humu.20009. [DOI] [PubMed] [Google Scholar]
- 25.Van de Water N, Williams R, Ockelford P, Browett P. A 20.7 kb deletion within the factor VIII gene associated with LINE-1 element insertion. Thromb. Haemost. 1998;79:938–942. [PubMed] [Google Scholar]
- 26.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst.) 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 27.Farkash EA, Prak ET. DNA damage and l1 retrotransposition. J. Biomed. Biotechnol. 2006 doi: 10.1155/JBB/2006/37285. 10.1155/JBB/2006/37285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 2006;34:1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eickbush TH. Repair by retrotransposition. Nat. Genet. 2002;31:126–127. doi: 10.1038/ng897. [DOI] [PubMed] [Google Scholar]
- 30.Martin SL, Li WL, Furano AV, Boissinot S. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogenet. Genome. Res. 2005;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 33.TCSAC. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 34.Rhesus Macaque Genome Sequencing and Analysis Consortium. The Rhesus macaque genome sequence informs biomedical and evolutionary analyses. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 35.Tremblay A, Jasin M, Chartrand P. A double-strand break in a chromosomal LINE element can be repaired by gene conversion with various endogenous LINE elements in mouse cells. Mol. Cell. Biol. 2000;20:54–60. doi: 10.1128/mcb.20.1.54-60.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruxton GD. The unequal variance t-test is an underused alternative to Student's t-test and the Mann–Whitney U test. Behav. Ecology. 2006;17:688–690. [Google Scholar]
- 38.Chalikian TV, Volker J, Plum GE, Breslauer KJ. A more unified picture for the thermodynamics of nucleic acid duplex melting: a characterization by calorimetric and volumetric techniques. Proc. Natl Acad. Sci. USA. 1999;96:7853–7858. doi: 10.1073/pnas.96.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Y, Waldman AS. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 2001;29:3975–3981. doi: 10.1093/nar/29.19.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ichiyanagi K, Nakajima R, Kajikawa M, Okada N. Novel retrotransposon analysis reveals multiple mobility pathways dictated by hosts. Genome Res. 2007;17:33–41. doi: 10.1101/gr.5542607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng SC, Kim B, Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 42.Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics. 2002;80:402–406. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- 43.Buzdin A, Ustyugova S, Gogvadze E, Lebedev Y, Hunsmann G, Sverdlov E. Genome-wide targeted search for human specific and polymorphic L1 integrations. Hum. Genet. 2003;112:527–533. doi: 10.1007/s00439-002-0904-2. [DOI] [PubMed] [Google Scholar]
- 44.Formosa T, Alberts BM. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 45.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ovchinnikov I, Troxel AB, Swergold GD. Genomic Characterization of Recent Human LINE-1 Insertions: Evidence Supporting Random Insertion. Genome Res. 2001;11:2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in l1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, Swergold GD, Henke J, Henke L, et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfeiffer P, Thode S, Hancke J, Vielmetter W. Mechanisms of overlap formation in nonhomologous DNA end joining. Mol. Cell. Biol. 1994;14:888–895. doi: 10.1128/mcb.14.2.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.IHGSC. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 51.Young JM, Trask BJ. The sense of smell: genomics of vertebrate odorant receptors. Hum. Mol. Genet. 2002;11:1153–1160. doi: 10.1093/hmg/11.10.1153. [DOI] [PubMed] [Google Scholar]
- 52.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 53.Voliva CF, Martin SL, Hutchison A, III, Edgell MH. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J. Mol. Biol. 1984;178:795–813. doi: 10.1016/0022-2836(84)90312-7. [DOI] [PubMed] [Google Scholar]
- 54.Edgell MH, Hardies SC, Loeb DD, Shehee WR, Padgett RW, Burton FH, Comer MB, Casavant NC, Funk FD, et al. The L1 family in mice. Prog. Clin. Biol. Res. 1987;251:107–129. [PubMed] [Google Scholar]
- 55.Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McVey M, Larocque JR, Adams MD, Sekelsky JJ. Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl Acad. Sci. USA. 2004;101:15694–15699. doi: 10.1073/pnas.0406157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smit AF, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 59.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mathews LM, Chi SY, Greenberg N, Ovchinnikov I, Swergold GD. Large differences between LINE-1 amplification rates in the human and chimpanzee lineages. Am. J. Hum. Genet. 2003;72:739–748. doi: 10.1086/368275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 62.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 63.Gasior SL, Wakeman TP, Xu B, Deininger PL. The Human LINE-1 Retrotransposon Creates DNA Double-strand Breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rapp A, Greulich KO. After double-strand break induction by UV-A, homologous recombination and nonhomologous end joining cooperate at the same DSB if both systems are available. J. Cell. Sci. 2004;117:4935–4945. doi: 10.1242/jcs.01355. [DOI] [PubMed] [Google Scholar]
- 65.Drouet J, Frit P, Delteil C, de Villartay JP, Salles B, Calsou P. Interplay between Ku, Artemis, and the DNA-dependent protein kinase catalytic subunit at DNA ends. J. Biol. Chem. 2006;281:27784–27793. doi: 10.1074/jbc.M603047200. [DOI] [PubMed] [Google Scholar]
- 66.Drouet J, Delteil C, Lefrancois J, Concannon P, Salles B, Calsou P. DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J. Biol. Chem. 2005;280:7060–7069. doi: 10.1074/jbc.M410746200. [DOI] [PubMed] [Google Scholar]
- 67.Downs JA, Jackson SP. Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol. Cell. Biol. 1999;19:6260–6268. doi: 10.1128/mcb.19.9.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell. Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 69.Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol. Biol. Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- 70.Lee J, Cordaux R, Han K, Wang J, Hedges DJ, Liang P, Batzer MA. Different evolutionary fates of recently integrated human and chimpanzee LINE-1 retrotransposons. Gene. 2007;390:18–27. doi: 10.1016/j.gene.2006.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.