Abstract
The genome of influenza A virus is organized into eight ribonucleoprotein complexes (RNPs), each containing one RNA polymerase complex. This RNA polymerase has also been found non-associated to RNPs and is possibly involved in distinct functions in the infection cycle. We have expressed the virus RNA polymerase complex by co-tranfection of the PB1, PB2 and PA genes in mammalian cells and the heterotrimer was purified by the TAP tag procedure. Its 3D structure was determined by electron microscopy and single-particle image processing. The model obtained resembles the structure previously reported for the polymerase complex associated to viral RNPs but appears to be in a more open conformation. Detailed model comparison indicated that specific areas of the complex show important conformational changes as compared to the structure for the RNP-associated polymerase, particularly in regions known to interact with the adjacent NP monomers in the RNP. Also, the PB2 subunit seems to undergo a substantial displacement as a result of the association of the polymerase to RNPs. The structural model presented suggests that a core conformation of the polymerase in solution exists but the interaction with other partners, such as proteins or RNA, will trigger distinct conformational changes to activate new functional properties.
INTRODUCTION
The influenza A viruses are negative-stranded RNA viruses of the family Orthomyxoviridae that contain a segmented genome consisting of eight RNA molecules (1). These are encapsidated into ribonucleoprotein particles (RNPs) by association to nucleoprotein (NP) monomers and to the polymerase complex. Each virion RNP (vRNP) is an independent template for transcription and replication in the nucleus of the infected cell (2). Transcription is initiated by cap-snatching primers from cellular pre-mRNAs (3) and is terminated by polyadenylation at an oligo-U signal before the end of the template (4,5). Conversely, replication involves initiation de novo and leads to complete copies of the template, which are encapsidated into complementary RNPs (cRNP). These act as replication intermediates to generate large amounts of progeny vRNPs (6).
Each virus RNP is a supercoiled ribbon-like structure (7,8) in which most of the RNA is covered by NP monomers, each one bound to ∼24 nt of the template (9). The polymerase complex can be detected at one end of the supercoil (10) and helps maintaining both RNA termini together (11). A more detailed structure of a vRNP has been obtained by 3D reconstruction of recombinant mini-RNPs in which the length of the template is limited and the configuration is circular (9,12).
The polymerase complex is a heterotrimer with an aggregate molecular mass of around 250 kDa, composed by the PB1, PB2 and PA subunits. The PB1 subunit is responsible for the polymerase and endonuclease activities. The PB2 protein has cap-binding activity and is essential for transcription initiation, while the PA protein is a phosphoprotein responsible for proteolytic activity and involved in RNA replication (2). Also, the phenotype of a number of mutants has shown that alterations in many of the subunits may alter either RNA transcription or replication (13,14). The structure of the polymerase included in these mini-RNPs has been also determined and appears as a tight complex in which the limits between the subunits are not apparent (15). Nevertheless, the position of specific domains of each polymerase subunit has been determined by 3D reconstruction of tagged RNPs or RNP-monoclonal antibody complexes. Likewise, the sites of polymerase interaction with the adjacent NP monomers in the RNP have been localized (15).
In addition to the polymerase complex present in the vRNPs, the polymerase heterotrimer, non-associated to RNPs, has also been detected in the nucleus of infected cells (16). These soluble heterotrimers should play a role in the recognition of the newly synthesized replication intermediate and progeny RNAs, as they bind specifically to the 5′-terminal conserved sequences and the 5′–3′ panhandle structure (17–19) (2). In fact, it has been proposed that binding to the polymerase complex is critical for the stabilization of the cRNA replication intermediate (20). Besides its function in RNA replication, two other potential roles have been proposed for the soluble polymerase heterotrimer: (i) modulation of the splice site selection during the processing of segment 7 primary transcripts (21) and (ii) protection of the viral mRNAs from cap snatching by the vRNPs (22). Hence, it is particularly relevant to gather structural information for the soluble form of the polymerase heterotrimer, non-associated to the vRNP and not bound to the template. In this report, we have addressed this problem by expression and purification of a recombinant polymerase complex in mammalian cells and its structural analysis by electron microscopy and 3D reconstruction after single-particle analysis (23).
MATERIALS AND METHODS
Biological materials
The COS-1 (24) and HEK 293T (25) cell lines were cultivated as described (26). The vaccinia recombinant virus vTF7-3 (27) was a gift of B. Moss. Plasmids pGPB1, pGPB2, pGPA have been described (28). The construction of plasmid pGPB2TAP, in which the TAP tag is fused to the C-terminus of the PB2 ORF was described previously (15). The origin of anti-PB1 serum and anti-PB2 and anti-PA monoclonal antibodies has been described (9,29,30).
Purification of polymerase
Cultures of COS-1 cells were infected with vTF7-3 virus at a moi of 5 pfu/cell and subsequently transfected with a mixture of plasmids containing (for a 100 mm dish) pGPB1 (3 µg), either pGPB2 or pGPB2TAP (3 µg) and pGPA (0.6 µg) using cationic liposomes (9). After 16–18 h incubation at 37°C, the cells were lysed for 2 h at 0°C in buffer A (10 mM Tris–HCl, 1 mM EDTA, 7.5 mM ammonium sulphate, 0.025% Igepal CA-630, 1 mM DTT, pH 7.9) containing RNAse A (400 µg/ml) and protease inhibitors (Roche, complete). The extract was centrifuged for 15 min at 10 000g and diluted 8-fold in IgG-binding buffer (10 mM Tris–HCl pH 8, 150 mM NaCl, 1% Igepal CA-630, -polyoxyethylene(9)nonylphenylether, Sigma-). The diluted extract was incubated overnight at 4°C with IgG-Sepharose beads (Amersham-Pharmacia). The resin was transferred to a column and washed with 50 volumes of the same buffer and further with 25 volumes of TEV cleavage buffer (10 mM Tris–HCl pH 8, 150 mM NaCl, 0.1% Igepal CA-630, 0.5 mM EDTA). The resin was incubated with the optimal amount of TEV protease for 3 h at room temperature and the supernatant was recovered. After dilution with calmodulin-binding buffer (10 mM Tris–HCl pH 8, 150 mM NaCl, 0.1% Igepal CA-630, 1 mM Imidazol, 1 mM Mg(AcO)2, 2 mM CaCl2), the sample was incubated overnight at 4°C with Calmodulin-agarose (Stratagene). The resin was washed with 200 volumes of the same buffer and the protein retained was eluted with calmodulin-elution buffer (10 mM Tris–HCl pH 8, 150 mM NaCl, 0.1% Igepal CA-630, 1 mM Mg(AcO)2, 2 mM EGTA).
Biochemical analyses
To test the activity of recombinant polymerase, HEK293T cells were infected with vTF7-3 virus and transfected as indicated above and extracts were prepared as described (31). In vitro synthesis was carried out in a reaction containing a panhandle analogue (5′-AGUAGAAACAAGGCC-3′ plus 5′- GGCCUGCUUUUGCU-3′), as described (31). In brief, the reactions contained, in a final volume of 20 µl, 10 µl of nuclear extract, 100 pmol of panhandle template and 20 µCi of α-32P-GTP in the presence of 50 mM Tris–HCl, pH 7.4, 50 mM KCl, 10 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1 mM ApG, 1 mM ATP, 1 mM UTP, 0.5 mM CTP, 1 µM GTP and 1 u/µl HPRI. After incubation for 1 h at 30°C, the reaction mixture was extracted with phenol, excluded in a Sephadex G25 column and analysed by electrophoresis in a 18% polyacrylamide–urea denaturing gel.
Western blot was carried out as described (32). Cell extracts were separated by polyacrylamide–SDS gel electrophoresis and transferred to nylon filters (Immobilon, Millipore). The membranes were saturated with 3% bovine serum albumin for 1 h at room temperature and incubated with either anti-PB1 serum, anti-PA or anti-PB2 monoclonal antibodies for 1 h at room temperature. The filters were washed with PBS containing 0.25% Tween 20 and incubated with either goat anti-rabbit IgG or goat anti-mouse IgG conjugated to horseradish peroxidase. After further washing as above, they were developed by enhanced chemiluminiscence. Silver staining was carried out as described (15).
Electron microscopy, image analyses and 3D reconstruction
Five microlitres of the purified polymerase preparations were applied to carbon-coated grids after glow-discharge and stained with 1% (w/v) uranyl acetate. Images were obtained using a JEOL 1230 electron microscope operated at 100 kV and a nominal magnification of X50 000, and recorded on Kodak 4489 film under low-dose conditions. To help increase the angular distribution of views from the polymerase, micrographs were collected both at 0° tilt and after tilting the specimen holder by 12°. Plates were digitized on a Dimage Scan Multi Pro scanner (Minolta) at 2400 dpi and down-sampled to a final sampling window of 4.2 Å/pixel at the specimen.
A collection of 5909 images of single molecules was extracted from the micrographs and used to reconstruct the 3D structure of the polymerase by angular refinement methods using the EMAN package (33). In order to minimize model bias, several initial volumes were created to act as initial references for the iterative cycles of refinement. Several featureless Gaussian blobs generated with the ‘makeinitialmodel.py’ command in EMAN were employed as starting models for image processing. After some cycles of refinement, all starting volumes were found to converge into similar structures and one of these was then taken for further refinement.
Furthermore, two different data sets, obtained in two instruments evolved to similar structures during refinement (data not shown). At the final step of the image processing there was a good agreement between the averages and the projections from the reconstruction and the 2D averages were found to cover the entire Fourier space, sufficient to perform a reliable 3D reconstruction of the polymerase. Resolution was estimated to be 26 Å by Fourier Shell Correlation of two independent volumes using half of the data set and the ‘eotest’ command in EMAN with a cut off value of 0.5. The handedness was selected to match the one already defined in the previous work (15), but the opposite handedness cannot be ruled out. Volumes were rendered to show 100% of their mass assuming an average density of 1.35 g/ml and displayed using UCSF Chimera (34) from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). Fitting between the reconstructions of the soluble polymerase (this work) and the RNP-associated protein (15) was performed using the ADP_EM platform (35).
RESULTS
Expression and purification of TAP-tagged influenza polymerase complex
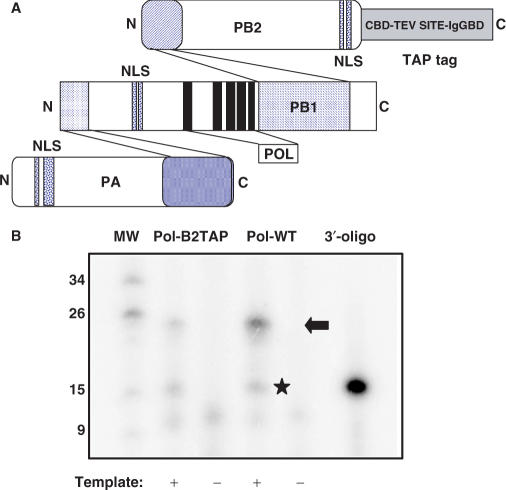
Earlier studies from our group led to the proposal of 3D models for recombinant influenza RNPs and the included polymerase complex (9,12,15). The biological materials used for these studies were RNPs amplified by replication in vivo, after transfection of cDNAs expressing the polymerase subunits, the NP and a model template vRNA. In order to study the isolated polymerase complex, we used a similar expression strategy but omitted the expression of the NP and the template. In addition, we switched from a His-tagged to a TAP-tagged polymerase (Figure 1A) to take advantage of the capacity for dual affinity chromatography implied in the TAP technology (36). The use of the TAP tag for the structural analysis of the polymerase had been previously validated, as complete RNPs with the TAP tag at the C-terminus of PB2 were able to replicate in vivo to an extent similar to wt RNPs (15) (E. Area, unpublished data). This is consistent with the activity of RNPs in which the PB2 subunit had been fused with GFP at its C-terminus (37). To verify further the biological activity of the TAP-tagged polymerase we measured the activity in vitro of cell extracts containing the TAP-tagged polymerase complex using a panhandle analogue as template. The results of one such an experiment are presented in Figure 1B and show the presence of ApG-dependent transcript (Figure 1B, star) as well as the cap-snatching transcript (Figure 1B, arrow), presumably derived from the utilization of endogenous cell mRNAs. Evaluation of a series of experiments indicated that the synthesis of the ApG transcript was similar for the wt or the TAP-tagged polymerase whereas the synthesis of the capped transcript was 2–3-fold less efficient in reactions with the tagged enzyme. The significance of this observation is not clear at this point in time but may reflect slight alteration in the cap-binding activity of the tagged polymerase.
Figure 1.
In vitro activity of TAP-tagged polymerase. (A) Diagram showing the general organization of the polymerase complex including interaction domains (52–56), the position of the subunit NLSs (57–60) and the polymerase motifs within PB1 (61). Also shown is the structure of the TAP tag, fused to the C-terminus of PB2, with the calmodulin-binding domain (CBD), the TEV cleavage site and the IgG-binding domain (IgGBD). (B) The RNA synthesis activity of wild-type (Pol-WT) or TAP-tagged (Pol-B2TAP) polymerase-containing cell extracts was determined in vitro in the presence (+) or absence (−) of panhandle template, as indicated under the Materials and Methods section. The reaction products were analysed on a 18% polyacrylamide–urea gel along with molecular weight markers (MW), whose length in nt are indicated to the left. The 3′-terminal oligonucleotide present in the panhandle (3′-oligo) was terminally labelled with polynucleotide kinase and loaded in parallel as a size marker. The arrow indicates the position of the cap-snatched transcript and the star shows the position of the ApG-dependent transcript.
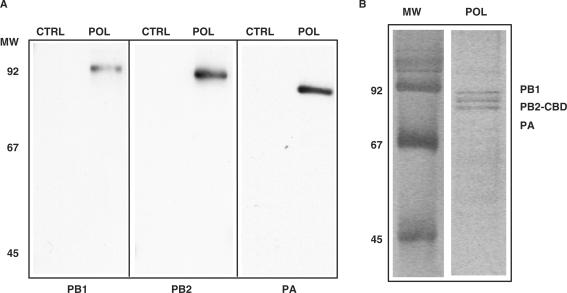
The TAP technique is especially appropriate for purification of intracellular complexes and was modified as indicated in the Materials and Methods section in an attempt to eliminate the host factors that co-purify with the polymerase complex. The material obtained after both chromatography steps was analysed by western blot with antibodies specific for each polymerase subunit and the results are presented in Figure 2A. All three subunits were present in the purified preparations (Figure 2A, POL), in contrast with the results obtained when an untagged polymerase was expressed and purified in parallel as a control (Figure 2A, CTRL). In agreement with the western-blot results, the analysis of the purified material by polyacrylamide gel electrophoresis and silver-staining indicated that the three subunits of the polymerase are present in equimolar amounts and, furthermore, that they are the most abundant proteins in the sample (Figure 2B, POL).
Figure 2.
Purification of soluble polymerase complexes. (A)Western blot analyses of purified polymerase complex (POL) using subunit-specific antibodies, as indicated below each panel. Control preparations (CTRL) consisted of parallel purifications of untagged polymerase expressed under the same conditions. The positions of molecular weight markers (MW) are indicated to the left. (B) Silver staining of purified polymerase complex. The position of the polymerase subunits (PB1, PB2-CBD, PA) is indicated on the right. The mobility of molecular weight markers is indicated on the left (MW).
3D reconstruction of a soluble polymerase heterotrimer
The 3D structure of the purified soluble polymerase heterotrimer was studied by electron microscopy of negatively stained samples. The observation of the polymerase at liquid nitrogen temperatures (cryo-EM) was hampered by the small size of this protein complex (∼250 kDa), in the lower range susceptible of these methods, and the low amount and concentration of the material recovered after purification. Micrographs revealed fields of clearly identified molecule images (Figure 3A), mostly displaying a rounded shape and dimensions compatible with individual molecules of the polymerase (diameter between ∼42 and 115 Å). Some very small particles were also found as a background in the micrographs but their sizes were too small to correspond to an assembled polymerase complex. They most likely represented single PB2-polymerase subunits since this subunit was the one tagged during purification, and they were not selected. Nevertheless, when the soluble polymerase was analysed in a gel filtration column most of the sample run as a ∼250-kDa trimeric complex indicating that unassembled subunits are a minority in the preparation. The soluble polymerase eluted after size exclusion chromatography was too diluted to be detected directly at the electron microscope.
Figure 3.
Electron microscopy of the soluble polymerase heterotrimer. (A) Image of an electron microscope field of purified polymerase, after negative staining. The bar indicates 250 Å. (B) Gallery of individual polymerase images. (C) Diagram of the Euler angular coverage of the collected data after refinement as displayed by EMAN (33), where each column within the triangle shows a height and grey level according to the number of images per class. It is revealed that most of the triangle is covered, indicating a good angular distribution, though some classes (therefore orientations) are more populated than others. (D) A few projections from the final 3D reconstruction (top row) and their corresponding class averages (bottom row) are shown.
A collection of 5909 images from individual polymerase complexes was extracted, a fraction of which were obtained after tilting the specimen holder to help increase the angular coverage in the different views of the protein (see a gallery of single images in Figure 3B). Processing of EM images derived from small asymmetric proteins still represents an important challenge (23) but advances in the methodology has allowed some proteins in the 100-kDa range to be solved at medium resolution with the help of staining agents (38,39). We have found that, because of the absence of symmetry and the small size of the molecules, the initial reference for refinement can introduce bias during the reconstruction procedure and lead towards a local minimum. The chances of getting into one of these local minima can be better avoided by using starting volumes that have no reminiscence to the actual protein apart from its overall size. Consequently and in order to avoid any possible bias from our previous work on the polymerase of influenza virus (15), images were processed from featureless blobs just accounting for the general size of the molecule (see the Materials and Methods section). At the final stage of refinement, there was a good agreement between the average images of the final classes and the corresponding projections of the reconstruction (see a sample of the 280 classes used for the reconstruction in Figure 3D). Refinement and reconstruction statistics are supplied in Table 1.
Table 1.
Statistics of the refinement and 3D reconstruction procedure
| Nominal magnification | 50 000 |
| Number of particles extracted from micrographs | 5909 |
| Number of particles used in the final reconstruction | 4326 |
| Resolution of scanning | 2400 dpi |
| Resolution of extracted particles | 2.1 Å/pixel |
| Resolution of extracted particles after down-sampling | 4.2 Å/pixel |
| Number of different 2D averages obtained after classification of the particles during refinement | 280 |
| Number of single particle images per 2D class average | Between 8 and 10 |
| Angular step in the last round of refinement | 6° |
| Resolution of 3D reconstruction at FSC 0.5 | 26 Å |
| Resolution of 3D reconstruction at FSC 0.14 | 19 Å |
| Cross-correlation with RNP-bound polymerase | 0.63 |
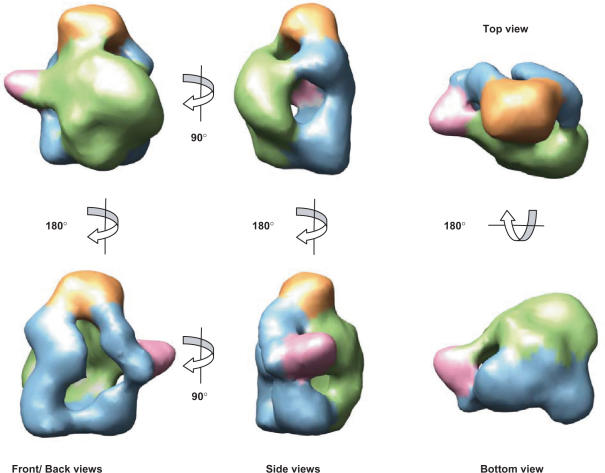
The final model obtained for the soluble polymerase had a resolution of 26 Å and displayed a hollow, globular conformation (Figure 4, structural regions have been coloured differently). The front view of the complex revealed a bulky domain (Figure 4—green) crowned at its top by a small globular head (Figure 4—orange), whereas a small horn projects outwards (Figure 4—pink). In contrast to the front side of the polymerase, its backside (Figure 4—blue) showed an aperture that leads to the interior of a small cavity. The head domain (Figure 4—orange) acted as a structural feature connecting both faces of the complex (Figure 4, top view). Side views revealed the clear transitions between the two faces of the complex.
Figure 4.
3D model for the soluble polymerase heterotrimer. Several views of the final model are presented with structural areas coloured differently. Protein density at the front has been coloured green whereas this at the back is coloured blue. The top small ‘head’ domain is shown in orange while a small outwards protrusion is coloured pink.
Comparison of the 3D model of the soluble complex with the structure of the polymerase in the viral RNP
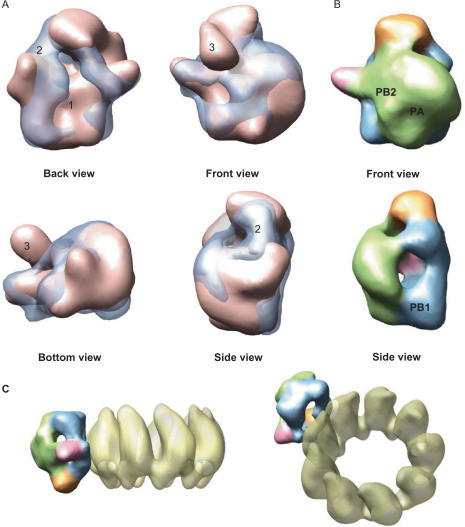
Visual inspection revealed that there was resemblance between the general structural features found in the soluble polymerase and those described previously in the protein incorporated to the RNP (15). In order to perform a more systematic examination of the similarities and differences between the polymerase in its free and RNP-bound conformation, the two reconstructions were fitted using a full 6D search and unbiased computational methods implemented in the new ADP_EM platform (35). The 3D reconstruction of the RNP-bound polymerase (Figure 5A—light pink) was used to scrutinize all possible fittings with that of the soluble protein (Figure 5A—blue transparency) by testing all the relative translations and rotations. The matches were evaluated by calculation of the cross-correlation coefficient and the best solutions revealed that the overall outline of the two structures was very similar. Actually, the cross-correlation coefficient between the two reconstructions (0.63) was a good indicative of such similarity. The resemblance between the two 3D reconstructions was therefore sufficient to correlate corresponding regions of each structure. This allowed us to map the location of the PB1 and PA subunits identified previously by antibody-labelling of the RNP-associated protein (15) in the soluble polymerase (Figure 5B). The position of the PB2 subunit is proposed to be at a slightly different place based on the conformational changes detected between the two reconstructions. In order to visibly reveal the areas of the soluble polymerase implicated in RNP binding, we also modelled the orientation of the soluble protein within a putative RNP (Figure 5C) using the published structure of RNPs (15) and substituting the polymerase in the compatible orientation according to the fitting results. As it is observed (Figure 5C), the backside of the polymerase (blue), that shows an aperture to an internal cavity, faces towards the RNP subunits. Importantly, despite the similitude, clear differences between the soluble and the RNP-bound structures were also found. They mainly focus in three areas (labelled as 1–3 in Figure 5A) and were analysed more thoroughly (Figure 6).
Figure 5.
Fitting between soluble and RNP-associated polymerase. (A) Fitting between the 3D reconstructions of the soluble (blue transparency) and the RNP-bound (pink solid density) polymerase, which are shown over-imposed. (B) Locations of the subunits in the soluble polymerase denoted as a result of its comparison with the model for the polymerase bound to the RNP (15) where these were mapped by antibody labelling. (C) Chimeric model using the 3D structures of an RNP (15) and of the soluble polymerase. The backside of the polymerase (blue) contains the sites of the interaction with the NP monomers whereas the front side (green) is opposite to the NP ring (15).
Figure 6.
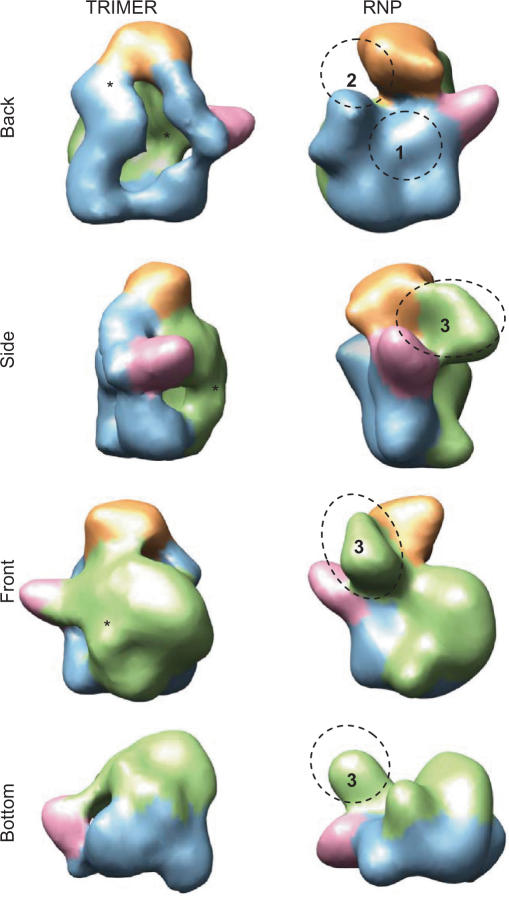
Comparison of the 3D structures of soluble and RNP-associated polymerase complexes. Views of the model for the isolated polymerase (TRIMER) and the RNP-associated polymerase (RNP) are compared. Structural features in the RNP-bound protein have been coloured as the corresponding areas in the soluble polymerase, revealed after the computational fitting between both reconstructions. Asterisks indicate areas of conformational changes found in the soluble protein and their corresponding location in the RNP-bound polymerase are denoted with numbers.
We investigated the similarities and differences between the soluble polymerase heterotrimer (Figure 6, labelled TRIMER) and that for the polymerase present in the RNP (Figure 6, labelled RNP). The general appearance of both models is similar and domains can be easily traced from one protein to the other also thanks to fitting performed with ADP_EM (35) (see for instance the coincidence in the front and bottom views in Figure 6 and the position of the ‘horn’, Figure 6—pink). Accordingly, structural features of the RNP-bound polymerase have been coloured following the same code as that used for the equivalent area in the soluble protein. The overall conformation of the RNP's polymerase appears more compact than the model for the soluble heterotrimer, as if the complex would adopt a more closed conformation when it is present in the RNP structure. This conformational difference is more evident in the back view of the proteins where there is an opening to an internal cavity and a connectivity in the soluble polymerase which are absent in the RNP-bound conformation (labelled with asterisks and as 1 and 2, respectively). Importantly, this region corresponds to the areas implicated in the interactions with the RNP (Figure 5C) (15).
Interestingly, a bulge detected in the front, side and bottom views of the RNP's polymerase (Figure 6, labelled as 3) where the N-terminal region of the PB2 subunit was mapped (15), is not present in the model for the soluble heterotrimer. On the other hand, the front view of the polymerase from the RNP shows a mass density depression that in the soluble protein is filled with a bulky mass (labelled with an asterisk in Figure 6, front view). Given that all other large densities in the polymerase seem to have stayed in similar positions, this change could be most likely interpreted as if the bulge would be folded down and leftwards, contributing to the massive aspect of the front view of the soluble heterotrimer. These structural differences are also clear in the bottom view (Figure 6, Bottom, labelled as 3).
DISCUSSION
At present, the molecular basis for the structural differences observed for the soluble versus RNP-associated polymerase is not clear, but some possibilities can be considered. First of all, the soluble heterotrimer whose structure is presented in this report lacks the template RNA, which is present in the recombinant RNP modelled previously (15). The vRNA promoter structure comprises both template termini and would include ∼15 nt from each end, i.e. a molecular mass of around 10 kDa (40)[reviewed in(2)]. The promoter-binding site in the polymerase includes not only residues of PB1 (41,42) but also from other subunits in the complex (43) and is probably located internally in the polymerase. Hence, one would expect to have an extra internal mass in the polymerase present in the RNP as compared to the soluble heterotrimer. The size of the promoter would not permit its direct mapping by electron microscopy, but the lack of a cavity in the RNP-bound conformation may result of the exclusion of the staining agent due to its partial occupancy by RNA. In addition, the interaction of the subunits with the promoter could induce conformational changes to adopt a more closed structure and, indeed, stabilization of the polymerase complex after binding to the promoter RNA panhandle structure has been reported (31). This proposal is not without precedents: (i) the recognition of a double-strand break during non-homologous end-joining DNA repair depends on the DNA-dependent protein kinase (DNA-PK), which contains a 460 kDa large catalytic subunit (DNA-PKcs). Its activity as protein kinase is greatly stimulated by interaction with DNA and such interaction is accompanied by a significant conformational change (44). Thus, DNA binding takes place in the cavity of the DNA-PKcs and correlates with large alterations in the relative positions of various domains in the protein and with the activation of the enzyme. (ii) A comparison of the T7 RNA polymerase structure with those of the initiation and elongation complexes indicates that considerable conformational changes occur during the switch from initiation to elongation of RNA synthesis (45). Thus, dissociation from the promoter region involves rotation and translation of protein domains as well as rearrangements of secondary structure in the polymerase, including the formation of an elongated RNA exit channel (46).
In addition to the absence of template binding, the soluble heterotrimer lacks the interaction with two adjacent NP monomers that are present in the recombinant RNP. These interactions are distinct (15) and probably involve the PB1 and PB2 subunits of the polymerase (47). Importantly, our model for the soluble polymerase shows extensive conformational rearrangements in the backside of the protein implicated in the contacts with the RNP (Figure 5C). It is fair therefore to assume that the absence of these contacts leads to alternative conformations of the domains implicated in these interactions and possibly also of distant regions of the polymerase complex. Furthermore, the polymerase described in the model for a recombinant RNP (12,15) reflects the situation of a resting RNP which is not in the process of RNA synthesis and may change as the polymerase complex reads out the sequence in the template RNA. Hence, the polymerase structure proposed in the model for the soluble heterotrimer could represent one of the potential conformations that could occur in the process of transcription or replication of a vRNP.
Intriguingly, a large domain containing the N-terminus of the PB2 subunit in the RNP-bound polymerase (Figure 6—label 3) is not detected in the soluble protein. A possibility could be that this domain has flipped back onto the body of the protein (Figure 6—label with an asterisk in the front view). Such a large displacement must certainly influence the function of the polymerase and it might reflect a distinct functional state. In this regard, the 3D model proposed for the soluble polymerase heterotrimer might be considered as the core conformation of a molecular machine able to perform various functions as a result of its interaction with a number of different partners. The interaction of this structure with distinct partners will trigger conformational changes that will drive the protein to an alternative conformation capable of performing a new function. For instance, the heterotrimer has to interact with the promoter region of the vRNA template during virus transcription, This allows recognition of cellular capped-RNAs and the generation of the capped-oligonucleotide primers (19, 42, 48, 49) and may imply a structural rearrangement of the protein complex. Such a minimal polymerase-template set is capable of elongation of an ApG or capped primer (50), but longer templates require an RNP structure for full-length transcription (51). Under these conditions, the conformational changes triggered after polymerase–NP interactions may be essential for the correct read-out of the template sequence and for the processivity of the enzyme. It is reasonable to assume that these polymerase–NP association may imply further structural alterations in the polymerase complex and, furthermore, that these should be reversible.
In addition to its obvious roles in RNA transcription and replication, the polymerase heterotrimer has been implicated in other unrelated functions. Thus, by means of its interaction with the virus-specific 5′-proximal sequences of viral mRNAs and the cap structure, the complex might be responsible for the protection of these transcripts from cap snatching by vRNPs (22). Furthermore, the specific complex between the polymerase heterotrimer and virus M1 mRNA might be responsible for the down-regulation of the splicing event leading to mRNA3 and the corresponding stimulation of M2 mRNA production during virus infection (21). These proposals would imply that the capped virus mRNA terminus binds the polymerase in a way different from cell pre-mRNA recognition during cap snatching, since it would occupy the cRNA template-binding site as well as the cap-binding site. Again, a yet different polymerase conformation might be responsible for this interaction, as compared to the basic heterotrimer structure or the polymerase present in the RNP.
In summary, we have generated a 3D model for the soluble influenza polymerase heterotrimer that should set the basis for future structural analyses. Our model could be used as a framework into which atomic structural information of specific polymerase subunits or domains could be docked.
ACKNOWLEDGEMENTS
We thank B. Moss, B. Seraphin, A. Portela, A. Nieto, for providing biological materials. The technical assistance of J. Fernández, D. Guilligay and N. Zamarreño is gratefully acknowledged. E.T. was a fellow from Instituto de Salud Carlos III. This work was supported by the Spanish Ministry of Education and Science (Ministerio de Educación y Ciencia) (grants BMC2001-1223 and BFU2004-491 to JO and SAF2005-00775 and GEN2003-20239-C06-06 to OLL), the European Vigilance Network for the Management of Antiviral Drug Resistance (VIRGIL) and the FLUPOL strep project (SP5B-CT-2007-044263). Funding to pay the Open Access publication charges for this article was provided by Grant BFU2004-00491.
Conflict of interest statement. None declared.
REFERENCES
- 1.Palese P, Shaw M. In: Fields Virology. Howley PM, editor. 2006. Vol. 1, 5th edn. Lippincott Williams & Wilkins, Philadelphia, pp. 1647–1689. [Google Scholar]
- 2.Elton D, Digard P, Tiley L, Ortín J. In: Current Topics in Influenza Virology. Kawaoka Y, editor. Norfolk: Horizon Scientific Press; 2005. pp. 1–92. [Google Scholar]
- 3.Krug RM, Broni BA, Bouloy M. Are the 5’-ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell. 1979;18:329–334. doi: 10.1016/0092-8674(79)90052-7. [DOI] [PubMed] [Google Scholar]
- 4.Robertson JS, Schubert M, Lazzarini RA. Polyadenylation sites for influenza mRNA. J. Virol. 1981;38:157–163. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon LLM, Pritlove DC, Fodor E, Brownlee GG. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 1999;73:3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hay AJ, Skehel JJ, McCauley J. Characterization of influenza virus RNA complete transcripts. Virology. 1982;116:517–522. doi: 10.1016/0042-6822(82)90144-1. [DOI] [PubMed] [Google Scholar]
- 7.Pons MW, Schulze IT, Hirst GK. Isolation and characterization of the ribonucleoprotein of influenza virus. Virology. 1969;39:250–259. doi: 10.1016/0042-6822(69)90045-2. [DOI] [PubMed] [Google Scholar]
- 8.Jennings PA, Finch JT, Winter G, Robertson JS. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1983;34:619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- 9.Ortega J, Martín-Benito J, Zürcher T, Valpuesta JM, Carrascosa JL, Ortín J. Ultrastructural and functional analyses of of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 2000;74:156–163. doi: 10.1128/jvi.74.1.156-163.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murti KG, Webster RG, Jones IM. Localization of RNA polymerases of influenza viral ribonucleoproteins by immunogold labeling. Virology. 1988;164:562–566. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 11.Klumpp K, Ruigrok RW, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martín-Benito J, Area E, Ortega J, Llorca O, Valpuesta JM, Carrascosa JL, Ortín J. Three dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2001;2:313–317. doi: 10.1093/embo-reports/kve063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gastaminza P, Perales B, Falcón AM, Ortín J. Influenza virus mutants in the N-terminal region of PB2 protein are affected in virus RNA replication but not transcription. J. Virol. 2003;76:5098–5108. doi: 10.1128/JVI.77.9.5098-5108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 2002;76:8989–9001. doi: 10.1128/JVI.76.18.8989-9001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Area E, Martín-Benito J, Gastaminza P, Torreira E, Valpuesta JM, Carrascosa JL, Ortín J. Three-dimensional structure of the influenza virus RNA polymerase: localization of subunit domains. Proc. Natl Acad. Sci. USA. 2004;101:308–313. doi: 10.1073/pnas.0307127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Detjen BM, St Angelo C, Katze MG, Krug RM. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J. Virol. 1987;61:16–22. doi: 10.1128/jvi.61.1.16-22.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiley LS, Hagen M, Mathews JT, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5’-end of the viral RNAs. J. Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González S, Ortín J. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 1999;18:3767–3775. doi: 10.1093/emboj/18.13.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MT, Klumpp K, Digard P, Tiley L. Activation of influenza virus RNA polymerase by the 5’ and 3’ terminal duplex of genomic RNA. Nucleic Acids Res. 2003;31:1624–1632. doi: 10.1093/nar/gkg253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vreede FT, Jung TE, Brownlee GG. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 2004;78:9568–9572. doi: 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shih SR, Nemeroff ME, Krug RM. The choice of alternative 5’ splice sites in influenza virus M1 mRNA is regulated by the viral polymerase complex. Proc. Natl Acad. Sci. USA. 1995;92:6324–6328. doi: 10.1073/pnas.92.14.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shih SR, Krug RM. Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology. 1996;226:430–435. doi: 10.1006/viro.1996.0673. [DOI] [PubMed] [Google Scholar]
- 23.Llorca O. Introduction to 3D reconstruction of macromolecules using single particle electron microscopy. Acta Pharmacol. Sin. 2005;26:1153–1164. doi: 10.1111/j.1745-7254.2005.00203.x. [DOI] [PubMed] [Google Scholar]
- 24.Gluzman Y. SV40 transformed simian cells support the replication or early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 25.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortín J, Nájera R, López C, Dávila M, Domingo E. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene. 1980;11:319–331. doi: 10.1016/0378-1119(80)90072-4. [DOI] [PubMed] [Google Scholar]
- 27.Fuerst TR, Earl PL, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mena I, de la Luna S, Albo C, Martín J, Nieto A, Ortín J, Portela A. Synthesis of biologically active influenza virus core proteins using a vaccinia-T7 RNA polymerase expression system. J. Gen. Virol. 1994;75:2109–2114. doi: 10.1099/0022-1317-75-8-2109. [DOI] [PubMed] [Google Scholar]
- 29.Bárcena J, Ochoa M, de la Luna S, Melero JA, Nieto A, Ortín J, Portela A. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J. Virol. 1994;68:6900–6909. doi: 10.1128/jvi.68.11.6900-6909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochoa M, Bárcena J, de la Luna S, Melero JA, Douglas AR, Nieto A, Ortín J, Skehel JJ, Portela A. Epitope mapping of cross-reactive monoclonal antibodies specific for the influenza A virus PA and PB2 polypeptides. Virus Res. 1995;37:305–315. doi: 10.1016/0168-1702(95)00039-s. [DOI] [PubMed] [Google Scholar]
- 31.Brownlee GG, Sharps JL. The RNA polymerase of influenza a virus is stabilized by interaction with its viral RNA promoter. J. Virol. 2002;76:7103–7113. doi: 10.1128/JVI.76.14.7103-7113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marión RM, Zürcher T, de la Luna S, Ortín J. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 1997;78:2447–2451. doi: 10.1099/0022-1317-78-10-2447. [DOI] [PubMed] [Google Scholar]
- 33.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 34.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Garzon JI, Kovacs J, Abagyan R, Chacon P. ADP_EM: fast exhaustive multi-resolution docking for high-throughput coverage. Bioinformatics. 2007;23:427–433. doi: 10.1093/bioinformatics/btl625. [DOI] [PubMed] [Google Scholar]
- 36.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 37.Fodor E, Smith M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 2004;78:9144–9153. doi: 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llorca O, Arias-Palomo E, Zugaza JL, Bustelo XR. Global conformational rearrangements during the activation of the GDP/GTP exchange factor Vav3. EMBO J. 2005;24:1330–1340. doi: 10.1038/sj.emboj.7600617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okorokov AL, Orlova EV, Kingsbury SR, Bagneris C, Gohlke U, Williams GH, Stoeber K. Molecular structure of human geminin. Nat. Struct. Mol. Biol. 2004;11:1021–1022. doi: 10.1038/nsmb835. [DOI] [PubMed] [Google Scholar]
- 40.Flick R, Neumann G, Hoffmann E, Neumeier E, Hobom G. Promoter elements in the influenza vRNA terminal structure. RNA. 1996;2:1046–1057. [PMC free article] [PubMed] [Google Scholar]
- 41.González S, Ortín J. Characterization of the influenza virus PB1 protein binding to vRNA: two separate regions of the protein contribute to the interaction domain. J. Virol. 1999;73:631–637. doi: 10.1128/jvi.73.1.631-637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li ML, Ramírez BC, Krug RM. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 1998;17:5844–5852. doi: 10.1093/emboj/17.19.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fodor E, Pritlove DC, Brownlee GG. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivera-Calzada A, Maman JD, Spagnolo L, Pearl LH, Llorca O. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) Structure. 2005;13:243–255. doi: 10.1016/j.str.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 45.Steitz TA. The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase. Curr. Opin. Struct. Biol. 2004;14:4–9. doi: 10.1016/j.sbi.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 47.Biswas SK, Boutz PL, Nayak DP. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 1998;72:5493–5501. doi: 10.1128/jvi.72.7.5493-5501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao P, Yuan W, Krug RM. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 2003;22:1188–1198. doi: 10.1093/emboj/cdg109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cianci C, Tiley L, Krystal M. Differential activation of the influenza virus polymerase via template RNA binding. J. Virol. 1995;69:3995–3999. doi: 10.1128/jvi.69.7.3995-3999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee MT, Bishop K, Medcalf L, Elton D, Digard P, Tiley L. Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 2002;30:429–438. doi: 10.1093/nar/30.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Honda A, Ueda K, Nagata K, Ishihama A. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J. Biochem. (Tokyo) 1988;104:1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- 52.Poole E, Elton D, Medcalf L, Digard P. Functional domains of the influenza A virus PB2 protein: identification of NP- and PB1-binding sites. Virology. 2004;321:120–133. doi: 10.1016/j.virol.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 53.González S, Zürcher T, Ortín J. Identification of two separate domains in the influenza virus PB1 protein responsible for interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 1996;24:4456–4463. doi: 10.1093/nar/24.22.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pérez DR, Donis RO. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J. Virol. 1995;69:6932–6939. doi: 10.1128/jvi.69.11.6932-6939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toyoda T, Adyshev DM, Kobayashi M, Iwata A, Ishihama A. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J. Gen. Virol. 1996;77:2149–2157. doi: 10.1099/0022-1317-77-9-2149. [DOI] [PubMed] [Google Scholar]
- 56.Zürcher T, de la Luna S, Sanz-Ezquerro JJ, Nieto A, Ortín J. Mutational analysis of the influenza virus A/Victoria/3/75 PA protein: Studies of interaction with PB1 protein and identification of a dominant negative mutant. J. Gen. Virol. 1996;77:1745–1749. doi: 10.1099/0022-1317-77-8-1745. [DOI] [PubMed] [Google Scholar]
- 57.Mukaigawa J, Nayak DP. Two signals mediate nuclear localization of influenza virus (A/WSN/33) polymerase basic protein 2. J. Virol. 1991;65:245–253. doi: 10.1128/jvi.65.1.245-253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nieto A, de la Luna S, Bárcena J, Portela A, Ortín J. Complex structure of the nuclear translocation signal of the influenza virus polymerase PA subunit. J. Gen. Virol. 1994;75:29–36. doi: 10.1099/0022-1317-75-1-29. [DOI] [PubMed] [Google Scholar]
- 59.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 2007;14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 60.Nath ST, Nayak DP. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1N1) Mol. Cell. Biol. 1990;10:4139–4145. doi: 10.1128/mcb.10.8.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biswas SK, Nayak DP. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 1994;68:1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]