Abstract
Autonomous parvoviruses induce severe morphological and physiological alterations in permissive host cells, eventually leading to cell lysis and release of progeny virions. Viral cytopathic effects (CPE) result from specific rearrangements and destruction of cytoskeletal micro- and intermediate filaments. We recently reported that inhibition of endogenous casein kinase II (CKII) protects target cells from parvovirus minute virus of mice (MVM)-induced CPE, pointing to this kinase as an effector of MVM toxicity. The present work shows that the parvoviral NS1 protein mediates CKII-dependent cytoskeletal alterations and cell death. NS1 can act as an adaptor molecule, linking the cellular protein kinase CKIIα to tropomyosin and thus modulating the substrate specificity of the kinase. This action results in an altered tropomyosin phosphorylation pattern both in vitro and in living cells. The capacity of NS1 to induce CPE was impaired by mutations abolishing binding with either CKIIα or tropomyosin. The cytotoxic adaptor function of NS1 was confirmed with fusion peptides, where the tropomyosin-binding domain of NS1 and CKIIα are physically linked. These adaptor peptides were able to mimic NS1 in its ability to induce death of transformed MVM-permissive cells.
Keywords: parvovirus minute virus of mice, protein–protein interactions, tropomyosin, protein kinase specificity
Rodent parvoviruses (PVs) are promising candidates for the virotherapy of cancer because they exert oncolytic action without showing pathogenicity (1). The PV minute virus of mice (MVM) consists of a small, icosahedral nonenveloped capsid containing a single-stranded 5.1-kb linear DNA. During productive infection, MVM induces dramatic morphological and physiological changes in transformed mouse fibroblasts, culminating in cell death and lysis. The cytotoxicity of MVM is attributed mainly to the large nonstructural protein NS1. This 83-kDa multifunctional viral product is endowed with enzymatic and nonenzymatic properties, enabling it to control various processes necessary for progeny particle production and spreading (for a review, see ref. 2). Independently of its functions directly involved in particle production, NS1 exerts specific activities that jeopardize the integrity and survival of infected cells (3, 4). The ability of NS1 to interact physically with host proteins is assumed to play an important role in its cytotoxicity. The interaction of NS1 with proteins involved in DNA replication and transcription suggests that scavenging mechanisms may contribute to NS1-triggered perturbation of host cells, but recent data indicate that NS1 could act in a more specific manner, e.g., by modulating the activity of some partner proteins. The cytotoxicity of NS1 correlates with its ability to bind casein kinase IIα (CKIIα), the catalytic subunit of the cellular casein kinase II, and suppression of CKII activity during infection protects cells from PV-induced cytopathic effects (CPE) (5). This correlation suggests that NS1-mediated modulation of cellular protein kinase (PK) activity could influence both the phosphorylation of NS1 itself (6) and multiple facets of cell metabolism, including signaling cascades and cytoskeleton dynamics, recently shown to be perturbed upon PV infection (5, 7, 8, 10; and S. Lachmann, S. Bär, J.R., and J.P.F.N., unpublished work).
To investigate this hypothesis, we have sought additional cellular partner proteins of NS1, associating with the wild-type (WT) toxic protein but having reduced affinity for nontoxic or less toxic NS1 variants. Using differential affinity chromatography, we have identified the cytoskeletal filament protein tropomyosin (TM) as having an NS1-binding capacity correlating with NS1 cytotoxicity. As a result of NS1 binding, TM becomes a target for the NS1–CKIIα complex, which leads to changes in its phosphorylation pattern. The impact of NS1-dependent retargeting of CKII to new substrates was further investigated in cells expressing a dominant-negative CKIIα mutant and with semisynthetic adaptor proteins mimicking NS1 in its ability to bring together CKIIα and TM. Our results support a model of NS1 cytotoxicity in which the viral protein brings together the catalytic subunit of CKII and new substrates thereof, thereby altering the phosphorylation of these substrates and causing cell disturbances and death.
Results
NS1 Interaction Partners Involved in MVM-Induced CPE.
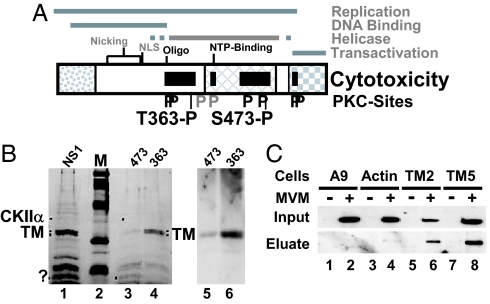
Because mutations abolishing NS1 toxicity appear to cluster in distinct areas of the viral protein (Fig. 1A), it was thought that NS1 might act as a complex with cellular partner proteins to perturb cells (2, 5). Recently, CKIIα was identified as a toxicity-mediating NS1 interaction partner on the basis of its binding to WT NS1 and its lack of affinity for the nontoxic NS1:S473A mutant (5). To identify additional cellular mediators of MVM cytotoxicity, the same approach was followed with a hypotoxic NS1 variant, NS1:T363A, mutated in a different domain of the protein (Fig. 1). To this end, a prefractionated A9 cell extract was subjected to differential affinity chromatography. In a first step, GST-NS1:T363A-coupled columns were used to retain proteins binding to NS1 despite the mutation, and in a second step, the flow-through was run through a column with WT GST-NS1 to capture cellular proteins whose interaction with NS1 is sensitive to T363A (i.e., correlates with NS1 cytotoxicity). Eluted proteins were then analyzed by SDS/PAGE. For comparison, similar experiments were performed without preadsorption or with nontoxic GST-NS1:S473A. Fig. 1B (Left) shows that multiple proteins, including CKIIα (1) and a prominent doublet migrating as proteins of ≈32 kDa, were able to bind to GST-NS1WT (lane 1). GST-NS1:S473A efficiently retained a number of these NS1 interaction partners, including the 32-kDa doublet (lane 3), but GST-NS1:T363A, although able to capture multiple proteins, including CKIIα, failed to bind the two 32-kDa proteins, making them available for WT NS1. Given their differential binding to WT and hypotoxic NS1, these 32-kDa cellular proteins stood as candidate mediators of NS1 cytotoxicity and were characterized by tandem mass spectrometry (MS/MS). Both polypeptides were identified as TM, a cellular cytoskeletal protein polymerizing into microfilaments. This identification was confirmed by Western blotting performed on GST-NS1WT eluates obtained after preadsorption with T363A vs. S473A (Right).
Fig. 1.
Determinants of NS1-induced cytotoxicity. (A) Domain structure of NS1 (5). The common N terminus of NS1 and NS2, the region of homology with the SV40 large T-antigen, and the trans activation domain are represented, respectively, as dotted, cross-hatched, and checkered boxes. The nucleotide-binding (NTP-binding) site, the oligomerization region (Oligo), the nuclear localization signal (NLS), and nicking motifs (metal coordination site and linkage tyrosine) are positioned. Black bars denote NS1 regions mediating cytopathogenicity. Consensus PKC-phosphorylation sites that modulate (black P) or do not modulate (gray P) CPE are indicated, including 363-P and 473-P targeted for differential affinity chromatography. (.B Left) SDS/PAGE and Coomassie blue staining of proteins eluted from GST-NS1 affinity columns. NS1, proteins in A9-P3-DE1 binding to GST-NS1WT; 473, proteins lacking affinity for GST-NS1:S473A and binding to GST-NS1WT; 363, proteins lacking affinity for GST-NS1:T363A and binding to GST-NS1WT. M, molecular mass markers (97, 66, 45, 30, and 20.1 kDa (Rainbow marker, Amersham). Proteins overrepresented in either 474 or 363 are marked with dots. The doublet marked TM was cut from the gel and subjected to MS/MS analysis. (Right) Western blot analyses of both flow-through samples with TM-specific antibodies. (C) Interaction of NS1 with TM in MVM-infected cells. Binding of NS1 to cytoskeletal proteins was estimated on the basis of its affinity for GST-tagged TM2, TM5, and β-actin. Cell extracts from (MVM-infected) A9 cells (lanes 1 and 2) or from derivatives expressing GST-tagged cytoskeleton proteins (β-actin, lanes 3 and 4; TM2, lanes 5 and 6; TM5, lanes 7 and 8) were run through glutathione–Sepharose columns specifically retaining GST-tagged proteins. Binding partners (e.g., NS1) of GST-tagged proteins were eluted with 700 mM NaCl (eluate) and analyzed by Western blotting with anti-NS1C compared with total amounts (input).
To determine which TM subtypes are expressed in A9 cells, RACE cDNA libraries were generated with primers corresponding to the conserved region of TM isoforms. Sequencing of six independent clones each led to the identification of TM2 and TM5. The determined amino acid sequences did not differ from the published fibroblast tropomyosin istotypes (TM2 accession no., NCBI NM 009416; TM5 accession no., NCBI NM 011628). Our screening of A9 cells failed to detect other isoforms such as TM1, a major component present in nontransformed fibroblasts (11). In agreement with our results, it was reported that transformed and normal fibroblasts can be distinguished by their TM isotypes (11).
To confirm the in vitro association of NS1 and TM under in vivo conditions, we generated stably transfected cell lines expressing full-length GST-tagged TM2 or TM5 driven by the PV P38 promoter. Transfectants were infected (or not) with MVM, then cell extracts were prepared and matched for the amount of GST-tagged protein because NS1 is able to activate this promoter (8). To detect the formation of NS1–GST-TM complexes, GST-TM and associated polypeptides were purified on glutathione–Sepharose columns. Proteins specifically binding GST-TM were eluted with high salt and tested for the presence of NS1 by Western blotting. Parental A9 cells and A9-P38:GST-β-actin transfectants served as negative controls. As shown in Fig. 1C, NS1 was detected in eluates from GST-TM5-loaded and, to a lesser extent, GST-TM2-loaded columns. NS1 did not bind nonspecifically to glutathione–Sepharose (A9[MVM]), and little NS1 was retained by GST-β-actin, which strongly suggests that the NS1–TM interaction actually occurs in the cell.
Changes in TM Phosphorylation by NS1–CKIIα.
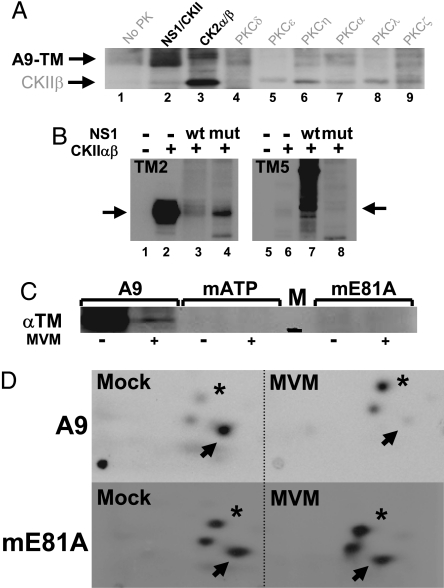
MVM infection is reported to protect microtubules from degradation through activation of PKCλ (10). We therefore tested whether TMs are also PK substrates and whether NS1 might modulate phosphorylation of these cytoskeletal proteins. We first subjected A9 cell-derived TM (A9-TM) (see Fig. 1B, lane 4) to an in vitro kinase assay. In addition to similarly prepared GST-NS1–CKIIα (see Fig. 1B, lane 3; ref. 5), various recombinant PKs were tested for their ability to phosphorylate A9-TM. As shown in Fig. 2A, A9-TM was phosphorylated by NS1–CKIIα (lane 2), whereas little or no phosphorylation was detected with the substrate alone (lane 1) or in the presence of various PKCs (lanes 4–9). Interestingly, recombinant CKIIαβ and NS1–CKIIα showed different substrate “preferences,” the former phosphorylating its regulatory subunit CKIIβ and (weakly) the TM isoform corresponding to the upper band of the A9-TM doublet (lane 3), the latter targeting mainly the faster-migrating TM species. This difference suggests that association of CKIIα with NS1 may modulate the substrate specificity of the PK.
Fig. 2.
In vitro and in vivo phosphorylation of A9 cell tropomyosin. (A and B) In vitro kinase assays of the indicated PK substrate combinations were performed in the presence of [γ-32P]ATP and analyzed by SDS/PAGE and autoradiography. (A) A9-TM purified by NS1 affinity (Fig. 1B lane 4), alone (no PK), or combined with similarly purified NS1–CKIIα (Fig. 1B lane 3) or with the indicated recombinant PK. Migrations of TM (A9-TM) and recombinant CKIIβ are indicated. (B) Phosphorylation of recombinant TM2 (Left) and TM5 (Right) without (lanes 1 and 5) or with CKIIαβ in the absence (lanes 2 and 6) or presence of WT (lanes 3 and 7) or mutant S473A (lanes 4 and 8) GST-NS1. The migration of monomeric TM is indicated by arrows. (C and D) MVM infection and functional CKIIα were tested for their impact on the in vivo phosphorylation of TM. A9 cells or derivatives expressing the dominant-negative CKII(mATP) or CKII(mE81A) were infected (or not) with MVM (30 pfu per cell) and subjected to metabolic 32P labeling. TM was isolated by immunoprecipation and analyzed by SDS/PAGE (C), with reference to a 30-kDa marker (black bar in lane M). Bands corresponding to phospholabeled TM were excised, and their tryptic phosphopeptide patterns were analyzed (D). Phosphopeptides characteristically overrepresented in mock- (arrow) or MVM (asterisk)-infected cells are marked.
This hypothesis was confirmed by subjecting purified bacterially expressed (and hence unmodified) TM2 and TM5 to in vitro kinase assays in which recombinant CKIIαβ was used either alone or in combination with purified WT GST-NS1 or S473A, a mutant impaired in interaction with CKIIα. Interestingly, TM2 emerged as a target of CKIIαβ phosphorylation (Fig. 2B, lane 2), but TM5 did not (lane 6); upon addition of GST-NS1, the CKIIαβ-mediated phosphorylation of TM2 was abolished (lane 3), and TM5 became a substrate for the NS1–CKIIα complex (lane 7). This specificity switch of CKII in the presence of the viral protein suggests that interaction of NS1 with CKIIα alters the substrate-recognition pocket of the PK, allowing it to target new phosphorylation sites. This interpretation is supported by the fact that GST-NS1:S473A failed to drive phosphorylation of TM5 by CKII (lane 8) and could not fully inhibit TM2 phosphorylation (lane 4).
The in vivo relevance of these in vitro data was tested by determining whether MVM infection affects TM phosphorylation in A9 fibroblasts. A9 cells or derivatives expressing dominant-negative CKIIα mutants (5), infected (or not) with MVM, were subjected to metabolic 32P labeling. TM proteins were isolated by immunoprecipitation and purified by SDS/PAGE (Fig. 2C), and their tryptic phosphopeptide patterns were analyzed (10). As shown in Fig. 2D (Upper), A9-TM characteristically resolved into three main 32P-labeled peptides. One of these peptides (marked by an arrow) was phosphorylated to a substantially higher extent in mock- vs. MVM-infected A9 cells, pointing to virus-induced down-regulation of one or more specific TM phosphorylation events. Conversely, a second phosphopeptide (marked with an asterisk) was overrepresented in MVM-treated A9 cells, indicating that infection also triggered one or more novel phosphorylations. Functional inactivation of CKII led to an overall reduction of TM phosphorylation (Fig. 2C), and the above-mentioned MVM-induced changes in the phosphorylation pattern were significantly less apparent (as documented for the mE81A mutant; Fig. 2D). These results confirm the involvement of CKII in both TM phosphorylation and its MVM-dependent modulation in living cells. Altogether, these in vitro and in vivo observations led us to conclude that NS1 induces a CKIIαβ substrate switch from TM2 to TM5.
Role of CKII in MVM-Induced Alteration of TM Filaments.
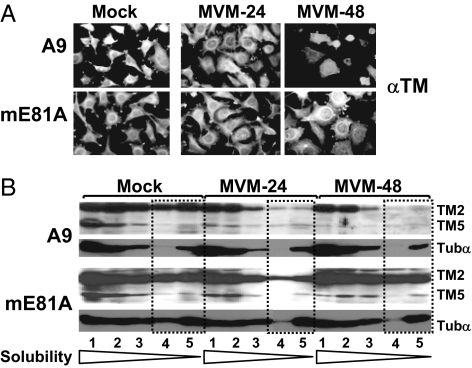
The NS1 interaction partners CKIIα and TM both appeared as candidate cellular mediators of MVM-induced CPE because NS1 mutants lacking affinity for these cellular proteins also show a reduced cytotoxic potential. To investigate the contribution of TM to MVM-induced CPE, the fate of TM filaments was monitored in MVM-infected cells in the presence and absence of functional CKII. A9 cells and A9-P38:CKII(E81A) cells were infected (or not) with 30 pfu MVM per cell and examined 24 and 48 h after infection for the integrity of the TM network by immunofluorescence microscopy (Fig. 3A) and biochemical fractionations (Fig. 3B) (10). In A9 cells, TM filaments became rearranged and degraded on infection, but expression of the dominant-negative CKII variant inhibited this process, protecting a large proportion of the infected A9-P38:CKII(E81A) cells from MVM-induced CPE. These observations were substantiated by analysis of the filament structure by extraction with increasing amounts of detergents. Because microtubules remain protected during MVM infection of A9 cells (10), tubulin α served as a control in these experiments. Upon MVM infection of A9 cells, the amount of TM present in fractions 3–5, corresponding to rigid fiber structures, was low, indicating that TM filaments were degraded under these conditions. In contrast, A9-P38:CKII(E81A) cells yielded abundant TM2 in fractions 4 and 5 and TM5 in fraction 3, even as late as 48 h after infection, suggesting the involvement of functional CKII in MVM-induced degradation of these microfilaments. Altogether, our results suggest that MVM-mediated alteration of TM phosphorylation leads to degradation of TM filaments and thereby contributes to morphological alterations in infected host cells.
Fig. 3.
Effect of CKIIα functional modifications on MVM-induced cytoskeleton alterations. A9 or A9:P38-CKII(mE81A) cells were infected (or not) with MVM (30 pfu per cell). (A) For immunofluorescence microscopy, cells were fixed with paraformaldehyde at the indicated time after infection, and the TM filament network was revealed by indirect fluorescence microscopy. (B) For biochemical characterization of cytoskeletal filaments, extracts were fractionated according to their differential solubility in detergent(s) as described in Materials and Methods,“ and the individual fractions (S1–S5) were analyzed for TM and tubulin α by Western blotting. Low-solubility fractions corresponding to rigid fiber structures are framed by dotted lines. The migration of TM2, TM5, and tubulin α (Tubα) are indicated.
Cytotoxic Activity of NS1-Like Adaptor Polypeptides Bridging CKIIα with TM.
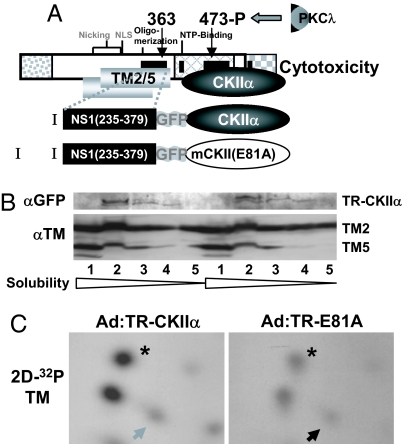
The above results led to the idea that MVM cytotoxicity might result from NS1-mediated targeting of CKII phosphorylation to new substrates, particularly TM. To test this hypothesis, constructs coding for the NS1 toxicity domain mediating TM binding [amino acids 235–379; called the targeting region (TR)] fused (by a GFP spacer) with either CKIIα or its dominant-negative E81A variant (see Fig. 4A) were expressed in A9 cells from adenovirus vectors (the corresponding fusion proteins were named TR-CKIIα and TR-E81A, respectively). The pattern of endogenous TM phosphorylation was determined in these cells by metabolic 32P-labeling and tryptic phosphopeptide analysis. Western blot analyses performed in parallel (Fig. 4B) revealed similar amounts of both effector (TR-CKIIα vs. TR-E81A) and target (TM2/TM5) proteins in the two cell populations. Interestingly, cells expressing TR-CKIIα showed a distinctive TM phosphorylation pattern (Fig. 4C Left) similar to that of MVM-infected cells (Fig. 2C), with its characteristic overrepresented (asterisk) and underrepresented (arrow) peptides. These distinctive features were much reduced in cells infected with Ad:TR-E81A (Fig. 4C Right), as in MVM-infected cells expressing the dominant-negative CKIIα variant (Fig. 2C). The TR domain of NS1 thus appears to target its fusion partner to TM, which supports the hypothesis that NS1 modulates the TM phosphorylation profile by acting as an adaptor protein between TM and CKII.
Fig. 4.
TM phosphorylation by adaptor molecules mimicking NS1-mediated targeting of CKIIα. (A) Schematic representation of NS1 and adaptor molecules (I and II). NS1 is shown with its cellular binding partners CKIIα and TM. The semisynthetic adaptor molecules consist of the NS1 TR (amino acids 235–379), a spacer element (GFP), and either WT or mutant (mE81A) CKIIα. (B and C) In vivo phosphorylation of TM by adaptor proteins 1 and 2. A9 cells were infected with recombinant adenoviruses expressing functional (Ad:TR-CKIIα) or dominant-negative (Ad:TR-mE81A) adaptor molecules. Infected cells were subjected (C) or not (B) to metabolic labeling and processed for protein analyses. The expression of effector proteins (TR-CKIIα) and substrates (TM2, TM5) was measured after cell fractionations by Western blotting with anti-GFP or anti-TM antiserum, respectively (B). (C) Tryptic phosphopeptide pattern of TM isolated from radiolabeled cell extracts. Phosphopeptides characteristically overrepresented in mock- vs. MVM-infected A9 cells and conversely are indicated, respectively, by an arrow and an asterisk.
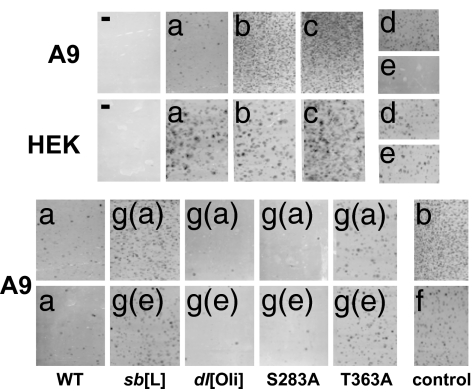
Because TR-CKIIα can mimic the effect of MVM on the TM phosphorylation pattern, it was further investigated whether this adaptor protein and others might jeopardize the survival of PV-permissive cells. A variety of constructs were made to express a WT or mutant NS1 TR domain, attached by a GFP spacer either to a WT or mutant CKIIα or to the DLEPDEELED sequence (BS), to which CKIIα is known to bind (12). These constructs were cloned into pCR3.1, and their toxic potential was assessed, after transfection, in colony-formation inhibition assays. As shown in Fig. 5(A9 b–d), coexpression of GFP-CKIIα, TR-GFP, or TR-mutCKII (a catalytically inactive CKIIα mutant) with the neoR gene gave rise to many colonies, similar in number to those obtained when cells were transfected with the empty pCR3.1 vector or with pCR3.1-GFP as an effector construct (data not shown). In contrast, cells expressing constructs coding for CKIIα linked by GFP to TR or for the TR-GFP-BS adaptor peptide yielded a dramatically reduced number of cell colonies (A9 a and e vs. b–d). No colonies were obtained with mock-transfected A9 or HEK293 cells (−). Thus, polypeptides in which a functional CKIIα- or a CKIIα-binding site is physically connected to the NS1 targeting region exert toxic effects that drastically reduce the viability of A9 cells. Interestingly, these polypeptides were toxic toward MVM-permissive A9 cells but harmless for nonpermissive HEK293 (HEK a and e vs. b–d). The impact of NS1-mediated targeting of CKIIα thus appears to vary according to the host cell.
Fig. 5.
Cytotoxic activity of adaptor molecules targeting CKIIα activity. Colony-formation inhibition assays are shown. A variety of adaptor constructs were made expressing the TM-interaction domain of NS1 (TR) and either CKIIα- or a CKIIα-binding site (BS, DLEPDEELED) in physically linked or dissociated forms, as WT or mutant polypeptides cloned into pCR3.1 containing the neoR expression cassette. A9 or HEK293 cells transfected with these constructs were tested for the ability to inhibit colony formation in the presence of G418, as visualized by crystal violet staining. (a) TR connected to WT CKIIα. (b) TR linked to dominant-negative CKII(mE81A). (c) GFP-WT CKIIα. (d) TR connected to GFP (without PK). (e) TR connected by GFP to CKIIα-BS recruiting endogenous CKIIα. (f) GFP-BS (GFP with a C-terminal CKIIαBS). [g(a)] Mutated NS1 targeting region (mTR) connected to functional CKIIα. [g(b)] mTR linked to CKIIαBS. The impact of structural elements in TR was investigated by testing the effects of the following mutations: sb[L], methionine-for-leucine substitutions in a leucine (L) stretch; dl[Oli], deletion of the oligomerization signal; S283A and T363A, alanine substitutions at consensus PKC phosphorylation sites.
In similar experiments, the TR domain of NS1was mutated to probe the link between TR-CKIIα-induced toxicity and NS1 functioning (3). TR mutants were linked to functional CKIIα [g(a)] BS [g(b)] and tested for the ability to inhibit colony formation by A9 cells. A leucine-rich region located between NS1 residues 288 and 358, seen as a possible structural element for protein–protein interaction, was altered by substituting methionines for leucines 322, 323, 327, 332, and 334 (sb[L]). Two consensus PKC site mutants previously shown to decrease (T363A) or slightly increase (S283A) NS1 cytotoxicity were also tested. Furthermore, given the importance of assembly into oligomers in NS1 functioning (13), the toxicity of TR-CKIIα and TR-BS constructs lacking the oligomerization signal in the N-terminal part of TR (dl[Oli]) was also tested. Because this deletion did not impair the toxicity of either construct (dl[Oli]), it seems that these adaptor peptides are toxic to cells as monomers. In contrast, the “leucine motif” proved essential to toxicity because its disruption led to a major increase in the number of surviving colonies (sb[L]). As for the PKC site substitutions, S283A caused a slight increase and T363A a partial suppression of the ability of TR-CKIIα and TR-BS constructs to inhibit colony formation (see, respectively, S283A and T363A vs. WT). This finding is in keeping with the respective effects of the S283A and T363A mutations on NS1 cytotoxicity. These correlations point further to a connection among NS1 targeting of CKII activity, CPE induction, and toxicity toward permissive cells.
Discussion
PVs are known for their tumor-suppressive effects. An important aspect of PV-promoted oncosuppression is the ability of these viruses to infect and kill transformed cells preferentially. The multifunctional NS1 protein has been identified as a major effector of PV cytotoxicity, jeopardizing the integrity and survival of PV-permissive host cells even in the absence of other viral constituents (3, 14, 15). Here we present the characterization of a viral trigger of cytotoxicity. By bridging CKIIα to a new substrate (e.g., TM), NS1 retargets the action of this cellular PK to that substrate and thereby modulates the properties of the target (i.e., induces degradation of TM filaments). Consequently, any impairment of the interaction of NS1 with CKII (3) or its substrates (3), or of the functioning of CKII (5), protects cells from PV-induced CPE and death. Furthermore, it is possible to obtain the same alterations of the TM phosphorylation pattern and the same effects on cell survival with adaptor polypeptides where a functional CKIIα is physically linked to the TM-targeting region of NS1. This interconnection among PV, PK, and cytoskeleton turnover calls to mind another finding that we have reported recently: MVM-induced activation of PKCλ and concomitant tubulin phosphorylation correlate with protection of the microtubule network against degradation (10).
We have thus highlighted an additional function of NS1: its ability to sequester the catalytic subunit of a cellular PK and thereby modulate its substrate specificity. Such a mechanism might have an impact on multiple aspects of cell metabolism because it is likely that NS1 targets CKIIα to other substrates besides TM. Candidates include regulators of cytoskeleton filaments (10) and participants in signaling cascades, which both appear to be modified after MVM infection (7, 8, and S. Lachmann, S. Bär, J.R., and J.P.F.N., unpublished work). Targeting to such components might be mediated by the TM-targeting region or by other domains of NS1. For example, the NS1 C terminus (which is differentially modified at late stages of infection and contains elements regulating NS1 toxicity) (4) or the NS1 helicase domain (interacting with two as-yet unidentified 18-/20-kDa polypeptides) (Fig. 1B) might bind specific partners and divert them from normal functions. The fact that mutations inhibiting the NS1–TM interaction did not fully abolish NS1 toxicity supports the hypothesis that additional NS1 partners might cooperate toward MVM-induced cell death.
NS1 induces death specifically in oncogene-transformed cells (15), making this viral protein an intriguing candidate oncotoxin for cancer therapy. On the basis of our data, we have designed semisynthetic adaptor molecules retaining three essential and related features of NS1: bridging of CKIIα to a target protein, modulation of the substrate specificity of CKIIα, and induction of toxicity to PV-sensitive host cells. The reason for this cell specificity of NS1-based toxins is presently a matter of speculation. It is noteworthy in this regard, however, that normal and transformed fibroblasts produce different TM isoforms (11), TM being shown here to mediate NS1-induced CPE. On the other hand, NS1 activity appears to depend on PKC phosphorylation of both the CKIIα-binding domain and the substrate recognition region (2). Because PKC activity is reported to be subject to alterations on transformation, the dependence of NS1on PKC phosphorylation might also contribute to exacerbating NS1 cytotoxicity rather toward transformed vs. normal cells. It might thus be possible to further engineer such adaptor peptides toward oncoselectivity and to use such components as anticancer agents.
Materials and Methods
Antibodies and Reagents.
Anti-TM antibodies (T3651) were from Sigma (St. Louis, MO). GFP was from Santa Cruz Biotechnology (Santa Cruz, CA), and αNS1C (16) was kindly provided by P. Tattersall and S.F. Cotmore (Yale University, New Haven, CT). Hi-Trap glutathione–Sepharose columns were from Amersham Pharmacia (Piscataway, NJ); recombinant protein kinases were from Roche (Mannheim, Germany) (CKIIαβ) or Sigma (PKC). Production and purification of WT and mutant GST-tagged NS1 proteins by means of recombinant vaccinia virus expression are described in ref. 5.
Cells and viruses.
A9 fibroblasts, derivatives thereof, and HEK293 cells (Clontech, Mountain View, CA) were maintained as monolayers in DMEM containing 10% FCS. MVM was propagated in A9 cells, and stocks were prepared by repeated freezing and thawing in TE (10 mM Tris/1 mM EDTA, pH 8.3). Recombinant adenoviruses were generated by using the Adeno-X system according to the manufacturer's recommendations (Becton-Dickinson, San Jose, CA) and propagated in HEK293 cells.
Plasmid Constructs.
Isolation of mouse TM cDNAs.
A9 cDNA libraries were generated from mRNA preparations by using SMART 5′- and 3′-RACE kits (Becton-Dickinson). Full-length TM2 and TM5 cDNAs were then generated by PCR amplification (8, 17). The following primers were used. For RACE reactions: 5′ terminus, 5′-TTGCTTCCTTTAGCTGGATTTCCTGGAGTTCCATCTTTTC-3′; 3′ terminus, 5′-GAAAAGATGGAACTCCAGGAAATCCAGCTAAAGGAAGCAA-3′. For full-length cDNAs: TM2, 5′-CCATGGACGCCATCAAGAAGAAGATGCAGATGCTGAAGCT-3′ and 5′-TCACATGTTGTTTAGCTCCAGTAAAGTCTGATCCAGCATC-3′; TM5, 5′-GATATCAGATCTCCATGGCCGGGACCACCACCATCGAGGC-3′ and 5′-GCGGCCGCAAGCTTCTACATCTCGTTCAGGTCCAGCAGGG-3′. PCR products were analyzed by agarose gel electrophoresis, excised from the gel, cloned directly into pCR2.1 vectors (Invitrogen, Carlsbad, CA), and analyzed by sequencing (Microsynth GmbH, Balgach, Switzerland). Candidate full-length TM2 and TM5 cDNA clones were further analyzed for expression of the recombinant protein, by using TNT kits (Promega, Madison, WI), and the resulting 35S-labeled proteins were analyzed by SDS/12% PAGE.
Generation and site-directed mutagenesis of fusion constructs.
Fusion constructs were generated by chimeric PCR (8) with primers overlapping by 20 nts each of the coding sequences to be connected. The following template plasmids were used: for NS1, pTM1-NS1 (18); for GST, pGEX-5X-2 (Amersham Pharmacia); for GFP, pEGFP (Clontech); for CKIIα and CKII(E81A) (5); for β-actin, pYFP-β-actin (Clontech). Site-directed mutagenesis was performed as described (8) by using complementary 40-mer primers harboring mutations in the middle of the sequence. End products generated by PCR were directly cloned into pCR2.1 (Invitrogen), characterized by sequencing, and subsequently transferred (using unique N- and C-terminal restriction sites) into the appropriate expression vectors.
Generation of expression constructs for stable cell transfection.
pP38:GST-actin, pP38:GST-TM2, and pP38:GST-TM5 were produced by cloning the respective tagged cDNAs as SmaI to NotI fragments into the HpaI- and NotI-cleaved pP38 vector (8).
For bacterial expression.
pQE-TM2 and pQE-TM5 were produced by in-frame insertion of SmaI- and HindIII-cleaved full-length TM2 and TM5 coding sequences into SmaI- and HindIII-cleaved pQE32 (Qiagen, Valencia, CA).
For colony-formation inhibition assays.
The various adaptor constructs generated by PCR were cloned either directly as PCR fragments into pCR3.1 (Invitrogen) or as EcoRI fragments from pCR2.1 into EcoRI-cleaved pCR3.1 and analyzed for proper orientation.
For recombinant adenoviral vectors.
Adaptor constructs TR-CKIIα and TR-CKII(E81A) were cloned as NheI and NotI fragments into similarly prepared pShuttle (Clontech), from which the whole expression cassette was transferred as an I-CeuI to PI-SceI fragment into Adeno. Before transfection into HEK293 cells, adenoviral sequences were excised by PacI digestion to prepare primary virus stocks.
Differential Affinity Purification of NS1-Interacting Partner Proteins (5).
Extracts prepared from A9 cells as nuclear squeezes into the cytoplasm were fractionated on phosphocellulose and DE52 columns to obtain A9-P3-DE1. For differential affinity column chromatography, Hi-Trap glutathione–Sepharose columns were coated with WT or mutant GST-NS1 expressed from a recombinant vaccinia virus to ensure proper phosphorylation necessary for protein–protein interactions (3, 5, 9). Proteins present in P3-DE1 derived from 1010 cells were loaded first on a GST-NS1:T363A or a GST-NS1:S473A column. The respective flow-throughs were collected and run over a GST-NS1WT column. Proteins retained by the latter column were eluted with 700 mM NaCl after extensive washes under isotonic conditions. Analysis and/or additional purification were performed by SDS/12% PAGE and Coomassie blue staining. Protein bands present in significantly different amounts in the two eluates were excised from the gel and subjected to MS/MS analysis (Keck Facility, Yale University, New Haven, CT).
Generation of Stably Transfected A9 Cell Lines.
Stable transfectants were generated as described previously (8) by using a pP38-X/pSV2neo molar ratio of 25:1. A9-P38:CKII(E81A) and A9-P38:CKII(mATP) have been described (5). To ensure optimal reproducibility, all transfectants were kept in culture for a limited time only (<25 passages).
NS1 Association with GST-TM.
Extracts prepared as nuclear squeezes into the cytoplasm from 2 × 108 A9 cells or derivatives thereof (A9-P38:TM2, A9-P38:TM5, A9-P38:β-actin) infected (or not) with 30 pfu MVM per cell were loaded onto 1-ml Hi-Trap glutathione–Sepharose columns under isotonic conditions (150 mM NaCl) and washed extensively. Proteins binding to the GST-tagged baits were then specifically eluted with 700 mM NaCl and analyzed by Western blotting compared with the total amount (input) present in the extracts (5).
In Vitro Kinase Reactions (5).
In vitro kinase reactions were performed as described (9) with the indicated PK and substrate combinations in 50 μl of 20 mM Hepes-KOH, pH 7.5/7 mM MgCl2/150 mM NaCl/1 mM DTT, by using 30 μCi of [γ-32P]ATP (3,000 mCi/mmol) and appropriate cofactors for 40 min at 37°C. Reactions were stopped, and the 32P-labeled reaction products were analyzed by SDS/PAGE and autoradiography.
Metabolic Labeling and Phosphopeptide Analyses.
Metabolic labeling and tryptic phosphopeptide analyses were performed as described previously (10). A9 cell cultures or derivatives were infected (or not) with MVM or a recombinant adenoviral vector (30 pfu per cell) and kept for 20 h before incubation for 4 h in labeling medium supplemented with 32Pi. Labeled cells were harvested directly in coimmunoprecipitation buffer, and immunoprecipitations were carried out with 10 μl of α-TM antiserum. Labeled TM proteins were purified by SDS/PAGE, and their tryptic phosphopeptide patterns were analyzed by two-dimensional thin-layer electrophoresis and chromatography.
Immunofluorescence Microscopy.
For immunofluorescence microscopy, cells were grown on spot slides, mock- or MVM-infected (30 pfu per cell), and fixed at the indicated time after infection with 3% paraformaldehyde. TM was detected with a polyclonal rabbit antiserum and FITC-conjugated anti-rabbit IgGs.
Biochemical Fractionation of Cell Extracts.
Fractionation according to different solubility in detergents was performed as described (10). Cells were harvested and suspended in proportional amounts of hypotonic buffer. Soluble proteins (S) were generated by repeated cycles of freezing and thawing. S1 was separated from insoluble material by centrifugation. S2 was then generated by extraction of the pellet in the same initial volume of 0.5% CHAPS. Fractions S3 and S4 were obtained similarly through successive extraction of the insoluble pellets with CHAPS/DOC buffer (supplemented with 0.2% sodium deoxycholate) and CHAPS/DOC/SDS buffer (containing 0.1% SDS), respectively. The remaining pellet was heated to 100°C in loading buffer before analysis by SDS/PAGE and Western blotting.
Colony-Formation Inhibition Assay.
To assess the toxicity of adaptor proteins, 105 A9 or HEK293 cells were transfected with 2 μg of the appropriate pCR3.1-X construct, by using 25 μl of Lipofectamine. Two days after transfection, cultures were split 1:10 and subjected to selection in 400 μg/ml G418 (Sigma) for 3 weeks. Colonies were fixed for 20 min at 4°C in acetic acid/ethanol (1:3 vol/vol) and stained with 100 mM citric acid containing 0.1% crystal violet for 1 h at room temperature.
Acknowledgments
We thank Claudia Plotzky for excellent technical assistance. We are also indebted to Drs. Jan J. Cornelis and Antionio Marchini for critical reading of the manuscript and to Drs. Peter Tattersall and Susan Cotmore for providing plasmid constructs and antisera.
Abbreviations
- CKII
casein kinase II
- CPE
cytopathic effects
- MS/MS
tandem mass spectrometry
- MVM
minute virus of mice
- PK
protein kinase
- PV
parvovirus
- TM
tropomysin
- TR
targeting region.
Footnotes
The authors declare no conflict of interest.
References
- 1.Geletneky K, Herrero YCM, Rommelaere J, Schlehofer JR. J Vet Med. 2005;52:327–330. doi: 10.1111/j.1439-0450.2005.00877.x. [DOI] [PubMed] [Google Scholar]
- 2.Nüesch JP. In: Parvoviruses. Kerr JR, Cotmore S, Bloom ME, Linden RM, Parrish CR, editors. London: Edward Arnold; 2006. pp. 275–290. [Google Scholar]
- 3.Corbau R, Duverger V, Rommelaere J, Nüesch JP. Virology. 2000;278:151–167. doi: 10.1006/viro.2000.0600. [DOI] [PubMed] [Google Scholar]
- 4.Daeffler L, Horlein R, Rommelaere J, Nüesch JP. J Virol. 2003;77:12466–12478. doi: 10.1128/JVI.77.23.12466-12478.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nüesch JP, Rommelaere J. J Virol. 2006;80:4729–4739. doi: 10.1128/JVI.80.10.4729-4739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbau R, Salomé N., Rommelaere J, Nüesch JP. Virology. 1999;259:402–415. doi: 10.1006/viro.1999.9786. [DOI] [PubMed] [Google Scholar]
- 7.Nüesch JP, Lachmann S, Corbau R, Rommelaere J. J Virol. 2003;77:433–442. doi: 10.1128/JVI.77.1.433-442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lachmann S, Rommeleare J, Nüesch JP. J Virol. 2003;77:8048–8060. doi: 10.1128/JVI.77.14.8048-8060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nüesch JP, Corbau R, Tattersall P, Rommelaere J. J Virol. 1998;72:8002–8012. doi: 10.1128/jvi.72.10.8002-8012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nüesch JP, Lachmann S, Rommelaere J. Virology. 2005;331:159–174. doi: 10.1016/j.virol.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharya B, Prasad GL, Valverius EM, Salomon DS, Cooper HL. Cancer Res. 1990;50:2105–2112. [PubMed] [Google Scholar]
- 12.Peyerin W, Ackermann K, Lorenz P. In: Protein Phosphorylation. Marks F, editor. Germany: VCH, Weinheim; 1996. pp. 117–147. [Google Scholar]
- 13.Pujol A, Deleu L, Nüesch JP, Cziepluch C, Jauniaux JC, Rommelaere J. J Virol. 1997;71:7393–7403. doi: 10.1128/jvi.71.10.7393-7403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caillet-Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mousset S, Ouadrhiri Y, Caillet-Fauquet P, Rommelaere J. J Virol. 1994;68:6446–6453. doi: 10.1128/jvi.68.10.6446-6453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore SF, Tattersall P. J Virol. 1988;62:851–860. doi: 10.1128/jvi.62.3.851-860.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mancini A, Borrelli A, Schiattarella A, Fasano S, Occhiello A, Pica A, Sehr P, Tommasino M, Nüesch JP, Rommelaere J. Int J Cancer. 2006;119:932–943. doi: 10.1002/ijc.21904. [DOI] [PubMed] [Google Scholar]
- 18.Nüesch JP, Cotmore SF, Tattersall P. Virology. 1992;191:406–416. doi: 10.1016/0042-6822(92)90202-z. [DOI] [PubMed] [Google Scholar]