Abstract
The circadian hormone melatonin is synthesized predominantly in the pineal gland by the actions of two pineal-specific enzymes: serotonin N-acetyltransferase (NAT) and hydroxyindole-O-methyltransferase (HIOMT). Pineal night-specific ATPase (PINA), another pineal- and night-specific protein we recently identified, is produced as a truncated form of the Wilson disease gene (Atp7b) product. To identify the regulatory elements required for pineal-specific gene expression, we isolated sequences upstream of the rat PINA gene and discovered a cis-acting element that is recognized by a novel pineal/retina-specific nuclear factor. This pineal regulatory element (PIRE) has a consensus of TAATC/T and is present in six copies in the 5′ regulatory region of the PINA gene, at least three copies in the rat NAT promoter, and at least one copy in each of the putative HIOMT promoters A and B. A recently identified retina-specific protein, cone rod homeobox (CRX), binds to PIRE in vitro and transactivates PIRE-reporter constructs. These data suggest that Crx may play a crucial role in regulating pineal gene expression through interactions with PIRE.
The pineal gland functions to transduce photoperiodic information into the rhythmic synthesis and release of the neurohormone melatonin. Melatonin, which also is expressed to a limited extent in the retina, is generated from serotonin by the sequential actions of two pineal-specific enzymes (1), serotonin N-acetyltransferase (NAT) and hydroxyindole-O-methyltransferase (HIOMT). Although the upstream regions of both rat NAT (2, 3) and human HIOMT (4) genes have been isolated, little is known about the molecular basis of their pineal-specific expression.
Using a night subtracted pineal cDNA library, we identified an alternatively spliced form (J.B., unpublished data) of Atp7b, a copper transporter disrupted in Wilson disease (5), a disorder of copper metabolism. This novel protein, designated pineal night-specific ATPase (PINA), is uniquely expressed in the pineal gland and shares an identical temporal expression pattern (J.B., unpublished data) with that of NAT (6). The dramatic diurnal variation of NAT and PINA expression during the circadian cycle imply a tightly regulated, pineal-specific transcriptional apparatus.
In the present study we have identified a conserved sequence TAATC/T, termed pineal regulatory element (PIRE), which is present in multiple copies in the upstream region of the PINA, NAT, and HIOMT genes. The PIRE sequence is recognized by a novel pineal/retina-specific nuclear protein, which is identical to the cone-rod homeobox (CRX), an otx-like homeobox protein isolated independently by Furukawa et al. (7), Chen et al. (8), and Freund et al. (9). In retina, CRX transactivates photoreceptor cell-specific genes and regulates photoreceptor differentiation (7, 8), and is crucial for photoreceptor survival (9, 10). Our data suggest that CRX also is involved in pineal-specific gene expression and may be important in circadian rhythm regulation.
MATERIALS AND METHODS
Experimental Animals.
Sprague–Dawley male rats (Zivic–Miller) were housed under light/dark 14:10 lighting condition (night from 21:00 to 07:00) for at least 1 week before use. Daytime animals were killed at 14:00; nighttime animals were killed in the dark under safe light between 01:00 and 05:00.
Electrophoretic Mobility Shift and DNase I Footprint Assays.
Nuclear extracts were prepared as described (8). Protein concentration was determined by using a Pierce protein assay kit. Probes for electrophoretic mobility shift assay (EMSA) were prepared according to Wang and Reed (11). EMSAs were performed by using a Stratagene Gelshift Assay kit. DNase I footprints were performed essentially as described previously (8), except that 32P-end-labeled DNA fragments were generated by phosphorylation using T4 polynucleotide kinase. A rat PINA upstream fragment (−238 to −1) was used as probe.
Northern Blot Analysis.
Total RNA was isolated from fresh rat tissues by using either TRIZOL reagent (GIBCO/BRL) or RNeasy kit (Qiagen). Northern analysis was performed as previously described (6).
PINA Promoter Analysis.
The DNA fragment between exon 8 and exon 9 of rat Atp7b (X.L. and J.B., unpublished data) was PCR-amplified by using rat genomic DNA as template, and used as probe to screen an EMBL3-SP6/T7 genomic phage library. A 4.5-kb EcoRI fragment was obtained. Primer extension (Promega Primer Extension kit) and 5′-rapid amplification of cDNA ends (CLONTECH) assays were performed according to the manufacturer’s instructions. A 238-bp PINA upstream fragment was subcloned into the KpnI–EcoRI sites of vector pSEAP2-Basic (CLONTECH) to generate pPINA-SEAP (secreted alkaline phosphatase). Double-stranded oligonucleotides containing three repeats of PIRE (5′-CACTAATCTCCCCACTAATCTCCCCACTAATCTCCC-3′) or PIRE mutant (5′-CACTCATCTCCCCACTCATCTCCCCACTCATCTCCC-3′) were cloned into the KpnI–BglII sites of vector pGL3-Promoter (Promega) to generate pPIRE-GL3P and pPIRE-M-GL3P. HEK293 cells were transfected with the constructs using Lipofectamine reagents (GIBCO/BRL) and assayed for alkaline phosphatase or luciferase activity 24 hr after transfection by using the CLONTECH Chemiluminescent SEAP Assay kit or Promega Dual-Luciferase Reporter Assay system.
RESULTS
Characterization of the PINA Upstream Regulatory Region.
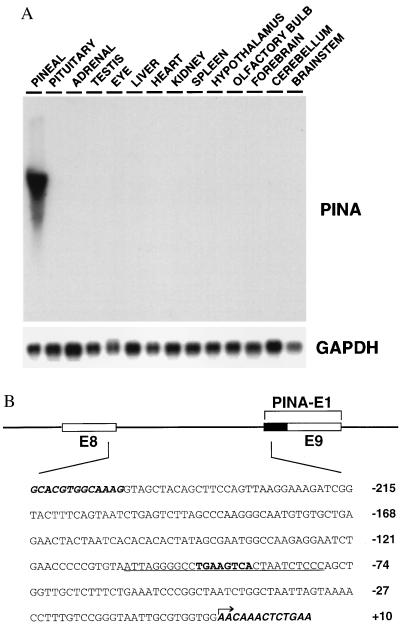
Recently we identified PINA as an alternatively spliced form of the Wilson disease protein. The diurnal variations in PINA expression closely resemble those of NAT with the nighttime peak of PINA expression being 100 times greater than its midday trough (J.B., unpublished data). Northern blot analysis of rat tissues at night reveals that PINA expression is essentially pineal specific (Fig. 1A), though low levels are detected in the eye (J.B., unpublished data).
Figure 1.
Tissue specificity of PINA and its upstream intronic sequence. (A) Northern blot analysis of a panel of rat tissue total RNAs (10 μg each) at night (02:00) with PINA cDNA as probe. Equal loading and quality of RNAs were confirmed by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) controls. (B) The PINA upstream intronic sequence. Schematic representation of the exon-intron organization of the PINA upstream region. Open boxes represent exons 8 and 9 of the rat Atp7b gene. The black box represents the part of the PINA exon 1 that belongs to intron 8 in Atp7b gene. The DNA sequence shown includes the PINA intronic promoter and small portions of flanking exons (bold italic). The arrow indicates the most upstream transcription initiation site as determined by primer extension and 5′-rapid amplification of cDNA ends (data not shown). The 28-bp sequence used as probe for EMSA (Fig. 2A) is underlined with the CRE in bold.
To uncover molecular mechanisms that determine the temporal and tissue specificities of PINA and NAT expression, we characterized the upstream regulatory sequence of the PINA gene. As determined by primer extension and 5′-rapid amplification of cDNA ends (data not shown), the transcription initiation sites of PINA reside in between exons 8 and 9 of the rat Atp7b gene. Thus, we speculated that the intronic fragment immediate upstream of PINA may contain elements directing night- and pineal-specific PINA expression. We isolated the PINA upstream region by screening a rat genomic library using a PCR-amplified intronic sequence between exons 8 and 9 of Atp7b as a probe. The position of this region in the Atp7b gene (Fig. 1B) was further confirmed by sequencing the isolated genomic clone. The 238-bp intronic sequence contains a cAMP response element (CRE)-like element (TGAAGTCA). EMSA demonstrates that this element specifically binds in vitro to the CRE-binding protein ICER (inducible cAMP early repressor) (data not shown). Because the temporal expression pattern of PINA resembles that of NAT, which is thought to be directed by elevated cAMP levels at night through the CRE element in its promoter (2, 3), the CRE element in PINA upstream region may function analogously.
Identification of PIRE Within the PINA Upstream Region.
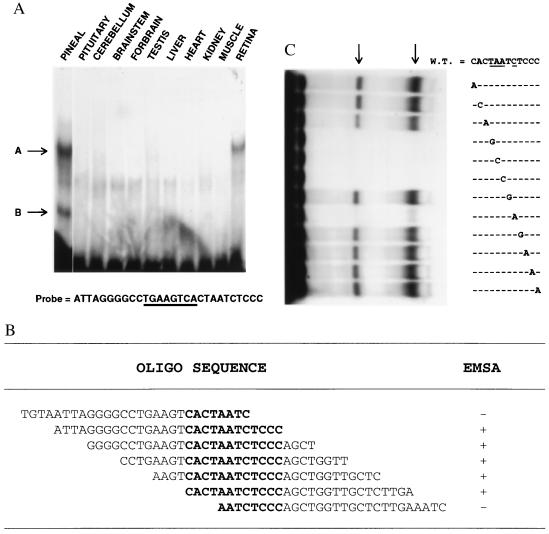
To isolate the molecular components that direct the tissue specificity of PINA expression, we performed EMSA with nuclear extracts from pineal and various other tissues. A 28-bp sequence from the PINA upstream region recognizes a pineal/retina-specific nuclear factor (Fig. 2A, band A). In the pineal, there is an additional complex observed (band B), which is not found in retina or any other tissue examined. Both complexes appear to recognize the same sequence. There are no obvious diurnal variations in levels of this pineal/retina-specific binding activity (data not shown).
Figure 2.
Identification of PIRE. (A) EMSA with a panel of rat tissue nuclear extracts (5 μg total protein each) and probe as indicated. The CRE site is underlined and the positions of shifted bands are indicated by arrows. (B) Result of EMSAs with pineal nuclear extract and a series of seven overlapping oligonucleotides. The sequence shared among the oligonucleotides used for EMSAs is in bold. (C) Mutation analysis of the 12 nucleotides sufficient for binding. All probes used have EcoRI sites at their 5′ termini and XbaI sites at their 3′ termini, plus a 12-nt sequence as indicated. The 12 nucleotides sufficient for binding were mutagenized one at a time, and each mutant was used as the core sequence of a probe. Arrows indicate the positions of shifted bands. The four nucleotides essential for binding are underlined.
To identify the sequence within the 28-bp probe that exhibits pineal/retina-specific binding, we conducted EMSA analysis with pineal nuclear extract and a series of seven overlapping probes from the PINA promoter (Fig. 2B). A 12-nt sequence, CACTAATCTCCC, appears to be sufficient for the binding activity. To ascertain the relative importance of the nucleotides within this sequence, we mutagenized each nucleotide and performed EMSA analysis on each with pineal extracts (Fig. 2C). Changes at four independent positions completely abolish the binding, indicating the importance of these nucleotides at the defined positions for binding activity. We have designated sequences capable of interacting with this pineal/retina-specific factor the PIRE, based on functional data below.
Multiple Functional PIRE Sites Occur in the Upstream Regions of PINA, NAT, and HIOMT.
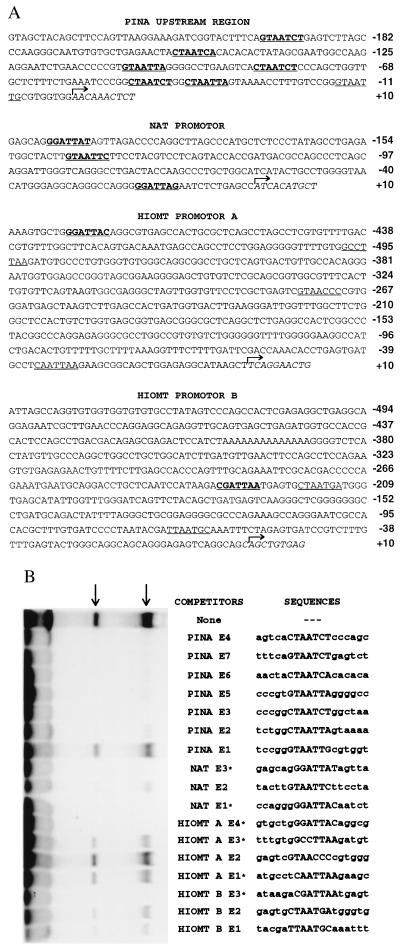
Examination of the upstream regions of PINA, NAT, and HIOMT (4) reveals multiple potential PIRE sites based on sequence homology to the PINA PIRE (Fig. 3A). The PINA upstream region contains seven putative sequences, the NAT promoter contains at least three, the HIOMT promoter A four, and the HIOMT promoter B three.
Figure 3.
Multiple PIRE sites in PINA, NAT, and HIOMT upstream regions. (A) Upstream sequences of rat PINA, NAT, and human HIOMT with putative PIREs underlined. Functional binding sites as determined by EMSA competition test (B) are further indicated in bold. Transcription initiation sites for PINA and NAT, as determined by primer extension and 5′-rapid amplification of cDNA ends (data not shown), and HIOMT (4) are indicated by arrows. (B) EMSA competition test with pineal nuclear extract and putative PIREs as competitors. The probe contains the 12-nt core sequence (CACTAATCTCCC) flanked by a 5′ EcoRI site and a 3′ XbaI site. The putative PIREs are numbered in order from 3′ to 5′. Sequences of double-stranded probes and competitors are indicated with the cores of PIREs in uppercase. Each competitor was in 100-fold excess of the concentration of probe. When no competitor was added, an equivalent amount of poly(dI-dC) was used as nonspecific competitor. ∗, PIREs as indicated are in reversed sequence.
To examine whether the potential sites are able to bind the same protein as the PINA PIRE, we tested each of them, together with their native flanking sequences, as competitors for PIRE binding to pineal nuclear extracts (Fig. 3B). When used as competitors, six of the seven sequences in PINA upstream region (E2-E7) abolish binding of the pineal nuclear protein to the labeled PIRE probe, and all three of the sequences in the NAT promoter compete for the binding. Elements E4 within the HIOMT promoter A and E3 within the HIOMT promoter B appear to compete effectively with the labeled probe for binding. Comparison of the competitor sequences from the PINA, NAT, and HIOMT promoters reveals a consensus binding site, TAATC/T. Thus, all the known pineal-specific genes contain at least one PIRE sequence, which suggests a role for PIRE in the coordinate regulation of pineal-specific genes.
CRX Accounts for the Pineal/Retina-Specific PIRE Binding Activity.
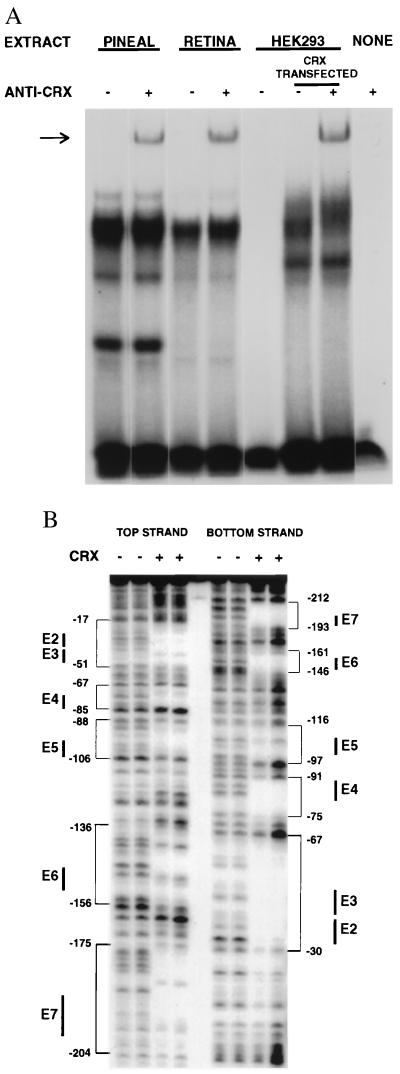
During the course of these studies, CRX was identified as a photoreceptor-specific transcription factor (7–9). The PIRE sequence is essentially the same as the CRX recognition site. To ascertain whether CRX accounts for PIRE binding activity in pineal extracts, we conducted a supershift assay using an antiserum raised against CRX (8) (Fig. 4A). The antiserum elicited a supershift with both pineal and retina extracts, indicating that at least part of the binding activity is attributable to CRX. To determine whether the partial supershift is caused by low antiserum titer or to other proteins capable of binding PIRE that comigrate with CRX, recombinant CRX protein produced in HEK-293 cells were used in a supershift analysis. Again only a partial shift is observed with a magnitude similar to the shift in pineal and retinal extracts, suggesting that the antibody is limiting and CRX may account for all of the PIRE binding activity in the pineal and retina extracts.
Figure 4.
CRX accounts for the pineal-retina specific binding activity. (A) Supershift assay with an anti-CRX antiserum. The same probe as in Fig. 3B was used. The antiserum was incorporated in EMSAs with a series of nuclear extracts indicated. The arrow indicates the position of supershifted band that is absent when no antibody or no nuclear extracts was incorporated. (B) DNase I footprint assay of PINA intronic promoter. A 32P end-labeled PINA promoter fragment was incubated with or without CrxHD-glutathione S-transferase fusion protein (8). Protected regions are indicated by brackets with the nucleotide positions indicated. The positions of PIREs are indicated by solid bars.
To ascertain whether CRX binds to the multiple PIRE sites within the 238-bp DNA fragment 5′ to the PINA gene, we conducted a DNase I footprint analysis (Fig. 4B). The sequences protected by CrxHD-glutathione S-transferase (8) correspond to the PIRE sequences in the PINA upstream region that compete for binding to pineal extracts (Fig. 3B). Thus, CRX binds to all six identified PIRE sites within the PINA upstream region.
Diurnal Variation of Crx Expression in the Pineal Gland.
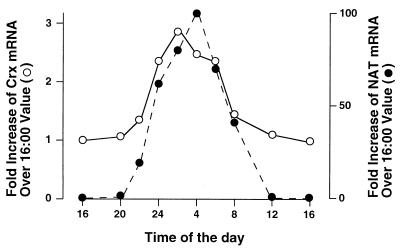
Northern blot analysis using Crx full-length cDNA as probe reveals a single band of identical mobility in both pineal and retina (data not shown). We examined pineal Crx expression over a 24-hr period (Fig. 5). Crx displays a diurnal variation with peak levels at 02:00 being 3-fold greater than levels at 16:00. The peak of Crx expression in pineal precedes the peak of NAT expression by 1–2 hr.
Figure 5.
Diurnal variations of Crx expression compared with that of NAT in pineal gland. Northern blot analysis of a panel of pineal total RNA isolated at different times over a 24-hr time course was quantified by PhosphorImaging (Molecular Dynamics). The Crx mRNA and NAT mRNA levels were normalized to those of glyceraldehyde-3-phosphate dehydrogenase mRNA and presented relative to their levels at 16:00.
CRX Transactivates Reporter Expression Through PIRE.
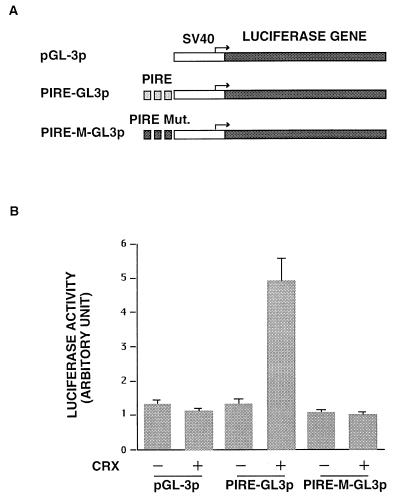
To examine the functional relevance of the CRX-PIRE interaction, a Crx expression construct was cotransfected into HEK293 cells with various reporter constructs (Fig. 6A), and reporter expression was assayed during the period of transient expression (Fig. 6B). First, we tested reporter plasmids containing the luciferase gene under the control of a simian virus 40 promoter linked to either three repeats of wild-type PIRE or PIRE mutant, which differs from PIRE by a single nucleotide and does not bind CRX (Fig. 2C). CRX elicits a 4- to 5-fold transactivation activity only in the presence of PIRE. When the reporter construct containing mutant PIRE is cotransfected with CRX, no activation is evident. In other experiments we have demonstrated that CRX also transactivates the 238-bp PINA upstream sequence in HEK293 cells (data not shown), indicating that the 238-bp intronic fragment upstream of the PINA transcription start site is a functional promoter. Our findings resemble observations of Furukawa et al. (7) and Chen et al. (8).
Figure 6.
Transactivation by CRX. (A) Reporter constructs for transactivation assays. The pGL3-promoter construct contains a simian virus 40 promoter upstream of the luciferase gene. The PIRE-GL3P construct contains three repeats of PIRE upstream of the simian virus 40 promoter whereas the PIRE-M-GL3P construct contains three repeats of PIRE mutant. (B) Luciferase assay results. Each reporter construct (25 ng) was cotransfected into HEK293 cells with 500 ng of either pcDNA 3.1/HisC expression vector alone (CRX −) or pcDNA3.1/HisC-Crx plasmid (CRX +) (8). Firefly luciferase activity was normalized by reference to renilla luciferase activity derived from the cotransfected internal control pRL-TK plasmid. These experiments were replicated four times.
DISCUSSION
We have identified a conserved nucleotide sequence, PIRE, in the upstream regions of three pineal-specific genes, NAT, HIOMT, and PINA. Binding activity for this sequence occurs in pineal and retina but in no other tissue examined. The newly discovered Crx gene product (7–9) binds to PIRE and is capable of activating transcription of PIRE-reporter construct. Crx mRNA expression is found abundantly during the day and displays a moderate diurnal rhythm in the pineal gland with elevated transcription at night. These results demonstrate that CRX is a pineal/retina-specific transcription factor that may be important in regulating the expression of PINA, NAT, and HIOMT. Its increased expression at night may facilitate the dramatic up-regulation of NAT and PINA.
The site of the Crx expression in pineal is likely to be the pinealocytes (8), the cell type expressing melatonin (12). Mammalian pinealocytes are phylogenetically derived from pineal photoreceptor cells. In lower vertebrates, pineal photoreceptors are similar to retinal cones and have outer and inner segments as well as synapses with afferent nerve fibers (13). During development, the pinealocytes of rodent pineal express a set of genes involved in phototransduction (14). Even in adult mammals, certain “photoreceptor-specific” genes still are found in abundance in the pineal (14, 15). Conversely, the known “pineal-specific” genes such as NAT (6, 16), PINA (J.B., unpublished data), and HIOMT (4, 17, 18) also are detected in adult retina, though at much lower levels. This cross-tissue expression suggests the existence of a common pineal/retina-specific transcriptional apparatus in which Crx may play a central role.
The extraordinarily precipitous rise and fall in NAT levels that parallel changes in NAT enzymatic activity (6, 19) indicate that transcriptional regulation is a primary determinant of NAT function in rat. The rhythmic nocturnal increase in NAT mRNA level is caused by nightly increase in pineal cAMP, which results in up-regulation of NAT enzyme activity and melatonin synthesis (20, 21). Consistent with the crucial role of cAMP in regulating NAT transcription, a CRE site, which is found in the promoter region of the rat NAT gene, appears to mediate cAMP-dependent modulation of NAT gene expression in pineal both in vitro (3) and in vivo (2). Thus cAMP-dependent transcriptional control seems to be necessary for the induction and attenuation of night-specific messages, including NAT. However, the cAMP inducibility of NAT gene expression is a unique feature of the pineal, as cAMP alone is not sufficient for induction of pineal genes in other systems. This finding indicates in addition to cAMP responsive nuclear factors such as CREB (cAMP response element binding protein) and ICER (inducible cAMP early repressor) (22), other nuclear factors function to activate tissue-specific transcription in the pineal. The unique tissue-specificity in Crx expression and the ability of Crx to activate PINA promoter-reporter expression suggest that Crx is one component of such machinery important in determining pineal-specific gene expression. The nocturnal expression of NAT and PINA may reflect a synergy between CRX and cAMP responsive nuclear factor(s), analogous to the synergy between CRX and neural retina leucine zipper protein (8) in transactivating opsin genes. Just as dramatic as the nighttime increase in pineal gene expression is the precipitous decline of the expression during the day. Crx levels are fairly substantial during the day in the pineal, suggesting that CRX cannot act alone to activate transcription of pineal-specific genes.
Recently mutations in Crx have been documented in human patients with cone-rod dystrophy (9, 10), an autosomal dominant inherited retina disease leading to progressive loss of photoreceptor cell function, suggesting that CRX is essential for the maintenance of mammalian photoreceptors. Analysis of pineal function in patients with cone-rod dystrophy and defined Crx mutations may clarify the in vivo role of Crx in regulating pineal-specific gene expression.
Unlike the retina, which is composed of several very different cell types, the pineal gland contains a largely homogeneous population of pinealocytes with a small number of astrocytes. In addition, both pineal cells and the entire gland can be cultured in vitro and expression of night-specific messages such as NAT can be induced in these preparations with agents activating the cAMP-dependent signal transduction pathway (ref. 19; J.B., unpublished data). Pineal preparations thus may be particularly useful in studying the role of Crx in coordinating gene expression.
Acknowledgments
We thank C. L. Cepko for sharing data before publication and J. Deng for technical support. We also thank M. M. Wang for helpful suggestions and discussions, and J. Nathans and S. Blackshaw for critical reading of this manuscript. This work was supported by United States Public Health Service Grant DA-00266, Research Scientist Award DA-00074, National Mental Health Institute Grant MH57299, National Eye Institute Grants EY09769 and EY01765, The Foundation Fighting Blindness, unrestricted funds from Research to Prevent Blindness, Inc., and Rebecca P. Moon, Charles M. Moon, Jr., and Dr. P. Thomas Manchester Research Fund. J.B. is a Merck fellow of the Life Science Research Foundation. S.C. is a recipient of an award from the Knights Templer Foundation. D.J.Z. is a recipient of a Career Development Award from Research to Prevent Blindness, Inc.
ABBREVIATIONS
- EMSA
electrophoretic mobility shift assay
- CRX
cone rod homeobox
- HIOMT
hydroxyindole-O-methyl transferase
- NAT
serotonin N-acetyltransferase
- PINA
pineal night-specific ATPase
- PIRE
pineal regulatory element
- CRE
cAMP response element
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF038389).
References
- 1.Axelrod J, Wurtman R J. Adv Pharmacol. 1968;6:157–166. doi: 10.1016/s1054-3589(08)61169-2. [DOI] [PubMed] [Google Scholar]
- 2.Foulkes N S, Borjigin J, Snyder S H, Sassone-Corsi P. Proc Natl Acad Sci USA. 1996;93:14140–14145. doi: 10.1073/pnas.93.24.14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baler R, Covington S, Klein D C. J Biol Chem. 1997;272:6979–6985. doi: 10.1074/jbc.272.11.6979. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez I R, Mazuruk K, Schoen T J, Chader G J. J Biol Chem. 1994;269:31969–31977. [PubMed] [Google Scholar]
- 5.Bull P C, Cox D W. Trends Genet. 1994;10:246–252. doi: 10.1016/0168-9525(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 6.Borjigin J, Wang M M, Snyder S H. Nature (London) 1995;378:783–785. doi: 10.1038/378783a0. [DOI] [PubMed] [Google Scholar]
- 7.Furukawa T, Morrow E M, Cepko C L. Cell. 1997;91:521–530. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 8.Chen S, Wang Q-L, Nie Z, Sun H, Lennon G, Copeland N G, Gilbert D J, Jenkins N A, Zack D J. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 9.Freund C L, Gregory-Evans C Y, Furukawa T, Papaioannou M, Looser J, et al. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 10.Swain P K, Chen S, Wang Q-L, Affatigato L M, Coats C L, Brady K D, Fishman G A, Jacobsen S G, Swaroop A, Stone E, et al. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- 11.Wang M M, Reed R R. Nature (London) 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 12.Freund D, Arendt J, Vollrath L. Cell Tissue Res. 1977;181:239–244. doi: 10.1007/BF00219983. [DOI] [PubMed] [Google Scholar]
- 13.Vollrath L. In: Mammalian Pinealocytes: Ultrastructural Aspects and Innervation. Vollrath L, Oksche A, editors. Berlin: Springer; 1981. pp. 9–23. [DOI] [PubMed] [Google Scholar]
- 14.Blackshaw S, Snyder S H. J Neurosci. 1997;17:8074–8082. doi: 10.1523/JNEUROSCI.17-21-08074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lolley R N, Craft C M, Lee R H. Neurochem Res. 1992;17:81–89. doi: 10.1007/BF00966868. [DOI] [PubMed] [Google Scholar]
- 16.Coon S L, Roseboom P H, Baler R, Weller J L, Namboodiri M A, Koonin E V, Klein D C. Science. 1995;270:1681–1683. doi: 10.1126/science.270.5242.1681. [DOI] [PubMed] [Google Scholar]
- 17.Gauer F, Craft C M. Brain Res. 1996;737:99–109. doi: 10.1016/0006-8993(96)00632-4. [DOI] [PubMed] [Google Scholar]
- 18.Wiechmann A F, Craft C M. Neurosci Lett. 1993;150:207–211. doi: 10.1016/0304-3940(93)90537-u. [DOI] [PubMed] [Google Scholar]
- 19.Roseboom P H, Coon S L, Baler R, McCune S K, Weller J L, Klein D C. Endocrinology. 1996;137:3033–3045. doi: 10.1210/endo.137.7.8770929. [DOI] [PubMed] [Google Scholar]
- 20.Sugden D, Vanecek J, Klein D C, Thomas T P, Anderson W B. Nature (London) 1985;314:359–361. doi: 10.1038/314359a0. [DOI] [PubMed] [Google Scholar]
- 21.Klein D C. In: Photoperiodism, Melatonin and the Pineal. Evered D, Clark S, editors. London: Pitman; 1985. pp. 38–56. [Google Scholar]
- 22.Foulkes N S, Borjigin J, Snyder S H, Sassone-Corsi P. Trends Neurosci. 1997;20:487–492. doi: 10.1016/s0166-2236(97)01109-0. [DOI] [PubMed] [Google Scholar]