Abstract
Inhibitors of the last steps of cholesterol biosynthesis such as AY9944 and BM15766 severely impair brain development. Their molecular target is the Δ7-sterol reductase (EC 1.3.1.21), suspected to be defective in the Smith–Lemli–Opitz syndrome, a frequent inborn disorder of sterol metabolism. Molecular cloning of the cDNA revealed that the human enzyme is a membrane-bound protein with a predicted molecular mass of 55 kDa and six to nine putative transmembrane segments. The protein is structurally related to plant and yeast sterol reductases. In adults the ubiquitously transcribed mRNA is most abundant in adrenal gland, liver, testis, and brain. The Δ7-sterol reductase is the ultimate enzyme of cholesterol biosynthesis in vertebrates and is absent from yeast. Microsomes from Saccharomyces cerevisiae strains heterologously expressing the human cDNA remove the C7–8 double bond in 7-dehydrocholesterol. The conversion to cholesterol depends on NADPH and is potently inhibited by AY9944 (IC50 0.013 μM), BM15766 (IC50 1.2 μM), and triparanol (IC50 14 μM). Our work paves the way to clarify whether a defect in the Δ7-sterol reductase gene underlies the Smith–Lemli–Opitz syndrome.
The pivotal importance of cholesterol biosynthesis for mammalian ontogenesis was recognized when newborns suffering from a syndrome of failure to thrive, psychomotor retardation, organ malformations, and feminization of male infants, known as Smith–Lemli–Opitz (SLO) syndrome, showed reduced cholesterol plasma levels. The discovery that in sera from these patients the intermediates 7-dehydrocholesterol and 8-dehydrocholesterol were increased rendered sterol metabolites a hallmark for diagnosis (1–3). In the liver of newborns with fatal SLO syndrome the in vitro activity of the enzyme Δ7-sterol reductase (EC 1.3.1.21) is reduced (4). This microsomal enzyme is found in plants and mammals and removes the C7–8 double bond in the B ring of sterols (Fig. 1A). The proposed connection between morphogenesis and sterol biosynthesis was supported by similar but more severe malformations in rodents treated with inhibitors of the Δ7-sterol reductase such as AY9944 and BM15766. Cholesterol supplementation reduced teratogenic drug effects, suggesting that malformations were indeed due to the inhibition of sterol biosynthesis (5–7). Beachy and coworkers (8, 9) recently identified an important link between cholesterol and tissue development when they discovered the covalent attachment of cholesterol to a hedgehog protein required for developmental patterning in brain and other embryonic structures. Strikingly, the holoprosencephaly in hedgehog knockout mice is the same observed on in utero exposure of rodents to AY9944 and BM15766.
Figure 1.
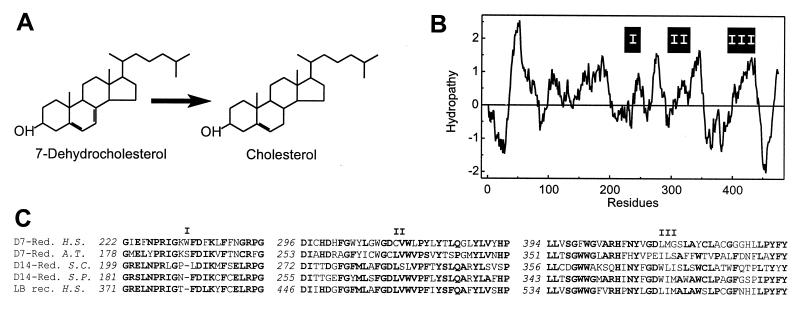
(A) NADPH-dependent reduction of 7-dehydrocholesterol. (B) Hydropathy analysis with an average window size of 19 amino acid residues plotted at one residue intervals. On the ordinate, hydrophobicity is indicated by positive numbers. The positions of segments highly conserved throughout sterol reductases and related proteins are shown (I, II, and III). (C) Partial alignment of highly conserved domains (I, II, and III) in the amino acid sequences of the Δ7-sterol reductases from man (D7-Red., H.S.) and Arabidopsis thaliana (D7-Red., A.T., GenBank accession no. U49398), the Δ14-sterol reductases from Saccharomyces cerevisiae (D14-Red., S.C., no. S69420) and Saccharomyces pombe (D14-Red., S.P., no. L36039), and the human lamin B receptor (LB rec., H.S., no. L25931). Gaps are indicated (–). Numbers in italics refer to the position in the amino acid sequence. Residues identical in the majority of sequences are shown in bold.
We recently purified and cloned the human sterol Δ8-Δ7 isomerase, which catalyzes the shift of the double bond in the B ring of sterols from C8–9 to C7–8 (10–12). This isomerase is a putative target of drugs effective in animal models of stroke (13). Surprisingly, the mammalian genome contains another protein without homology to the human sterol Δ8-Δ7 isomerase that shows significant pharmacological and structural similarities with the sterol Δ8-Δ7 isomerase of Saccharomyces cerevisiae (14–17). We now report the cloning of the ultimate enzyme of mammalian sterol biosynthesis, the Δ7-sterol reductase. This enzyme removes the C7–8 double bond introduced by the sterol Δ8-Δ7 isomerases. Because of its role in drug-induced malformations and its suspected deficiency in SLO syndrome, this enzyme is of outstanding pharmacological and medical significance.
EXPERIMENTAL PROCEDURES
Materials.
The following chemicals were obtained from the indicated sources: Bradford protein reagent and molecular weight markers, Bio-Rad; Marathon-Ready cDNA, Multiple tissue Northern blots and human RNA master blot, CLONTECH; AY9944, P. Benveniste (Strasbourg, France); CDP-Star and BM15766, Boehringer Mannheim; 9E10 c-myc antibody, Oncogene Science; EST clones [I.M.A.G.E. Consortium (LLNL) cDNA Clones (18)], Resource Centre/Primary Database (Berlin, Germany); and all other chemicals, Sigma. Yeast strain JB811 was obtained from K. Nasmyth (Vienna, Austria).
Molecular Cloning and PCR.
Partial human cDNA clones [GenBank accession no. H09710 (infant brain, I.M.A.G.E. Consortium Clone ID46546), AA017586 (adult retina, I.M.A.G.E. Consortium Clone ID361378), H04989 (infant brain, I.M.A.G.E. Consortium Clone ID43848), and R61101 (infant brain, I.M.A.G.E. Consortium Clone ID42337)] homologous to the Δ7-sterol reductase from Arabidopsis thaliana [GenBank accession no. U49398 (19)] were identified with the TBLASTN algorithm in the expressed sequence tag database and sequenced. The 5′ end of the cDNA was amplified with PCR by using Marathon-Ready cDNA from human liver and the antisense oligonucleotide GCAGCGTGTAAAGATAAGGC. The full-length cDNA was constructed in pBluescript SK by using a unique SacI site at position 975 of the cDNA sequence. DNA sequencing of both strands was performed according to standard procedures. Alignments and hydrophobicity analysis were calculated with the Wisconsin Sequence Analysis Package according to the algorithms of Smith and Waterman and Kyte and Doolittle, respectively (Genetics computer group, programs bestfit and pepplot).
Northern Blotting.
Human multiple tissue Northern blot and human RNA master blot (CLONTECH) were probed with 32P-labeled cDNA of the human Δ7-sterol reductase or the human sterol isomerase (GenBank accession no. Z37986). They were quantitatively analyzed with a FujiX Bas 1000 phosphorimager and the program tina.
Expression in S.
cerevisiae. Protein expression in S. cerevisiae was performed as described (11, 14, 15). The 5′ noncoding region was removed with oligonucleotides ACGCGTCGACGTCATGGCTGCAAAAATGCAACCC and ACGCGTCGACAGATCTTGCTGCAAAATTGCAACCCAAC introducing 5′ SalI (D7-ORF) and BglII (mycD7-ORF) restriction sites. These sites were used in conjunction with 3′ NotI and SpeI sites, respectively, for subcloning in the yeast expression plasmids Yep351ADC1 or c-myc-Yep351ADC1 (14). Lithiumacetate transformation of yeast strain JB811 (ade2–1 leu2–3,112 pep4–3 trp1–289 ura3–52), isolation of microsomal proteins and immunoblotting with a 9E19 c-myc antibody were performed as described (11, 14, 15). Protein concentrations were determined according to (20) by using BSA as a standard.
Δ7-Sterol Reductase Assay.
7-dehydrocholesterol, ergosterol, or lathosterol (dissolved in Tween 80 at a weight ratio of 70:1, detergent:sterol) were added at a final concentration of 300 μM to 1 ml of 20% (wt/vol) glycerol, 0.1 M Tris⋅HCl, pH 7.4 (37°C), 1 mM glutathione, 0.5 mM EDTA, 2 mM NADPH, 140 mM glucose, and 20 units glucose oxidase that had been preincubated under N2 atmosphere at 37°C for 4 min (21). Incubation at 37°C with 0.5–2 mg of microsomal protein was carried out anaerobically in sealed glass flasks and terminated by the addition of 1 ml methanolic KOH followed by 10 min under reflux. Sterols were extracted with 3 ml of petroleum ether, dried under N2 gas, and analyzed by gas–liquid chromatography (22). Enzymatic activity was calculated from the relative amounts of substrate and product in incubated samples compared with unincubated controls. For the in vitro inhibition experiments, drugs were dissolved in dimethyl sulfoxide. Incubation time for inhibition and substrate saturation experiments was 20 min, in which the initial reaction velocity was linear. The final dimethyl sulfoxide concentration was less than 0.3% (vol/vol), which did not affect catalytic activity.
RESULTS AND DISCUSSION
The Human Δ7-Sterol Reductase Is Structurally Related to Other Sterol Reductases.
We isolated a 2,597-bp cDNA containing an ORF for a protein with 475 amino acid residues and 35% identity and 60% similarity with the Δ7-sterol reductase from A. thaliana. Fig. 1B shows the hydropathy plot of the amino acid sequence predicting six to nine putative transmembrane α-helices. Amino acid sequence segments highly conserved throughout the plant Δ7-sterol reductase, Δ14-sterol reductases from fungi and the human lamin B receptor, a nuclear protein of unknown role in sterol metabolism, are shown in Fig. 1C. The Δ7-sterol reductase as well as the Δ14-sterol reductases require NADPH as a cofactor. Supposedly, they share a common reaction mechanism involving the formation of a short-lived carbocationic reaction intermediate by enzyme-mediated protonization followed by hydride ion transfer from NADPH (23). Similar hydropathy plots (19) and highly conserved amino acid sequence segments (Fig. 1C) suggest that the same protein platforms and catalytic scaffolds are used to catalyze identical reactions in substrates, differing only in the position of the double bond. In contrast to vertebrates and plants, yeast lacks the Δ7-sterol reductase activity. It is therefore conceivable that the Δ7-sterol reductase in multicellular organisms evolved from the Δ14-sterol reductases of unicellular ancestors.
Inhibition of the Heterologously Expressed Δ7-Sterol Reductase by Drugs.
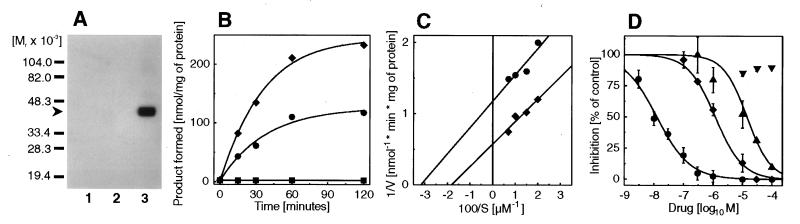
To establish its Δ7-sterol reductase activity we expressed the cDNA without the 5′ noncoding region (D7-ORF) and a derivative with an N-terminal 9E10 c-myc epitope (mycD7-ORF) in Saccharomyces cerevisiae. Expression of mycD7-ORF was verified by immunoblotting (Fig. 2A). Its apparent molecular mass in 12% SDS/polyacrylamide gels of 40 kDa differed from the molecular mass calculated from the amino acid sequence of 54,516 Da for the native protein and of 55,672 Da for mycD7-ORF. For the structurally related lamin B receptor a similar discrepancy between the mass of 73,375 Da calculated from the amino acid sequence and of 58 kDa estimated from SDS/PAGE was also observed and attributed to aberrant migration (24). Microsomes prepared from strains transformed either with D7-ORF or a derivative with an N-terminal 9E10 c-myc epitope (mycD7-ORF) were able to convert 7-dehydrocholesterol to cholesterol (Fig. 1A), whereas no such activity was found in microsomes from strains transformed with the plasmid alone (Fig. 2B). 7-Dehydrocholesterol was preferred as a substrate when compared either with ergosterol or lathosterol (specific activities 2.60, 0.11, and 0.05 nmol/min per mg of protein, respectively). The reaction required NADPH as a cofactor (data not shown). The Lineweaver Burk plot revealed a higher specific activity of the tagged Δ7-sterol reductase (mycD7-ORF) as compared with the nontagged version D7-ORF (Vmax 1.65 and 0.85 nmol/min per mg of protein, respectively, Fig. 2C) in line with the previous observation of increased protein expression in yeast upon epitope tagging (14). As expected, the recombinant human Δ7-sterol reductase (D7-ORF) was inhibited by AY9944 (IC50 = 0.013 μM), BM15766 (IC50 = 1.2 μM), and triparanol (IC50 = 14 μM) but not by the antiestrogene tamoxifen, a triparanol analogue (Fig. 2D). The IC50 value for BM15766 was in excellent agreement with a value reported previously for the rat isoenzyme (25).
Figure 2.
Heterologous expression of the 5′ truncated Δ7-sterol reductase cDNA (D7-ORF) and a myc-tagged derivative (mycD7-ORF) in S. cerevisiae strain JB811 (ade2–1 leu2–3,112 pep4–3 trp1–289 ura3–52). (A) For Western blotting, 5 μg of microsomal protein was separated on a 12% (wt/vol) SDS gel under reducing conditions (sample buffer containing 10 mM DTT), immunostained with 20 ng/ml 9E10 c-myc antibody as in ref. 15, and developed with CDP-Star chemiluminescent reagent. The arrow indicates the migration of a c-myc immunoreactive band with a migration corresponding to 40 kDa (lane 1, mock; lane 2, D7-ORF; lane 3, mycD7-ORF). (B) Time course of Δ7-sterol reductase activity. Microsomes (0.5 mg) prepared from strains transformed with the vector without cDNA (mock, ▪), D7-ORF (•), and mycD7-ORF (⧫) were incubated anaerobically for the indicated times with 7-dehydrocholesterol in the presence of 2 mM NADPH. Mean values from duplicate determinations are shown. (C) Lineweaver Burk plot from yeast microsomes expressing D7-ORF (•) and mycD7-ORF (⧫). From this experiment Km values of 30 μM and 50 μM, respectively, were determined for D7-ORF and mycD7-ORF (Vmax 0.85 and 1.65 nmol/min per mg of protein, respectively). (D) Inhibition of the Δ7-sterol reductase (D7-ORF) by AY9944 (•, IC50 = 0.013 μM), BM15766 (⧫, IC50 = 1.2 μM), triparanol (▴, IC50 14.2 μM), and tamoxifen (▾, IC50 > 100 μM). Data shown are mean ± SD (n = 3).
The Δ7-Sterol Reductase Transcripts Are Ubiquitously Expressed.
In Northern blots the 2.9-kb mRNA was most abundantly expressed in liver and brain. A second 2.3-kb Δ7-reductase transcript was found to a variable extent (Fig. 3A). A poly(A) tail at position 2,048 of the cDNA sequence in one clone (GenBank accession no. H09710) and at position 2,581 in three other clones (GenBank accession no. AA017586, H04989, R61101) was in line with two putative polyadenylation sites at positions 2,018 (AAATAAAA) and 2,561 (ATATTAAA), respectively.
Figure 3.
Tissue distribution of the two Δ7-sterol reductase transcripts. Northern Blots with 2 μg of poly(A)+ RNA (Multiple tissue Northern blot, CLONTECH) were probed with the 32P-labeled human Δ7-sterol reductase (A) and human sterol Δ8-Δ7 isomerase cDNAs (B), respectively, and exposed for 16 hr. The arrows indicate the migration of the 2.9-kb and 2.3-kb Δ7-sterol reductase (A) and the 1.3-kb sterol isomerase (B) mRNAs, respectively.
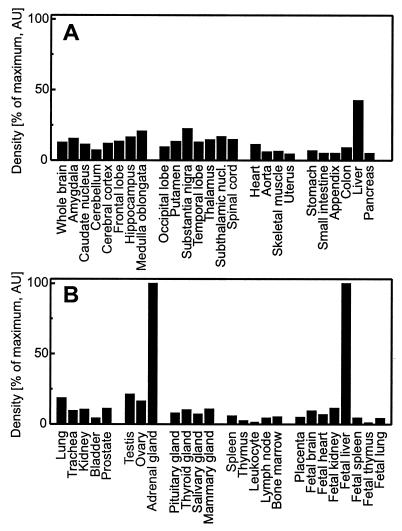
Surprisingly, the tissue distribution of the Δ7-sterol reductase transcripts was not identical with that of the 1.3-kb mRNA of the human sterol Δ8-Δ7 isomerase (Fig. 3B). Whereas the sterol isomerase is expressed to a higher extent in liver, the Δ7-sterol reductase is relatively more abundant in brain. A quantitative analysis of dot blots from multiple human tissues revealed ubiquitous expression of the Δ7-sterol reductase mRNA in adult and fetal tissues with the highest densities in adult adrenal gland, liver, testis, and brain (Fig. 4 A and B). The two enzymes Δ7-sterol reductase and sterol Δ8-Δ7 isomerase are supposed to work sequentially. It is therefore intriguing for us to find that they are differentially expressed (Fig. 3 A and B). The sigma1 receptor is a mammalian protein abundant in brain with striking structural and pharmacological similarities to the yeast sterol isomerase (14–16). The surprisingly high expression level of the Δ7-sterol reductase mRNA in adult brain lends support to our recent proposal that the sigma1 receptor is a brain-specific sterol Δ8-Δ7 isomerase (17). The regulation of these enzymes catalyzing the last steps of the de novo sterol biosynthesis as well as the biological significance of their substrates zymosterol and 7-dehydrocholesterol remain to be clarified.
Figure 4.
Ubiquitous expression of the Δ7-sterol reductase mRNA. Northern dot blots with 100–500 ng poly(A)+ RNA normalized to ubiquitin (Human RNA master blot, CLONTECH) were probed with the 32P-labeled human Δ7-sterol reductase cDNA and exposed for 4 hr. Hybridization signals were quantified with a phosphorimager and normalized to adult adrenal gland and fetal liver (100%), respectively. Data shown are the mean from two different blots. Variability was less than 10%.
The Δ7-Sterol Reductase and the SLO Syndrome.
Sterols are key players not only for the synthesis of bile acids, cholecalciferol, and steroid hormones but also for plasma membrane function. The importance of cholesterol biosynthesis for morphogenesis is illustrated by an inborn failure in the 7-dehydrocholesterol-to-cholesterol conversion. This deficiency is thought to cause the malformations characteristic of the SLO syndrome (2). Either a defect of the Δ7-sterol reductase itself, of a cofactor, or of a regulatory protein could be responsible for the observed tissue accumulation of 7-dehydrocholesterol (2, 26). A 263-bp sequence-tagged sequence (WI-16597), identical with the human Δ7-sterol reductase cDNA sequence, was assigned to human chromosome 11 between D11S913 and D11S1337 (27). Some patients with the SLO syndrome carry a translocation on chromosome 7q, but no well documented families are known (28–30). The cDNA for the Δ7-sterol reductase therefore will be important to clone the Δ7-sterol reductase gene and to identify additional proteins possibly required for the conversion of 7-dehydrocholesterol. This will help to clarify whether inherited abnormalities in this or other genes cause the disorder. With the isolation of the first mammalian cDNA for the enzyme, the cloning of the murine orthologue is feasible and will allow targeted mutations in mice to investigate treatment strategies. These mutant mice will also help to further investigate the molecular mechanism by which inhibitors of the Δ7-sterol reductase impair morphogenesis.
Acknowledgments
This work was supported by grants from the Korean Science and Engineering Foundation through Bioproducts Research Center at Yonsei University (9514–0401-00–12-3 to Y.K.P.), Fonds zur Förderung der wissenschaftlichen Forschung (S6601 and P11636 to H.G.), the Legerlotz Foundation (to F.F.M.), and the Österreichische Nationalbank (P6515 to H.G.). We thank B. Fiechtner for outstanding technical assistance.
ABBREVIATIONS
- SLO syndrome
Smith–Lemli–Opitz syndrome
- AY9944
1,4-bis-(2-chlorobenzylaminomethyl)cyclohexane
- BM15766
4-(2-(4-(4-cinnamyl)piperazine-7-yl)ethyl)-benzoic acid
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF034544).
References
- 1.Smith D W, Lemli L, Opitz J M. J Pediatr. 1964;64:210–217. doi: 10.1016/s0022-3476(64)80264-x. [DOI] [PubMed] [Google Scholar]
- 2.Tint G S, Irons M, Elias E R, Batta A K, Frieden R, Chen T S, Salen G. N Engl J Med. 1994;330:107–113. doi: 10.1056/NEJM199401133300205. [DOI] [PubMed] [Google Scholar]
- 3.Batta A K, Tint G S, Shefer S, Abuelo D, Salen G. J Lipid Res. 1995;36:705–713. [PubMed] [Google Scholar]
- 4.Shefer S, Salen G, Batta A K, Honda A, Tint G S, Irons M, Elias E R, Chen T C, Holick M F. J Clin Invest. 1995;96:1779–1785. doi: 10.1172/JCI118223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llirbat B, Wolf C, Chevy F, Citadelle D, Bereziat G, Roux C. J Lipid Res. 1997;38:22–34. [PubMed] [Google Scholar]
- 6.Dehart D B, Lanoue L, Tint G S, Sulik K K. Am J Med Genet. 1997;68:328–337. doi: 10.1002/(sici)1096-8628(19970131)68:3<328::aid-ajmg15>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 7.Wolf C, Chevy F, Pham J, Kolf-Clauw M, Citadelle D, Mulliez N, Roux C. J Lipid Res. 1996;37:1325–1333. [PubMed] [Google Scholar]
- 8.Porter J A, Young K E, Beachy P A. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 9.Chiang C, Litingtung Y, Lee E, Young K E, Corden J L, Westphal H, Beachy P A. Nature (London) 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 10.Moebius F F, Hanner M, Knaus H-G, Weber F, Striessnig J, Glossmann H. J Biol Chem. 1994;269:29314–29320. [PubMed] [Google Scholar]
- 11.Hanner M, Moebius F F, Weber F, Grabner M, Striessnig J, Glossmann H. J Biol Chem. 1995;270:7551–7557. doi: 10.1074/jbc.270.13.7551. [DOI] [PubMed] [Google Scholar]
- 12.Silve S, Dupuy P H, Labit-Lebouteiller C, Kaghad M, Chalon P, Rahier A, Taton M, Lupker J, Shire D, Loison G. J Biol Chem. 1996;271:22434–22440. doi: 10.1074/jbc.271.37.22434. [DOI] [PubMed] [Google Scholar]
- 13.Moebius F F, Burrows G G, Striessnig J, Glossmann H. Mol Pharmacol. 1993;43:139–148. [PubMed] [Google Scholar]
- 14.Hanner M, Moebius F F, Flandorfer A, Knaus H-G, Striessnig J, Kempner E, Glossmann H. Proc Natl Acad Sci USA. 1996;93:8072–8077. doi: 10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moebius F F, Bermoser K, Reiter R J, Hanner M, Striessnig J, Glossmann H. Biochemistry. 1996;35:16871–16878. doi: 10.1021/bi961996m. [DOI] [PubMed] [Google Scholar]
- 16.Moebius F F, Reiter R J, Hanner M, Glossmann H. Br J Pharmacol. 1997;121:1–6. doi: 10.1038/sj.bjp.0701079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moebius F F, Striessnig J, Glossmann H. Trends Pharmacol Sci. 1997;18:67–70. doi: 10.1016/s0165-6147(96)01037-1. [DOI] [PubMed] [Google Scholar]
- 18.Lennon G, Auffray C, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 19.Lecain E, Chenivesse X, Spagnoli R, Pompon D. J Biol Chem. 1996;271:10866–10873. doi: 10.1074/jbc.271.18.10866. [DOI] [PubMed] [Google Scholar]
- 20.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Lee, J. N. & Paik, Y.-K. (1998) J. Biochem. Mol. Biol. 30, in press.
- 22.Bae S-H, Paik Y-K. Biochem J. 1997;326:609–616. doi: 10.1042/bj3260609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer E I. Lipids. 1991;26:584–597. doi: 10.1007/BF02536422. [DOI] [PubMed] [Google Scholar]
- 24.Worman H J, Evans C D, Blobel G. J Cell Biol. 1990;111:1535–1542. doi: 10.1083/jcb.111.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis D, Galczenski H, Needle S, Tang S-Y, Amin D, Gleason M, Bilder G, Perrone M, Merkel L, Rojas C. Steroids. 1995;60:475–483. doi: 10.1016/0039-128x(95)00054-t. [DOI] [PubMed] [Google Scholar]
- 26.Kelley R I. Am J Med Genet. 1997;68:251–256. doi: 10.1002/(sici)1096-8628(19970131)68:3<251::aid-ajmg1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 27.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 28.Berry R, Wilson H, Robinson J, Sandlin C, Tyson W, Campbell J, Porreco R, Manchester D. Am J Med Genet. 1989;34:358–365. doi: 10.1002/ajmg.1320340312. [DOI] [PubMed] [Google Scholar]
- 29.Alley T L, Gray B A, Lee S-H, Scherer S W, Tsui L-C, Tint G S, Williams C A, Zori R, Wallace M R. Am J Hum Genet. 1995;56:1411–1416. [PMC free article] [PubMed] [Google Scholar]
- 30.Alley T L, Scherer S W, Huizenga J J, Tsui L-C, Wallace M R. Am J Med Gen. 1997;68:279–281. [PubMed] [Google Scholar]