Abstract
By interference of the yeast pheromone mitogen-activated protein kinase (MAPK) pathway with an alfalfa cDNA expression library, we have isolated the MP2C gene encoding a functional protein phosphatase type 2C. Epistasis analysis in yeast indicated that the molecular target of the MP2C phosphatase is Ste11, a MAPK kinase kinase that is a central regulator of the pheromone and osmosensing pathways. In plants, MP2C functions as a negative regulator of the stress-activated MAPK (SAMK) pathway that is activated by cold, drought, touch, and wounding. Although activation of the SAMK pathway occurs by a posttranslational mechanism, de novo transcription and translation of protein factor(s) are necessary for its inactivation. MP2C is likely to be this or one of these factors, because wound-induced activation of SAMK is followed by MP2C gene expression and recombinant glutathione S-transferase–MP2C is able to inactivate extracts containing wound-induced SAMK. Wound-induced MP2C expression is a transient event and correlates with the refractory period, i.e., the time when restimulation of the SAMK pathway is not possible by a second stimulation. These data suggest that MP2C is part of a negative feedback mechanism that is responsible for resetting the SAMK cascade in plants.
Keywords: signal transduction, stress
The mitogen-activated protein kinase (MAPK) cascade is a universal signal transduction pathway found in both unicellular and multicellular eukaryotes (1). Cells have multiple MAPK pathways that are differentially activated by a variety of external stimuli including growth factors, UV radiation, and osmotic stress. MAPK cascades are usually composed of three protein kinases. By phosphorylation of conserved threonine and tyrosine residues, a MAPK becomes activated by a specific MAPK kinase (MAPKK) (2, 3). A MAPKK kinase (MAPKKK) activates MAPKK through phosphorylation of conserved threonine and/or serine residues (4).
Inactivation of MAPK pathways can be mediated through dephosphorylation of MAPKs by the dual-specificity phosphatases, such as murine MKP-1 (5) or the yeast MSG5 product (6), or tyrosine phosphatases, encoded by the yeast PTP2 and PTP3 genes (7). Serine/threonine phosphatases, such as mammalian protein phosphatase 2A (PP2A) (8), and yeast protein phosphatases type 2C (PP2Cs) can also function as negative regulators of MAPK pathways (9–11).
By suppression of the budding yeast MAPK pathway that mediates pheromone signaling, we have identified the alfalfa MP2C gene, encoding a PP2C. Genetic analysis indicated that the molecular target of MP2C is the MAPKKK, encoded by the yeast STE11 gene. Several lines of evidence indicate that MP2C also acts as a negative regulator in plants that may be important for resetting a stress-signaling MAPK pathway.
MATERIALS AND METHODS
Yeast Strains and Techniques.
For suppression analysis of MP2C in the pheromone pathway, K2149 (MATa, HMLa, HMRa, ho-βgal, ura3, HIS4, ade2–1, can1–100, met, his3, leu2–3, 112, trp1–1, bar1∷HISG) cells were transformed with pGal-STE4 (12), pGal-STE20ΔN (13), pGal-STE11–4 (14), pGal-STE12 (15), pGAL-MP2C (pYEUra3-MP2C), or vector (pYEUra3).
For testing the ability of MP2C to inactivate the HOG1 pathway, TM222 (MATa, ura3, leu2, trp1, his3, lys2) was transformed with pGAL-SSK2ΔN or pGAL-PBS2D,D, carrying dominant active alleles of the MAPKKK and MAPKK, respectively (7, 16), and pGal-MP2C (YCPlac111 containing GAL1/10p-MP2C from pYEura3-MP2C) or vector (YCPlac111).
The diploid strain GA2201, carrying the MATa and silent a mating locus gene, was derived by mating strains CY2 (MATα∷sup40, HMLa, HMRa, ho-βgal, ura3, HIS4, ade6-O, ade2–1, can1–100, met, his3, leu2–3, 112, trp1–1, swi6∷TRP1, bar1∷HISG) and K2149 (MATa, HMLa, HMRa, ho-βgal, ura3, HIS4, ade2–1, can1–100, met, his3, leu2–3, 112, trp1–1, bar1∷HISG). GA2201 was transformed with an alfalfa cDNA expression library (17) that was constructed in the EcoRI site of the pYEUra3 plasmid (CLONTECH).
Expression of Alfalfa MP2C in Bacteria.
To study the MP2C phosphatase, the alfalfa gene was cloned in the pGEX-3x vector (Pharmacia). The reading frame was inserted into pGEX3x via EcoRI. After transforming Escherichia coli with the construct, positive clones containing the fragment in the correct orientation were isolated and sequenced to verify that no mutations had occurred in the ORF. Preparation of the glutathione S-transferase (GST) fusion protein and affinity purification was done according to the manufacturer’s instructions (Pharmacia). Protein concentration was determined with a Bio-Rad detection system (Bio-Rad). Proteins were collected in 50-μl aliquots, frozen in liquid nitrogen, and stored at −80°C. The quality of the purification was checked on denaturing 10% polyacrylamide gels.
Phosphatase Assays.
Phosphatase activity of GST–MP2C was determined with radiolabeled casein as described (18). Briefly, dephosphorylated bovine casein (Sigma, product C-4765) was 32P-labeled with bovine heart cAMP-dependent protein kinase (Sigma, product P-2645). Radiolabeled casein was precipitated with 20% trichloroacetic acid. After washing three times with 20% trichloroacetic acid and once with acetone, the casein was dissolved in 0.2 M Tris⋅HCl (pH 8.0). The phosphatase assay was done in a total volume of 50 μl containing 60 mM Tris⋅HCl (pH 7.0), 0.1 mM EGTA, 10 mM MgCl2, 10 mM 2-mercaptoethanol, 500 nM okadaic acid, and 100,000 cpm of radiolabeled casein. Bead-bound recombinant GST–MP2C (15 μl) was incubated with the mixture for various times at room temperature. The reaction was terminated by the addition of 0.75 ml of an acidic charcoal mixture [0.9 M HCl/90 mM sodium pyrophosphate/2 mM NaH2PO4/4% (vol/vol) Norit A]. Samples were centrifuged, and the amount of radioactivity in 0.4 ml of supernatant was determined by scintillation counting.
Plant Culture Conditions.
Alfalfa (Medicago sativa ssp. varia, cv. Rambler, line A2) plants were grown in soil or under sterile conditions on hormone-free MS medium (19).
Immunokinase Assays.
Alfalfa leaves were mechanically wounded by cutting the lamina with a razor blade to strips of 1 mm2. Leaf extracts were prepared at various times after wounding in extraction buffer [25 mM Tris⋅HCl, pH 7.5/15 mM MgCl2/5 mM EGTA/75 mM NaCl/1 mM DTT/0.1% Nonidet P-40/15 mM p-nitrophenyl phosphate/60 mM β-glycerophosphate/15 mM p-nitrophenyl phosphate/60 mM β-glycerophosphate/0.1 mM NaVO3/1 mM NaF/1 mM phenylmethylsulfonyl fluoride/leupeptine (10 μg/ml)/aprotinin (10 μg/ml)/soybean trypsin inhibitor (10 μg/ml)/antipain (5 μg/ml)/chymostatin (5 μg/ml)/pepstatin (5 μg/ml)]. After the centifugation at 100,000 × g for 1 h, the supernatant was collected and used for further investigations.
Leaf extracts containing 100 μg of total protein were immunoprecipitated with 5 μg of protein A-purified M7 antibody, which was produced against synthetic peptide encoding the carboxyl-terminal 10 amino acids of the alfalfa stress-activated MAPK (SAMK) (20). The immunoprecipitated protein was washed three times with buffer I (20 mM Tris⋅HCl/5 mM EDTA/100 mM NaCl/1% Triton X-100), once with the same buffer but containing 1 M NaCl, and once with kinase buffer (20 mM Hepes, pH 7.5/15 mM MgCl2/5 mM EGTA/1 mM DTT). Kinase reactions of the immunoprecipitated protein were performed in 15 μl of kinase buffer containing myelin basic protein at 0.5 mg/ml, 0.1 mM ATP, and 2 μCi of [γ-32P]ATP (1 Ci = 37 GBq). The protein kinase reaction was carried out at room temperature for 30 min. The reaction was stopped by addition of SDS sample buffer. The phosphorylation of myelin basic protein was analyzed by autoradiography after SDS/PAGE.
In Vitro Inactivation of SAMK Pathway.
To study the factors involved in the inactivation of the SAMK pathway, protein extracts were prepared from leaves 10 min after wounding, a time, when the SAMK activity was decreasing. The initial extraction was performed in a buffer containing no phosphatase inhibitors [25 mM Tris⋅HCl, pH 7.5/15 mM MgCl2/15 mM EGTA/75 mM NaCl/1 mM DTT/0.1% Nonidet P-40/1 mM phenylmethylsulfonyl fluoride/leupeptine (10 μg/ml)/aprotinin (10 μg/ml)/soybean trypsin inhibitor (10 μg/ml)/antipain (5 μg/ml)/chymostatin (5 μg/ml)/pepstatin (5 μg/ml)]. After the centifugation at 100,000 × g for 1 h, aliquots of the supernatant were incubated for 30 min at room temperature in the absence or presence of GST–MP2C and various combinations of the following inhibitors: 500 nM okadaic acid and 0.1 mM NaVO3 (+Inh) and/or 10 mM EDTA (+EDTA). After incubation, the activation state of the SAMK pathway was determined by in vitro kinase assays of the immunoprecipitated SAMK.
RNA Gel Blot Analysis.
RNA was extracted from leaves as described (17). Two micrograms of poly(A)+ RNA was separated per lane on a denaturing formaldehyde gel. After blotting to nylon membranes, the blot was sequentially hybridized with radiolabeled fragments, containing the MP2C cDNA, the 3′ nontranslated region of the SAMK gene, and the coding region of the MsWIP gene (21). As a control, the blot was hybridized with a radiolabeled EcoRI–XhoI fragment of the constitutively expressed Msc27 gene (22).
RESULTS
MP2C Suppresses the Pheromone-Induced MAPK Signal Transduction Pathway in Yeast.
To identify plant genes that are involved in the regulation of MAPK signaling pathways, we conditionally expressed a plant cDNA library in a pheromone-hypersensitive yeast strain and selected for cells that could override pheromone-induced cell cycle arrest and form colonies in the presence of pheromone (17). One of the cDNAs putatively encodes a PP2C and was, therefore, denoted as MP2C for Medicago phosphatase 2C.
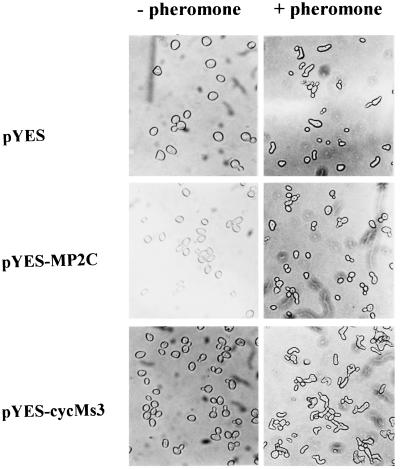
The addition of a pheromone has multiple effects on yeast: it results in the activation of a MAPK pathway inducing a transcriptional program with characteristic morphological changes (projection formation called shmooing) and an arrest in the G1 phase of the cell cycle. These two responses are thought to be due to a bifurcation of the pathway at the level of the Fus3 and Kss1 MAPKs. In the absence of α-pheromone, yeast cells expressing the previously identified cyclin cycMs3 (17) or MP2C cDNA did not show significant morphological differences compared with nontransformed cells (Fig. 1). Yeast cells expressing cycMs3 responded to α-pheromone by exhibiting projection formation (shmooing) but no cell cycle arrest (Fig. 1), indicating that cycMs3 acts downstream of the bifurcation point. In contrast, expression of MP2C suppressed both projection formation and cell cycle arrest (Fig. 1). According to the model described above, these results suggest that MP2C acts at or upstream of the Fus3 and Kss1 kinases of the pheromone signaling pathway.
Figure 1.
MP2C suppresses pheromone-induced cell cycle arrest and projection formation. The diploid strain GA2201 (17) was transformed with pYES, pYES-MP2C, and pYES-cycMs3. Yeast cells grow normally in the absence of pheromone (− pheromone) but show cell cycle arrest and projection formation in its presence (+ pheromone). Cells expressing MP2C grow normally in the absence and presence of pheromone. In contrast, GA2201 cells, expressing the cycMs3 cyclin, grow normally in the absence of pheromone, but addition of pheromone induces projection formation but not cell cycle arrest.
MP2C Encodes a Plant PP2C.
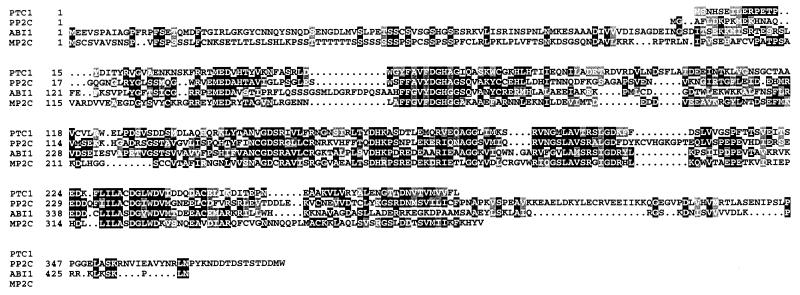
Sequence analysis of the MP2C cDNA revealed an ORF of 1160 nucleotides potentially encoding a 45-kDa protein. MP2C showed significant homology to PP2Cs such as Ptc1 from yeast (23), ABI1 from Arabidopsis (24, 25), and human PP2C (26). The similarity between these proteins is restricted to the catalytic domains (Fig. 2), and MP2C has a unique amino terminus that shows no similarity with any known protein.
Figure 2.
Sequence alignment of predicted protein sequences of yeast PTC1 (7), human PP2C (26), Arabidopsis ABI1 (24, 25), and alfalfa MP2C. Gaps were introduced for optimal alignment and are shown by dashes. Identical amino acid residues are indicated with black boxes, and similar amino acids are indicated with shaded boxes.
MP2C Encodes a Functional PP2C.
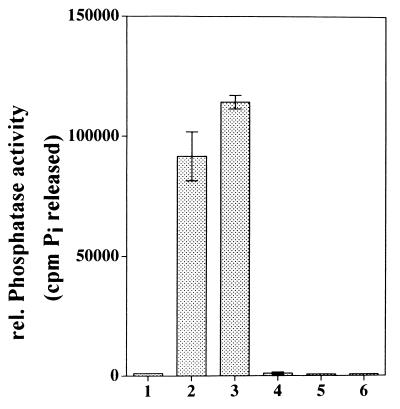
Protein phosphatases can be divided into two major classes: protein serine/threonine phosphatases and protein tyrosine phosphatases. Protein serine/threonine phosphatases are further categorized into four groups according to their substrate specificities, divalent cation requirements, and sensitivity to inhibitors: types 1 (PP1), 2A (PP2A), 2B (PP2B), and 2C (PP2C) (27). Whereas PP1, PP2A, and PP2B share approximately 40% identity in their catalytic domains, the encoded PP2C proteins have no apparent sequence homology to the other classes of phosphatases (28). PP2C enzymes also have the unique feature of occurring as monomers, whereas the other classes form oligomeric complexes with regulatory subunits, which are thought to regulate activity, substrate specificity, and cellular localization of the phosphatases (27, 28). In many respects, PP2C also differs from the other serine/threonine phosphatases in that PP2C requires Mn2+ and/or Mg2+ and is insensitive to the phosphatase inhibitor okadaic acid (27). To test whether the MP2C cDNA encodes a functional enzyme that has the properties of PP2C enzymes, we expressed MP2C as a GST fusion protein in E. coli. The phosphatase activity of affinity-purified GST–MP2C protein was determined with 32P-phosphorylated casein as substrate. GST–MP2C was able to dephosphorylate casein, and this ability was strictly dependent on the presence of Mg2+ or Mn2+ (Fig. 3, lanes 2 and 3, respectively). As a control, GST was incubated with 32P-labeled casein but was unable to release Pi (Fig. 3, lane 1). The ABI1 gene encodes a PP2C that has an EF hand motif that is thought to regulate the activity of the enzyme in a Ca2+-dependent manner. Although no such motif was found in the MP2C amino acid sequence, we also tested an influence of Ca2+ on the phosphatase activity of GST–MP2C. GST–MP2C was completely inactive when Mg2+ or Mn2+ was replaced by Ca2+ (Fig. 3, lane 4). Moreover, the addition of Ca2+ to GST–MP2C containing Mg2+ or Mn2+ did not affect the phosphatase activity of MP2C (data not shown). PP2Cs are Mg2+- or Mn2+-dependent enzymes and can be inhibited by the chelating agent EDTA. When 10 mM EDTA was added to active GST–MP2C containing Mg2+ or Mn2+, the dephosphorylation of 32P-labeled casein was completely inhibited (Fig. 3, lanes 5 and 6, respectively). Protein serine/threonine phosphatases other than PP2C are sensitive to okadaic acid (28). Addition of 500 nM okadaic acid did not inhibit the phosphatase activity of GST–MP2C in vitro and was, therefore, included in all PP2C assays (Fig. 3, lanes 1–6). These data indicate that MP2C encodes an enzyme that has all the properties of type 2C protein phosphatases.
Figure 3.
MP2C encodes an active PP2C. The MP2C gene was expressed as a GST fusion protein in E. coli. Affinity-purified GST–MP2C was incubated with 32P-labeled casein, and the released 32Pi was taken as a measure for the phosphatase activity. 32Pi released by incubation of 32P-labeled casein with GST containing Mg2+ (bar 1), GST–MP2C containing Mg2+ (bar 2), GST–MP2C containing Mn2+ (bar 3), GST–MP2C containing Ca2+ (bar 4), GST–MP2C containing Mg2+ and 10 mM EDTA (bar 5), GST–MP2C containing Mn2+ and 10 mM EDTA (bar 6) is shown.
The Molecular Target of MP2C in Yeast Is the Ste11 Kinase.
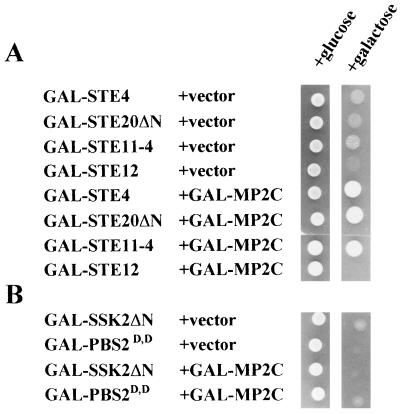
To define the target of MP2C in the pheromone pathway, suppression analysis of MP2C was performed in yeast cells that conditionally overexpressed genes encoding either the Gβ protein subunit Ste4 (12), the transcription factor Ste12 (15), or dominant active forms of the MAPKKK Ste11 (Ste11–4) (14) or MAPKKK activator Ste20 (Ste20ΔN) (13). Whereas MP2C expression suppressed the lethal phenotype mediated by ectopic expression of the STE4, STE20ΔN and STE11–4 genes, no suppression was obtained in yeast that overexpressed STE12 (Fig. 4A). These results indicate that MP2C acts at or below the Ste11 kinase but above the Ste12 transcription factor.
Figure 4.
Inactivation of the Ste11 MAPKKK by MP2C. (A) MP2C suppresses the lethal phenotype caused by overexpression of STE4, STE20ΔN, and STE11–4 but not STE12. For suppression analysis of MP2C in the pheromone pathway, K2149 cells were transformed with pGal-STE4, pGal-STE20ΔN, pGal-STE11–4, pGal-STE12, pGAL-MP2C, or vector. All strains containing the vector are viable on glucose but show galactose-induced lethality. MP2C can suppress the galactose-induced lethality of STE4, STE20ΔN, and STE11–4, but not that of STE12. (B) For testing the ability of MP2C to inactivate the HOG1 pathway, TM222 was transformed with pGAL-SSK2ΔN or pGAL-PBS2D,D, carrying dominant active alleles of the MAPKKK and MAPKK, respectively, and pGal-MP2C or vector. All strains containing the vector are viable on glucose but show galactose-induced lethality. MP2C expression was unable to suppress the galactose-induced lethality.
Because Ste11 activates both the Ste7 kinase of the pheromone pathway and the Pbs2 kinase of the osmosensing HOG1 pathway (7, 16), suppression of the dominant active Ste11–4 allele by MP2C would be possible only if MP2C acted on Ste11 itself or on downstream components of both the pheromone and osmosensing pathways. To distinguish between these possibilities, yeast cells that conditionally overexpressed a constitutively active SSK2 allele (SSK2ΔN) were analyzed for the ability of MP2C to inactivate the downstream target Pbs2 or Hog1. MP2C was not able to suppress the SSK2ΔN-mediated lethal phenotype (Fig. 4B), indicating that it does not act on Pbs2 or other downstream components of the Hog1 cascade. In agreement with these results, MP2C was also not able to suppress a constitutively active PBS2 allele (PBS2D,D), indicating that MP2C does not act on Hog1 or other downstream components of the Hog1 pathway (Fig. 4B). Thus, the genetic analysis suggests that MP2C-mediated inactivation of the pheromone and osmosensing pathways is mediated via the Ste11 kinase.
MP2C Gene Expression Is Induced by Wounding and Correlates with Inactivation of the SAMK Pathway.
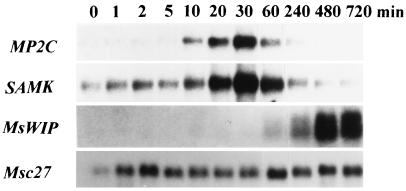
Because MP2C appears to act as a negative regulator on the pheromone MAPK pathway in yeast, we tested the possibility that MP2C might also be involved in regulation of a plant MAPK pathway. The stress-induced SAMK pathway is a MAPK cascade that is activated in response to cold, drought, touch, and wounding, leading to the transient activation of SAMK (20, 21, 29). The transient activation of SAMK was shown to be dependent on two opposing processes: on an activating posttranslational mechanism and on inactivating protein factor(s) that is synthesized de novo (21). To test whether the MP2C gene expression pattern might qualify for the negative regulatory process that is responsible for the inactivation of the SAMK pathway, gel blot analysis was performed with RNA extracted from leaves before and after wounding. MP2C gene expression was induced within 10 min after wounding (Fig. 5). After maximal expression at 30 min, MP2C transcript amounts gradually decreased to nondetectable levels. Transcript amounts of the SAMK gene also showed transient accumulation with maximal levels at 30 min (Fig. 5). In contrast to SAMK transcript, no MP2C mRNA was detected in nonwounded leaves (Fig. 5, at 0 min). Whereas the MP2C and SAMK genes are rapidly and transiently induced, the systemically expressed MsWIP gene, encoding a Bowman–Birk type protease inhibitor (21), was only induced at 60 min after wounding and showed gradual transcript accumulation over many hours (Fig. 5). Hybridization of the same filter with a radiolabeled fragment of the constitutively expressed Msc27 gene (Fig. 5) showed that the fluctuation of MP2C and SAMK transcripts was not due to differences in the loading of the RNA gel blot. The similar kinetics of the wound-induced expression of the SAMK and MP2C genes and the fact that these genes were induced shortly after activation of SAMK suggest that the expression of these genes might be regulated by the SAMK pathway.
Figure 5.
MP2C gene is transiently induced after wound-induced SAMK activation. Alfalfa plants were grown under sterile conditions on hormone-free MS medium (19). RNA was extracted from leaves as described (17) before (0 min) and 10, 30, 60, 120, and 240 min after wounding. Poly(A)+ RNA (2 μg) was separated per lane on a denaturing formaldehyde gel. After blotting to nylon membranes, the blot was sequentially hybridized with fragments containing the coding regions of the MP2C and MsWIP genes, and the 3′ nontranslated region of the SAMK gene, respectively (21). As a control, the blot was hybridized with a radiolabeled fragment of the constitutively expressed Msc27 gene (22).
MP2C Inactivates the SAMK Pathway.
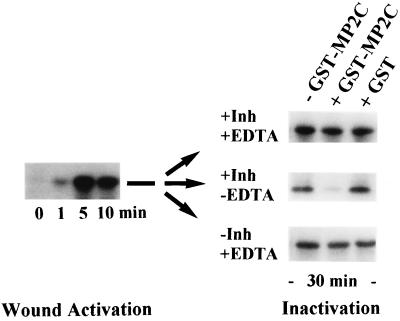
MP2C expression is correlated with the inactivation of the SAMK pathway in plants. To test whether MP2C can inactivate the SAMK pathway in a direct way, wound-induced leaf extracts were incubated in the absence or presence of active recombinant GST–MP2C. After 30 min of incubation of the extracts, the activity of the SAMK was determined by immunocomplex kinase assays (Fig. 6). Different MAPKs can be inactivated by the serine/threonine-specific phosphatase PP2A (8), tyrosine-specific phosphatases (7), and dual-specificity phosphatases (5, 6). PP2A is highly sensitive to okadaic acid, and the tyrosine-directed phosphatases can be inhibited by vanadate (27). Because MP2C is insensitive to these drugs, leaf extracts were incubated with GST–MP2C in the presence (+Inh) or absence (−Inh) of okadaic acid and vanadate. In the presence of okadaic acid and vanadate, GST–MP2C still showed significant inactivation of SAMK (Fig. 6, +Inh −EDTA). However, when EDTA was included in the reaction buffer, the MP2C-dependent inactivation was completely blocked (Fig. 6, +Inh +EDTA). Although it can be argued that EDTA might affect other enzymes besides MP2C, the addition of GST alone showed no inactivation of SAMK under these conditions (Fig. 6). These results strongly support the idea that MP2C is a negative regulator of the SAMK pathway, suggesting that the wound-induced expression of the MP2C gene constitutes a feedback mechanism for resetting this stress signaling pathway.
Figure 6.
MP2C inactivates the SAMK pathway. MP2C-mediated inactivation of the SAMK pathway was determined by incubating wound-induced extract that was prepared 10 min after wounding and contained 100 μg of total protein for 30 min at room temperature with GST–MP2C, no protein, or GST and various combinations of the following phosphatase inhibitors: 500 nM okadaic acid, 0.1 mM NaVO3 (+Inh), and 10 mM EDTA (+EDTA). After incubation, the activity of the SAMK was determined by immunocomplex kinase assays.
DISCUSSION
PP2Cs: Common Negative Regulatory Elements of Eukaryotic MAPK Pathways.
Signal transduction processes are based on the reversible phosphorylation of signaling components and MAPK cascades are key players in several of these pathways. To identify and study the function of negative regulatory components of MAPK pathways, we have used a heterologous approach that makes use of the pheromone-mediated MAPK cascade of yeast cells. Interference of this yeast signaling cascade by expression of an alfalfa cDNA library led to the isolation of the MP2C gene that encodes a PP2C. A common function of yeast PP2Cs from fission yeast and budding yeast is their action on MAPK pathways that are responsible for adaptation to abiotic stresses, such as heat shock and hyperosmolar conditions (9–11). Although the MP2C gene was identified in a screen for genes that suppress pheromone signaling, genetic analysis indicated that the target of MP2C is Ste11, a MAPKKK that plays a role in both the pheromone and the osmosensing yeast MAPK cascade. Furthermore, we provide evidence that MP2C has a negative function on a plant MAPK cascade, suggesting that PP2Cs might be common negative regulators of eukaryotic MAPK pathways.
MP2C Is a Negative Regulator of the Stress-Activated MAPK Pathway in Plants.
Abiotic stresses such as cold, drought, touch, and wounding induce the activation of the SAMK pathway (20, 21, 29). The stress-induced activation of SAMK is a transient process, indicating the operation of activating and inactivating mechanisms. We have previously shown that the activation of the SAMK cascade is a posttranslational event but that the inactivation requires the transcription and translation of a protein factor(s). Several lines of evidence strongly indicate that MP2C might be this or one of the factors responsible for the inactivation of the SAMK pathway. (i) Active recombinant GST–MP2C was able to inactivate the SAMK pathway in vitro. (ii) MP2C gene expression occurred rapidly and transiently after wounding. (iii) The presence of MP2C transcript was correlated with the length of the refractory period, i.e., when an inactivation mechanism prohibits reactivation of the SAMK pathway by a second stimulus.
SAMK-Dependent MP2C Expression Provides a Regulatory Mechanism to Reset the Stress Signaling Pathway.
A cell requires sensing mechanisms to monitor various extracellular parameters, such as environmental conditions and nutrients, and potential enemies, such as microbial pathogens. After activation of a plasma-membrane-located sensor, the information is transmitted through intracellular signaling mechanisms to distinct effectors, in many cases transcription factors in the nucleus where expression of specific target genes is induced. Because a cell requires continuous monitoring of the extracellular environment, a mechanism must exist that turns the signal transduction cascade off again. One way to do this is through inactivation of the MAPK by dephosphorylation. In yeast HOG1 pathway, the tyrosine phosphatases Ptp1 and Ptp2 appear to inactivate Hog1 by a posttranslational mechanism (7). However, as shown for for the dual-specificity phosphatase genes MSG5 and MKP-1 (5, 6), MAPK cascades can also be inactivated through transcriptional induction of their respective phosphatase genes. The plant SAMK pathway is inactivated through the transcriptional induction of negative regulatory gene(s) and our evidence indicates that MP2C belongs to this class of genes. As long as the MP2C gene is expressed, de novo activation of the pathway will be counteracted by the accumulating MP2C phosphatase, resulting in the inability to transmit further incoming signals. This situation is known as a refractory period, when restimulation of a pathway is not possible, but an intriguingly simple solution to this inherent dilemma is provided by coupling MP2C gene expression to the activity state of the SAMK. Under these conditions, MP2C-mediated inactivation of the SAMK pathway will also lead to a halt in MP2C gene expression, allowing the reactivation and transduction of further signals. In the SAMK pathway, such a mechanism appears to operate, because restimulation is generally found to be completely inhibited 30 min after applying the first stimulus. Thereafter, it usually takes another 30 min to obtain a full response upon restimulation (21). Not only does the refractory period coincide with the time of MP2C gene expression, but the amounts of MP2C transcript also tightly correlate with the activation levels of the SAMK. Because MP2C can inactivate MAPK pathways in yeast and plants, it would not be surprising to find that PP2Cs are common negative players in other MAPK cascades.
Acknowledgments
This work was supported by grants from the Austrian Science Foundation (P10394-MOB and P12188-GEN), the Austrian National Bank (Grants 6431 and 6159), the Human Capital and Mobility (CHRX-CT94-0656), and the Training and Mobility of Researchers (ERB 4061 PL 95-0114) programmes of the European Union.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; MAPKKK, MAPKK kinase; PP2C, protein phosphatase 2C; PP2A, protein phosphatase 2A; SAMK, stress-activated MAPK; GST, glutathione S-transferase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y11607).
References
- 1.Waskiewicz A J, Cooper J. Curr Opin Cell Biol. 1995;7:798–802. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 2.Anderson N G, Maller J L, Tonks N K, Sturgill T W. Nature (London) 1990;343:651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- 3.Posada J, Sanghera J, Pelech S, Aebersold R, Cooper J. Mol Cell Biol. 1991;11:2517–2528. doi: 10.1128/mcb.11.5.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alessi D R, Saito Y, Campbell D G, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall C J, Cowley S. EMBO J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun H, Charles C H, Lau L F, Tonks N K. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 6.Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, Matsumoto K. EMBO J. 1994;13:61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wurgler-Murphy S M, Maeda T, Witten E A, Saito H. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alessi D R, Gomez N, Moorhead G, Lewis T, Keyse S M, Cohen P. Curr Biol. 1995;5:283–295. doi: 10.1016/s0960-9822(95)00059-5. [DOI] [PubMed] [Google Scholar]
- 9.Maeda T, Wurgler-Murphy S M, Saito H. Nature (London) 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 10.Shiozaki K, Akhavian-Niaki H, McGowan C H, Russell P. Mol Cell Biol. 1994;14:3742–3751. doi: 10.1128/mcb.14.6.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiozaki K, Russell P. EMBO J. 1995;14:492–502. doi: 10.1002/j.1460-2075.1995.tb07025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole G M, Stone D E, Reed S I. Mol Cell Biol. 1990;10:510–517. doi: 10.1128/mcb.10.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramer S W, Davis R W. Proc Natl Acad Sci USA. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolan J W, Fields S. Genes Dev. 1990;4:492–502. doi: 10.1101/gad.4.4.492. [DOI] [PubMed] [Google Scholar]
- 15.Stevenson B J, Rhodes N, Errede B, Sprague G F. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 16.Maeda T, Tsai A Y M, Saito H. Mol Cell Biol. 1993;13:5408–5417. doi: 10.1128/mcb.13.9.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meskiene I, Bögre L, Dahl M, Pirck M, Ha D T H, Swoboda I, Heberle-Bors E, Ammerer G, Hirt H. Plant Cell. 1995;7:759–771. doi: 10.1105/tpc.7.6.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Streuli M, Krueger N X, Thai T, Tang M, Saito H. EMBO J. 1990;9:2399–2407. doi: 10.1002/j.1460-2075.1990.tb07415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murashige T, Skoog F. Physiol Plant. 1962;15:473–479. [Google Scholar]
- 20.Jonak C, Kiegerl S, Ligterink W, Barker P J, Huskisson N S, Hirt H. Proc Natl Acad Sci USA. 1996;93:11274–11279. doi: 10.1073/pnas.93.20.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bögre L, Ligterink W, Meskiene I, Barker P J, Heberle-Bors E, Huskisson N S, Hirt H. Plant Cell. 1997;9:75–83. doi: 10.1105/tpc.9.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pay A, Heberle-Bors E, Hirt H. Plant Mol Biol. 1992;19:501–503. doi: 10.1007/BF00023399. [DOI] [PubMed] [Google Scholar]
- 23.Maeda T, Tsai A Y M, Saito H. Mol Cell Biol. 1993;13:5408–5417. doi: 10.1128/mcb.13.9.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung J, Bouvier-Durnad M, Morris P-C, Guerrier D, Chefdor F, Giraudat J. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- 25.Meyer K, Leube M P, Grill E. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- 26.Mann D J, Campbell D G, McGowan C H, Cohen P T W. Biochim Biophys Acta. 1992;1130:100–104. doi: 10.1016/0167-4781(92)90471-b. [DOI] [PubMed] [Google Scholar]
- 27.Cohen P. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 28.Cohen P, Cohen P T W. J Biol Chem. 1989;264:21435–21438. [PubMed] [Google Scholar]
- 29.Bögre L, Ligterink W, Heberle-Bors E, Hirt H. Nature (London) 1996;383:489–490. doi: 10.1038/383489a0. [DOI] [PubMed] [Google Scholar]