Abstract
Photosynthetic membranes of higher plants contain specific nonphosphorous lipids like the sulfolipid sulfoquinovosyl diacylglycerol in addition to the ubiquitous phospholipid phosphatidylglycerol. In bacteria, an environmental factor that drastically affects thylakoid lipid composition appears to be the availability of phosphate. Accordingly, we discovered an increase in the relative amount of sulfolipid and a concomitant decrease in phosphatidylglycerol in Arabidopsis thaliana grown on medium with reduced amounts of phosphate, as well as in the pho1 mutant of A. thaliana deficient in phosphate transport. To investigate the molecular basis of the observed change in lipid composition, we isolated a cDNA of A. thaliana, designated SQD1, that encodes a protein involved in sulfolipid biosynthesis as suggested by three lines of evidence. First, the cDNA shows high sequence similarity to bacterial sqdB genes known to be essential for sulfolipid biosynthesis; second, the SQD1 gene product is imported into chloroplasts where sulfolipid biosynthesis takes place; and third, transgenic plants expressing SQD1 in antisense orientation show a reduction in sulfolipid content. In the pho1 mutant as well as in wild-type plants grown under reduced phosphate availability, increased amounts of SQD1 mRNA and SQD1 protein are detected, suggesting that the increase in sulfolipid content under phosphate limitation is the result of an increased expression of at least one gene required for sulfolipid biosynthesis in A. thaliana. It is suggested that a certain amount of anionic thylakoid lipid is maintained by substituting sulfolipid for phosphatidylglycerol under reduced phosphate availability.
Oxygenic photosynthesis of higher plants depends on highly organized pigment protein complexes that are embedded in the polar lipid matrix of thylakoid membranes inside chloroplasts. The lipid composition of thylakoids is distinct from that of other cell membranes, but it is also largely conserved between different plants, suggesting that the structure of thylakoid lipids is important for photosynthesis. Four glycerolipids (monogalactosyl diacylglycerol, digalactosyl diacylglycerol, sulfoquinovosyl diacylglycerol, and phosphatidylglycerol) provide the bulk of the lipid matrix of the thylakoids (1). Although the overall lipid composition of photosynthetic membranes generally appears to be under tight homeostatic control, we discovered that phosphate limitation can cause drastic changes in photosynthetic bacteria. In experiments with the purple bacterium Rhodobacter sphaeroides, we observed a decrease in the relative amounts of all phospholipids and an increase in the amounts of nonphosphorous lipids in response to phosphate limitation (2). Most notably was the accumulation of a novel dihexosylglycerolipid and a betaine lipid, and a drastic increase in the amount of sulfolipid (3). More recently, we observed an increase in the relative amounts of digalactosyl diacylglycerol and sulfolipid in the cyanobacterium Synechococcus sp. PCC7942 grown under phosphate limitation (4).
With the isolation of sqd genes required for sulfolipid biosynthesis in R. sphaeroides (5–7) and in Synechococcus sp. PCC7942 (4), it was possible to create sulfolipid-deficient null-mutants by gene disruption. The complete lack of sulfolipid in mutants of R. sphaeroides and of Synechococcus sp. PCC7942 did affect neither the growth nor photosynthesis of cells grown under optimal conditions, but caused reduced growth under phosphate limitation (2, 4). The relative amount of phosphatidylglycerol was increased in the mutants and did not drastically decrease under reduced phosphate availability. Taken together, these observations led to the hypothesis that, although sulfolipid presumably has no specific role in anoxygenic and oxygenic photosynthesis in bacteria, it acts as a surrogate to replace the anionic phosphatidylglycerol under conditions of reduced phosphate availability (2, 4). The question remained whether this hypothesis can be extended to algae and higher plants. Unlike bacteria, the photosynthetic apparatus of eukaryotes is organized in a subcellular compartment, the chloroplast, and the light harvesting antenna system of higher plants is different from those of photosynthetic bacteria (8). Recently, a sulfolipid-deficient mutant of the unicellular alga Chlamydomonas reinhardtii has been isolated that shows altered chlorophyll fluorescence and changes in the organization of the photosynthetic apparatus (9). This result suggests that sulfolipid may be more crucial to photosynthesis in this organism compared with photosynthetic bacteria.
In the present study, we demonstrate that similar changes in the thylakoid lipid composition occur in A. thaliana in response to reduced phosphate availability as in the photosynthetic bacteria. To understand the regulatory basis for this apparent adaptation, we isolated and characterized a SQD gene encoding a protein involved in sulfolipid biosynthesis in higher plants and studied its expression in response to phosphate availability.
MATERIALS AND METHODS
A. thaliana Growth Conditions.
For routine growth, plants of A. thaliana ecotype Columbia (Col-2) were raised either on sterile Murashige–Skoog medium (10) containing 0.8% (wt/vol) agarose, 1% (wt/vol) sucrose, and 25 μg/ml hygromycin B as required, or in soil (Einheitserde Type P/Einheitserde Type T/sand (2:1:1); Gebrüder Patzer, Sinntal-Jossa, Germany). For phosphate limitation experiments, a sterile mineral solution was used as described by Estelle and Somerville (11), but at half concentration and containing 0.8% agarose, 1% sucrose, 20 mM Hepes (pH 6.0), and KH2PO4 as indicated. Irrespective of the medium, plants were exposed to a 16-h light/8-h dark regime at 100 μmol photons m−2 sec−1. Seeds were surface-sterilized according to Estelle and Somerville (11).
cDNA Isolation and Analysis.
The SQD1 cDNA was isolated by screening 2.4 million plaque-forming units of the PRL2 cDNA library (12) by heterologous DNA hybridization (13) using a 400-bp XhoI–EcoRV fragment of the rice EST D46477 (14). Hybond N+ (Amersham, Braunschweig, Germany) membranes were used, and hybridization was performed at 53°C in 0.25 M sodium phosphate buffer (pH 7.2) containing 7% (wt/vol) SDS, 1 mM EDTA, and 1% (wt/vol) BSA. After hybridization, the membrane was washed twice for 20 min in 2× SSPE, 0.1% (wt/vol) SDS at 53°C. The stock solutions were prepared following standard procedures (15). Recombinant DNA techniques and DNA sequencing were done as described (5, 6). The plasmid containing a full-length cDNA, which was used for sequence analysis, was designated pSQD1. For sequence comparison the Wisconsin Genetics Computer Group programs pileup and prettybox (16) were used.
Construction of Transgenic Lines.
For antisense expression, the central 699-bp long EcoRI–HindIII fragment of the cDNA insert contained in pSQD1 first was cloned into pBluescript II-SK+ (Stratagene) and excised by digestion with KpnI and XbaI. This fragment was inserted into the corresponding sites of the binary vector pBINAR-Hyg that is derived from pBIB-Hyg (17) by insertion of the HindIII–EcoRI fragment from pA7 (18). The resulting binary plasmid pαSQD1 contained the central portion of the SQD1 ORF in antisense orientation under the control of the 35S cauliflower mosaic virus promoter. This construct was introduced into Agrobacterium tumefaciens C58C1 and used to transform A. thaliana Col-2 plants via vacuum infiltration (19, 20). Transformed seedlings (T1 generation) were selected on Murashige–Skoog medium containing 0.8% (wt/vol) agarose 1% (wt/vol) sucrose, and 25 μg ml−1 hygromycin B (Calbiochem).
RNA Manipulation.
Total RNA isolated from A. thaliana tissues according to Logemann et al. (21) was separated and transferred to Hybond N+ membranes (Amersham). The blots were probed by using standard procedures (15) with a 699-bp EcoRI–HindIII fragment of pSQD1.
In Vitro Translation and Chloroplast Import Experiment.
To obtain a C-terminal-truncated in vitro SQD1 translation product with a molecular mass of 30 kDa, a 1,009-bp SalI–BamHI fragment was prepared by PCR and subsequent restriction from pSQD1 using the T7 primer and a reverse primer with a sequence of CAGGATCCTCAAGTAAAAGCAATGTT. This fragment was ligated into pBluescript II-SK+ cut with the respective enzymes giving rise to plasmid pSQD1–30. The [35S]methionine (Amersham) labeled in vitro translation product was prepared by using T7 polymerase and rabbit reticulocyte lysate (Promega). Chloroplasts were prepared from 30 g of 6-day-old pea seedlings kept in the dark for 12 hr (Pisum sativum convar. medullase, “Progress No. 9,” Walz-Samen, Stuttgart, Germany), and uptake experiments were performed by using standard procedures (22). Thermolysin (Boehringer-Mannheim) was added (0.1 mg/ml) as indicated. Labeled proteins were analyzed by SDS/PAGE according to Laemmli (23) and visualized by fluorography using Amplify (Amersham).
Lipid and Phosphate Analysis.
Rosette leaves were frozen in liquid nitrogen, and lipids were extracted as previously described (24). Lipid extracts were analyzed on activated ammonium sulfate impregnated silica gel TLC plates (5), except that the solvent system was replaced by acetone-toluene-water (91:30:8, vol/vol). Lipids were visualized with iodine vapor and identified by cochromatography with lipid extracts of known composition. For quantitative analysis, individual lipids were isolated from TLC plates and used to prepare fatty acid methyl esters (5). The methyl esters were quantified by GLC using myristic acid as internal standard (25).
Total phosphate in plant samples was determined by the method of Ames (26).
Production of Antibodies Against Recombinant SQD1 Protein and Immunoblotting.
To produce recombinant SQD1 protein in Escherichia coli, we cloned a 1,199-bp fragment of pSQD1 into the HIS-tag expression vector pQE30 (Qiagen, Hilden, Germany) by using a PCR-based strategy. For this purpose, we used as the forward primer AAAGGATCCCGTGTTATGGTCATTGG and as the reverse primer CAGCCTAGGAATACACCAGTACCTGA such that BamHI sites were provided for cloning into pQE30 and the N-terminal 84 amino acids containing the presumed signal peptide were removed. The resulting plasmid pSQD1-TP allowed the expression of the recombinant protein in E. coli and the purification of the protein because of six N-terminal histidine residues on Ni-NTA agarose (Qiagen) following the instructions by the manufacturer. Approximately 1 mg of the purified protein was sent to EUROGENTEC (Parc scientific du Sart Tilman, Belgium) to produce polyclonal antibodies in rabbits. For the immunopurification of antibodies, recombinant SQD1 protein was coupled to Affi-Gel 10 gel (Bio-Rad), and antibodies were eluted by using standard procedures (27).
For immunoblotting, 0.1 g of frozen leaf tissue was extracted with 0.5 ml buffer containing 20 mM Tris⋅HCl (pH 7.5), 10% (vol/vol) glycerol, 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride. After centrifugation for 5 min at 10,000 × g the proteins in the supernatant were separated by SDS/PAGE (23). Immunoblotting and detection using anti-rabbit alkaline phosphatase-coupled antibodies (Promega) were performed by using standard procedures (27).
RESULTS
Altered Lipid Composition in the pho1 Mutant and Phosphate-Deprived Wild Type.
In the pho1 mutant of A. thaliana, the inorganic phosphate content in leaves is reduced to approximately 5% of wild type because of a deficiency in phosphate loading of the xylem (28). It previously has been shown that the expression of RNS1 encoding a RNase is increased in the pho1 mutant to a similar extent as in the wild type after phosphate deprivation (29). Thus, the pho1 mutant provides a genetic model to study the causal relationship between the decrease in the amount of leaf inorganic phosphate and subcellular processes sensitive to the availability of inorganic phosphate. To investigate the effect of decreased phosphate in leaves of A. thaliana on the complex lipid composition of the membranes, we compared lipids extracted from the wild type and the pho1 mutant by TLC and quantified the lipids by GLC of the corresponding fatty acid methyl esters (Table 1). In the pho1 mutant, the relative amounts of all phospholipids decreased approximately to 50%, whereas the relative amounts of the nonphosphorous sulfolipid and digalactosyl diacylglycerol increased to 300% and 160%, respectively. The relative amount of the third nonphosphorous lipid, monogalactosyl diacylglycerol, did not change within the statistical limitation of the experiment.
Table 1.
Quantitative analysis of leaf lipid extracts from wild type and the pho1 mutant of A. thaliana grown on soil for 14 days
| Amount, mol %
|
||
|---|---|---|
| Lipid | Wild type | pho1 mutant |
| MGDG | 39.1 ± 1.1 | 40.0 ± 6.0 |
| DGDG | 16.4 ± 0.5 | 24.7 ± 3.9 |
| SQDG | 4.1 ± 0.9 | 13.4 ± 0.7 |
| PG | 16.4 ± 2.3 | 5.1 ± 2.2 |
| PE | 10.9 ± 2.1 | 5.9 ± 0.6 |
| PC | 17.6 ± 1.0 | 10.9 ± 1.9 |
DGDG, digalactosyl diacylglycerol; MGDG, monogalactosyl diacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol. Values represent the means ± SD of three independent measurements.
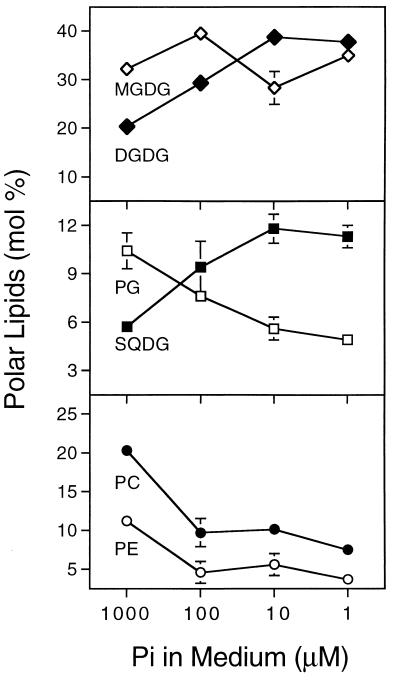
Raising the wild type on defined medium with decreasing amounts of phosphate led to a reduction in the inorganic phosphate content of leaves and reduced growth as indicated by the fresh weight of 15-day-old seedlings (Table 2). Lipids were extracted from these plants, separated by TLC and quantified by GLC of the corresponding fatty acid methyl esters. The result is shown in Fig. 1. With the decrease of inorganic phosphate in the leaves (Table 2), the relative amounts of digalactosyl diacylglycerol and sulfolipid increased, whereas that of all phospholipids decreased, and the relative amount of monogalactosyl diacylglycerol did not change within the experimental limitations. Thus, like in the pho1 mutant, there is a correlation between the internal concentration of inorganic phosphate and the observed change in membrane lipid composition. Most strikingly, there appears to be an inverse relationship between the relative amount of the two anionic lipids phosphatidylglycerol and sulfolipid such that the sum of both remains constant at about 16% of total lipids.
Table 2.
Fresh weight and inorganic phosphate content of leaves of wild-type A. thaliana grown for 14 days on agar-solidified medium supplemented with different amounts of phosphate
| Phosphate in medium, μM | Fresh weight of plant, mg | Pi in leaves, μg g−1 fresh weight |
|---|---|---|
| 1,000 | 11.8 ± 1.2 | 19.4 ± 2.8 |
| 100 | 3.3 ± 0.7 | 1.9 ± 0.2 |
| 10 | 1.0 ± 0.1 | 1.4 ± 0.4 |
| 1 | 0.8 ± 0.1 | 0.6 ± 0.4 |
Values represent the means ± SD of at least three independent measurements.
Figure 1.
Changes in relative amounts of complex lipids of wild-type A. thaliana in response to changing phosphate concentration. The plants were germinated and grown for 15 days on agar-solidified mineral medium as described under methods. Values represent the means ± SD of at least three independent measurements. Error bars are indicated, if they are not smaller than the symbol size. DGDG, digalactosyl diacylglycerol; MGDG, monogalactosyl diacylglycerol; PC, phosphatidylcholine; PE, phosphatidyl ethanolamine; PG, phosphatidylglycerol; SQDG, sulfoquinovosyl diacylglycerol.
Isolation of an A. thaliana cDNA with Sequence Similarity to Bacterial sqdB Genes.
To investigate the molecular basis and physiological relevance of the phenomenon described above, we began to isolate A. thaliana genes encoding enzymes involved in the head group biosynthesis of thylakoid lipids. Searching the dbEST database of expressed sequence tags (30) with the predicted amino acid sequence of the bacterial sqdB genes by using the algorithm tblastn (31), we were able to identify a partial rice cDNA (D46477) encoding a putative protein with high sequence similarity to the bacterial sqdB gene products. We used part of this cDNA as a probe to screen an A. thaliana cDNA library and isolated several clones, of which the longest with an insert of 1,799 bp was sequenced (GenBank accession no. AF022082). The ORF beginning at nucleotide 169 encodes a putative protein with a calculated molecular mass of 53.1 kDa. In-frame stop codons in the first 169 nucleotides of the sequence suggested that the cDNA is complete. Fig. 2 shows a comparison of the deduced amino acid sequences of the A. thaliana cDNA and the sqdB gene of Synechococcus sp. PCC7942. This comparison revealed a sequence identity of 42% and provided indirect evidence that the cDNA derived from A. thaliana may encode a protein involved in sulfolipid biosynthesis. Therefore, we tentatively designated the corresponding genetic locus of A. thaliana SQD1 and the plasmid containing the cDNA pSQD1. At the amino acid level, the partial rice sequence (D46477) was 86% identical to the SQD1 sequence of A. thaliana, but apparently contains some sequencing errors as evident from the sequence alignment (not shown). In comparison to the bacterial sqdB gene product, the sequence of A. thaliana showed a N-terminal extension of 84 amino acids. This peptide sequence exhibited features similar to chloroplast targeting peptides, with a high proportion of hydroxylated amino acids such as serine and threonine (28%), and a low proportion of acidic amino acids (2%) such as aspartate and glutamate (32). However, a putative cleavage site could not be determined by comparison to the proposed consensus sequence (33).
Figure 2.
Comparison of the amino acid sequences predicted from the sqdB gene of the cyanobacterium Synechococcus sp. PCC7942 (Syne) and the SQD1 cDNA of A. thaliana (Arab). Identical amino acid residues are marked by black boxes.
Probing a Southern blot of wild-type DNA restricted with eight different enzymes with the SQD1 cDNA under stringent conditions revealed one signal for six, and two signals for two of the restriction digests (data not shown). This result suggested that SQD1 represents a single copy gene.
The SQD1 Protein Is Imported and Processed by Isolated Chloroplasts.
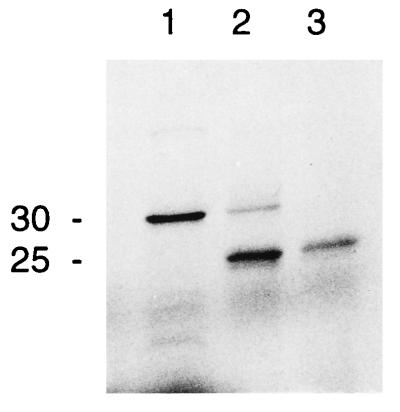
Since the first demonstration of the precursor processing and transport using isolated chloroplasts (34), an improved reconstitution assay has been developed that faithfully mimics intracellular targeting and transport into chloroplasts (35). To demonstrate that the SQD1 protein contains a chloroplast targeting peptide and that the mature protein is located in the chloroplast, we used this in vitro reconstitution assay. For the ease of handling we used isolated chloroplasts of peas instead of A. thaliana, assuming that the import machinery is conserved among higher plant chloroplasts. Furthermore, we engineered a C-terminally truncated form the SQD1 protein with a molecular mass of 30 kDa to avoid coelectrophoresis with the abundant large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase with a molecular mass of 50 kDa. As shown in Fig. 3, the 30-kDa SQD1 precursor was converted to the lower molecular weight form with a molecular mass of approximately 25 kDa, and this processed form was resistant to thermolysin treatment, indicating that it was located inside the chloroplasts. This result strongly suggested that the SQD1 protein contains an N-terminal transit peptide with a molecular mass of approximately 5 kDa and that the mature SQD1 protein is imported into the chloroplast. Furthermore, this observation is consistent with a chloroplast association of the SQD1 gene product and would be expected for an enzyme involved in sulfolipid biosynthesis, because isolated chloroplasts can incorporate sulfate into sulfolipid and thus should contain all enzymes involved in sulfolipid biosynthesis (36).
Figure 3.
Uptake and processing of a carboxy-terminally truncated SQD1 preprotein of A. thaliana by isolated pea chloroplasts. Lane 1, translation product; lane 2, translation product incubated in the presence of pea chloroplasts; lane 3, translation product incubated in the presence of pea chloroplasts and thermolysin. Proteins were labeled with [35S]methionine and visualized by autoradiography after SDS/PAGE. The approximate molecular mass (kDa) of the proteins is indicated.
Transgenic Plants Expressing SQD1 in Antisense Orientation Contain Less Sulfolipid.
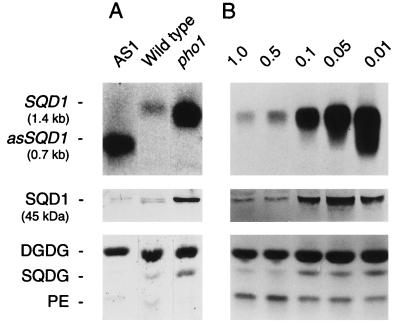
To provide functional proof that the SQD1 gene encodes a protein involved in sulfolipid biosynthesis in A. thaliana, we generated transgenic lines with the central portion of the SQD1 cDNA placed in antisense orientation under the control of the 35S cauliflower mosaic virus promoter (see Materials and Methods for details). It was expected that strong antisense lines showed a reduction in sulfolipid content, if the amount of SQD1 protein is limiting for sulfolipid biosynthesis. Lipid extracts prepared from single leaves of 50 kanamycin-resistant transformants (T1) were analyzed by TLC. Several transgenic lines with a reduction in sulfolipid content were identified by visual examination of the developed thin-layer plates. Because these lines were derived from different plants, they can be considered independent. As an example, the result for the best antisense line AS1 is shown in Fig. 4A. Northern blot analysis revealed a strong signal presumably for the antisense RNA, which is expected to be shorter than the mRNA, but only a barely detectable signal for the SQD1 mRNA. Nevertheless, using affinity purified antibodies raised against the N-terminally truncated recombinant SQD1 protein (see Materials and Methods for details), a protein with a molecular mass of approximately 50 kDa still could be detected in extracts of the AS1 line. The immunoblot result obtained for the wild type revealed two crossreacting proteins of similar molecular mass (Fig. 4A) that may represent two forms of the SQD1 protein after secondary modification. In AS1 the lower molecular weight form was preferentially reduced. By TLC analysis of lipids extracted from the antisense line AS1 (Fig. 4A) we still could detect a weak band comigrating with sulfolipid, and the subsequent quantification of lipids by GLC of fatty acid methyl esters prepared from extracts of different offsprings of the line AS1 confirmed an approximate 30% reduction in sulfolipid content regardless of the phosphate concentration in the medium (data not shown). This result was in agreement with the immunoblotting data, suggesting that the SQD1 protein is reduced, but still present in AS1 plants. Within the statistical limitations of the experiment, the relative amounts of phosphatidylglycerol were indistinguishable in the transgenic line and the wild type (data not shown).
Figure 4.
Expression of the SQD1 gene in leaves of A. thaliana. (A) Wild type, the strongest SQD1 antisense line AS1, and the pho1 mutant grown for 14 days in soil. (B) Wild type germinated and raised for 7 days on agar-solidified Murashige–Skoog medium and subsequently grown for 7 days on mineral medium containing a concentration of Pi (mM) as indicated. (Upper) SQD1 mRNA (SQD1) or the presumed antisense RNA (asSQD1) detected by hybridization with the SQD1 cDNA (the approximate sizes are indicated). (Middle) SQD1 protein (SQD1) detected by immunoblotting (the apparent molecular mass is indicated). (Lower) Central part of iodine-stained lipid chromatograms (DGDG, digalactosyl diacylglycerol; PE, phosphatidylethanolamine; SQDG, sulfoquinovosyl diacylglycerol). RNA, protein, and lipids were isolated from identical batches of plants. Equal amounts of total RNA, protein, or lipids were loaded based on visual examination of stained gels or TLC plates.
To test whether the observed reduction in sulfolipid content would affect photosynthetic performance, we compared the growth of wild-type and AS1 plants in different media under optimal light conditions. No differences in the growth rates of plants raised in soil or on agar plates with different concentrations of phosphate could be detected (data not shown). Furthermore, the overall appearance of AS1 plants was indistinguishable from that of the wild type.
The Expression of SQD1 Is Increased in the pho1 Mutant and in Phosphate-Deprived Wild Type.
To understand the molecular basis of the increase in relative amounts of sulfolipid in the pho1 mutant and phosphate-deprived wild type, we determined the amounts of SQD1 mRNA, and SQD1 protein. The result is shown in Fig. 4. The amount of SQD1 mRNA in the pho1 mutant (Fig. 4A) and in the wild type raised on decreasing amounts of phosphate (Fig. 4B) was drastically increased. Quantification by PhosphorImaging revealed a 15-fold increase in the amount of SQD1 mRNA in wild type grown in the presence of 0.01 mM phosphate compared with wild type grown in the presence of 1 mM phosphate. Furthermore, the signal presumed to be specific for the SQD1 protein as detected by immunoblotting (Fig. 4, Middle) was increased in the pho1 mutant and in wild-type plants raised on decreasing amounts of phosphate. The increase in SQD1 protein appears to be proportional to the increase in SQD1 mRNA. The analysis of lipid extracts prepared from the same plant batches used for RNA and protein preparation showed a corresponding increase in the relative amounts of sulfolipid (Fig. 4, SQDG). Taken together, these results provide strong evidence that a decrease in the internal phosphate concentration, regardless whether imposed by a specific mutation in phosphate transport in the pho1 mutant or by limiting the external supply of phosphate to the wild type (see Table 2), leads to an increase in the expression of SQD1, a gene encoding a protein involved in sulfolipid biosynthesis. This increase in the abundance of the SQD1 gene product can be correlated to the increase in the relative content of sulfolipid, suggesting that the SQD1 protein may catalyze a limiting reaction in sulfolipid biosynthesis, which is regulated by changes in the expression of the SQD1 gene in response to altered availability of inorganic phosphate.
DISCUSSION
It was the aim of this study to test in a higher plant the concept of sulfolipid function under phosphate-limiting growth conditions that has been derived from experiments with photosynthetic bacteria (2–4). We chose A. thaliana, because it can be easily grown on defined medium, and the pho1 mutant with a low internal concentration of inorganic phosphate (28) was available, thus, allowing us to use two completely independent approaches of imposing phosphate deprivation. Like the two investigated photosynthetic bacteria, R. sphaeroides (2, 3) and Synechococcus sp. PCC7942 (4), A. thaliana wild type grown on medium with reduced inorganic phosphate shows a drastically altered lipid composition. The overall changes are similar in the two bacteria and A. thaliana, with an increase in the relative amounts of nonphosphorous sulfolipid and dihexosyl lipids, such as digalactosyl diacylglycerol, and a decrease in phospholipids. The relative amounts of the two anionic lipids phosphatidylglycerol and sulfolipid show an inverse relationship as a function of the phosphate concentration in the medium in all three organisms. Moreover, the pho1 mutant reveals the same changes in lipid composition as the A. thaliana wild type grown in medium with reduced phosphate, thereby providing corroborating evidence that indeed the internal inorganic phosphate concentration and not secondary effects, like altered pH, buffer capacity, or ionic strength of the phosphate starvation medium trigger the change in lipid composition. These results demonstrate that defined changes in lipid composition in response to inorganic phosphate deprivation are not restricted to photosynthetic bacteria, but represent a more widespread phenomenon extending also to higher plants.
To distinguish between an active adaptation in response to reduced phosphate availability or a more passive process, i.e., the dilution of phospholipids after the cessation of phospholipid biosynthesis, we examined the expression of the SQD1 gene for which we isolated a cDNA. Several lines of evidence suggest that this gene encodes a protein (SQD1), which is required for sulfolipid biosynthesis in A. thaliana: (i) the SQD1 gene shows high sequence similarity to the bacterial sqdB genes known to be essential for sulfolipid biosynthesis (Fig. 2); (ii) the protein is imported into the chloroplast (Fig. 3); and (iii) antisense expression in transgenic plants causes a reduction of sulfolipid content (Fig. 4A). Under phosphate deprivation, the expression of the SQD1 gene is increased in the wild type (Fig. 4). Likewise, the expression is increased in the pho1 mutant, providing corroborating evidence that the effect is indeed caused by low levels of internal inorganic phosphate. Based on this observation we conclude that the increase in the relative amount of sulfolipid in response to phosphate deprivation is at least in part the result of an active regulation of the transcription of a gene involved in sulfolipid biosynthesis.
The presumed function of the observed changes in lipid composition would be the conservation of phosphorus, a substituent of many essential metabolites, by reducing the amount of phosphorus bound in membrane lipids without compromising the function of photosynthetic membranes. Apparently, to maintain the proper function of the thylakoid membranes, the anionic sulfolipid substitutes for the anionic phosphatidylglycerol among other adjustments, such as the increase of the relative amount of the bilayer forming lipid digalactosyl diacylglycerol. Taking into consideration that about one-third of organic phosphate in leaves of A. thaliana is bound each in phospholipids, nucleic acids, and other phosphor esters (28), reducing membrane-bound phosphorus would be a reasonable measure to conserve phosphate under conditions of limited phosphate availability, frequently encountered by plants in nature. Apparently, the increased expression of RNases (29) or phosphate transporters (37) under phosphate deprivation serves the same purpose by reducing the amount of organic phosphorus bound in the nucleic acid fraction, or by enhancing the uptake of phosphate from the medium, respectively. If the hypothesis on sulfolipid function elaborated above is correct, one would expect that a strong reduction in sulfolipid content caused by the antisense expression of the SQD1 cDNA may hamper growth, or that the overexpression of the SQD1 cDNA and the resulting increase in sulfolipid content may stimulate growth under phosphate-limiting conditions. Although the second experiment has not yet been performed, the growth of the best available antisense line AS1 was not affected under different conditions. It seems likely that the moderate reduction (30%) in sulfolipid content in AS1 is not sufficient to show an effect on growth. Thus, further testing of the hypothesis has to await the construction of sulfolipid-deficient antisense lines or genetic mutants completely lacking sulfolipid, and the construction of sulfolipid overproducing plants.
The reaction catalyzed by the SQD1 protein or the corresponding bacterial sqdB gene products remains elusive. However, their amino acid sequences are remarkably conserved, suggesting that this enzyme class may catalyze a unique reaction of sulfolipid biosynthesis common to all sulfolipid containing organisms. Like the bacterial sqdB gene products, the SQD1 protein shows sequence similarity to sugar nucleotide modifying enzymes such as UDP-glucose-4-epimerases or nucleotide-hexose-4,6-dehydratases (4, 6). Based on sequence similarity one may assume that this class of enzymes is involved in the biosynthesis of UDP-sulfoquinovose from UDP-glucose, the known precursor of sulfolipid biosynthesis (7). It is expected that the analysis of the SQD1 protein of A. thaliana as well as of the corresponding bacterial proteins finally will allow us to solve the pathway of sulfolipid biosynthesis.
Acknowledgments
We would like to thank the members of the Japanese Rice Genome Research Program for providing us with the EST clone D46477 and Dr. Y. Poirier for the pho1 mutant. Financial support in parts by the Bundesministerium für Bildung und Forschung (Grant 0311024) and the Deutsche Forschungsgemeinschaft (Grant BE 1591/1-2) is gratefully acknowledged.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF022082).
References
- 1.Webb M S, Green B R. Biochim Biophys Acta. 1991;1060:133–158. [Google Scholar]
- 2.Benning C, Beatty J T, Prince R C, Somerville C R. Proc Natl Acad Sci USA. 1993;90:1561–1565. doi: 10.1073/pnas.90.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benning C, Huang Z H, Gage D A. Arch Biochem Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- 4.Güler S, Seeliger A, Härtel H, Renger G, Benning C. J Biol Chem. 1996;271:7501–7507. doi: 10.1074/jbc.271.13.7501. [DOI] [PubMed] [Google Scholar]
- 5.Benning C, Somerville C R. J Bacteriol. 1992;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benning C, Somerville C R. J Bacteriol. 1992;174:6479–6487. doi: 10.1128/jb.174.20.6479-6487.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossak M, Tietje C, Heinz E, Benning C. J Biol Chem. 1995;270:25792–25797. doi: 10.1074/jbc.270.43.25792. [DOI] [PubMed] [Google Scholar]
- 8.Kühlbrandt W. Curr Opin Struct Biol. 1994;4:519–528. [Google Scholar]
- 9.Sato N, Sonoike K, Tsuzuki M, Kawaguchi A. Eur J Biochem. 1995;234:16–23. doi: 10.1111/j.1432-1033.1995.016_c.x. [DOI] [PubMed] [Google Scholar]
- 10.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 11.Estelle M A, Somerville C R. Mol Gen Genet. 1987;206:200–206. [Google Scholar]
- 12.Newman T, de Brujn F J, Green P, Keegstra K, Kende H, et al. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed K C, Mann D A. Nucleic Acids Res. 1985;13:7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki T, Song J, Koga-Ban Y, Matsui E, Fang F, et al. Plant J. 1994;6:615–624. doi: 10.1046/j.1365-313x.1994.6040615.x. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker D. Nucleic Acids Res. 1990;18:203. doi: 10.1093/nar/18.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Schaeven A. Ph.D. thesis. Freie Universität Berlin; 1989. [Google Scholar]
- 19.Bechtold N, Ellis J, Pelletier G. C R Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- 20.Bent A F, Kunkel B N, Dahlbeck D, Brown K L, Schmidt R, Giraudat J, Leung J, Staskawicz B J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 21.Logemann J, Schell J, Willmitzer L. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 22.Bruce B D, Perry S, Froehlich J, Keegstra K. In: Plant Molecular Biology Manual. Gelvin S B, Schilperoort R A, editors. Boston: Kluwer; 1994. Sect. J, pp. 1–15. [Google Scholar]
- 23.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. Plant Cell. 1995;7:1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossak M, Schäfer A, Xu N, Gage D A, Benning C. Arch Biochem Biophys. 1997;340:219–230. doi: 10.1006/abbi.1997.9931. [DOI] [PubMed] [Google Scholar]
- 26.Ames B N. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 27.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 28.Poirier Y, Thoma S, Somerville C R, Schiefelbein J. Plant Physiol. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bariola P A, Howard C J, Taylor C B, Verburg M T, Jaglan V D, Green P J. Plant J. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- 30.Boguski M S, Lowe T M J, Tolstoshev C M. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 31.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.von Heijne G, Steppuhn J, Herrmann R G. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- 33.Gavel Y, von Heijne G. FEBS Lett. 1990;261:455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- 34.Highfield P E, Ellis R J. Nature (London) 1978;271:420–424. [Google Scholar]
- 35.Grossman A R, Bartlett S G, Schmidt G W, Mullet J E, Chua N-H. J Biol Chem. 1982;257:1558–1563. [PubMed] [Google Scholar]
- 36.Haas R, Siebertz H P, Wrage K, Heinz E. Planta. 1980;148:238–244. doi: 10.1007/BF00380033. [DOI] [PubMed] [Google Scholar]
- 37.Muchal U S, Pardo J M, Raghothama K G. Proc Natl Acad Sci USA. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]