Abstract
Regulation of a variety of cellular contractile events requires that vertebrate smooth and non-muscle myosin II can achieve an “off” state. To examine the role of the myosin rod in this process, we determined the minimal size at which a myosin molecule is capable of regulation via light chain phosphorylation. Expressed smooth muscle myosin subfragments with as many as 100 amino acids of the coiled-coil rod sequence did not dimerize and were active independently of phosphorylation. To test whether dimerization per se restores regulation of ATPase activity, mutants were expressed with varying lengths of rod sequence, followed by C-terminal leucine zippers to stabilize the coiled-coil. Dimerization restored partial regulation, but the presence of a length of rod approximately equal to the myosin head was necessary to achieve a completely off state. Partially regulated short dimers could be converted into fully regulated molecules by addition of native rod sequence after the zipper. These results suggest that the myosin rod mediates specific interactions with the head that are required to obtain the completely inactive state of vertebrate smooth and non-muscle myosins. If these interactions are prohibited under cellular conditions, unphosphorylated crossbridges can slowly cycle.
The structure of the myosin II molecule is distinguished from other members of the myosin superfamily in that the head, which consists of the motor domain and light chain binding region, is followed by a 150-nm extended coiled-coil rod. The rod sequence has a repeated pattern of 7 amino acids (approximately 3.5 amino acids per turn), with hydrophobic residues in the “a” and “d” positions, so that the two α-helical chains interact along this hydrophobic interface. Although the conventional role of the myosin rod is to mediate filament assembly, a number of studies have also focused on the possibility that a helix–coil transition in the rod may contribute to myosin’s ability to produce force (1). The latter role is now in question, because force measurements on single motor molecules have shown that the myosin head and even the motor domain can generate force and move actin (2). These results with minimal domains of myosin have led to the belief that the rod is primarily a passive structural component.
Another consequence of the coiled-coil is that myosin II molecules have two heads. Because some element of both heads is required to achieve the enzymatically “off” state, the myosin rod is indirectly implicated in regulation (3). Recent work has reinforced the need for head–head interactions in regulation. A proteolytically prepared myosin molecule with an intact rod, but only a single myosin head, was poorly regulated by phosphorylation (4). Although this study clearly demonstrates that two heads are necessary for regulation, it does not address whether they are sufficient. Thus this study left unanswered the important question of whether the rod is also an essential component to ensure complete regulation. Herein we provide evidence from a series of expressed mutant heavy meromyosins (HMMs) that a specific region of the rod mediates an interaction between the heads that is essential to achieve an inhibited state. When these interactions occur, then phosphorylation is necessary and sufficient to regulate cellular processes. Alternatively, if this interaction is perturbed, a second regulatory system would be necessary to completely suppress activity.
MATERIALS AND METHODS
Construction of Heavy Chain cDNAs.
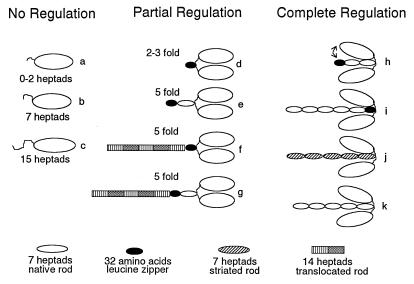
The different DNA constructs were created using a variety of strategies, alone or in combination, including ligation of PCR products and annealed synthetic oligonucleotides. Each construct is given both a descriptive name and a name such as “construct a,” which refers to the letter it was assigned in Fig. 4. For the truncation constructs without leucine zippers or an antibody tag (constructs a–c, Fig. 4), a stop codon was introduced following the codon that specified an amino acid that completed an integral number (i.e., 2, 7, or 15) of heptad (7-aa) repeats following Pro-849 (d position). The 37-heptad construct ended with amino acid 1112 (construct k, Fig. 4). In the 2-, 7-, and 15-heptad/C-terminal leucine zipper constructs (constructs d, e, and h in Fig. 4, respectively), the 32-aa leucine zipper sequence followed an integral number of heptads. The sequence of the GCN4 leucine zipper, with position “d” of the heptad repeat underlined, is: MKQLEDKVEELSKNYHLENEVARLKKLVGER (5). For the 0-heptad C-terminal leucine zipper (construct d, Fig. 4), the leucine zipper sequence follows Arg-855. After the zipper sequence, a segment of the rod that contains the epitope for antibody S2.2 (Gln-1081 through Arg-1175) was included to provide an attachment site for motility. For the internal leucine zipper constructs, codons specifying the 32-aa GCN4 sequence (Met-1 through Arg-32) was inserted into the 37-heptad rod sequence. For the construct in which the spatial positions of the rod elements were preserved (construct i, Fig. 4), amino acids 867–898 (inclusive) were replaced by the leucine zipper sequence. The last residue in the zipper (Arg) was changed to Leu, because the native rod residue at position 898 is Leu in the d position of the heptad. A second internal zipper construct, in which some rod elements were spatially displaced (translocated rod; construct f, Fig. 4), was made by inserting the leucine zipper sequence between amino acids Glu-866 and Leu-867 of the native rod sequence. The third internal zipper sequence with rod elements displaced (construct g, Fig. 4) was constructed by following native sequence amino acid Lys-901 with the leucine zipper, which was followed by native amino acids Gln-899 to Gln-1112.
Figure 4.
Schematic diagram grouping the constructs that show no regulation, partial regulation (<10-fold), or complete regulation (>10-fold) by light chain phosphorylation as determined by steady-state actin-activated ATPases. All monomeric constructs (a–c), regardless of length, show no regulation with phosphorylation. Constructs that show partial regulation include C-terminal zippers after 0, 2, or 7 heptads of native sequence (d and e), and constructs in which a piece of rod is translocated from its normal position relative to the invariant proline (f and g). Among the constructs showing full regulation are 15-heptad/zipper (h), a 37-heptad construct with the leucine zipper replacing amino acids 867–898 (i), a smooth head/striated rod chimera (j), and a 37-heptad wild-type construct (k). Dimers that are fully regulated are shown with their heads bent down toward the rod to indicate that specific sequences in the rod mediate head–rod or head–head interactions. The head–rod interaction is likely to be most stable in 10S myosin, which has the lowest rate of product release. The changes in head disposition that occur in HMM may be more subtle than implied by this diagram, which was drawn to emphasize a head–rod interaction.
Infection of Sf9 Cells and Biochemical Analysis of Expressed Constructs.
Recombinant baculovirus was isolated by conventional protocols (6). Sf9 cells in suspension culture were coinfected with viral particles containing the HMM heavy chain construct and a virus expressing both the smooth muscle regulatory light chain (RLC) and the essential light chain. The cells were harvested at 65–75 h, and the recombinant HMM molecules were purified as described (7). The expressed HMMs were phosphorylated by addition of Ca2+, calmodulin, and myosin light chain kinase, and dephosphorylated by addition of protein phosphatase 1-M (gift from T. Haystead, University of Virginia, Charlottesville, VA). Immunoblots of glycerol gels were used to verify phosphorylation of the RLC (7).
Native gels were prepared and run as described in (8). Rotary shadowed platinum images were obtained in 0.5 M ammonium acetate/66% glycerol and observed with a Philips EM301 electron microscope operated at 60 kV (9). Actin-activated ATPase assays were performed in 10 mM imidazole, (pH 7), 8 mM KCl, 1 mM MgCl2, and 1 mM EGTA at 37°C. The molar concentration of actin/gizzard tropomyosin was 1:5. Inorganic phosphate was determined colorimetrically (10) at six time points per actin concentration.
RESULTS AND DISCUSSION
Monomeric Constructs.
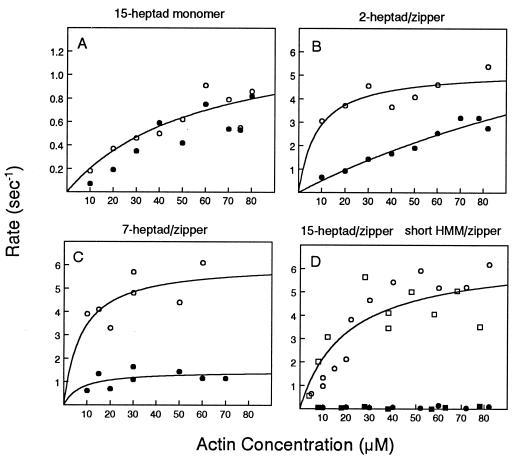
To investigate whether the myosin rod has a functional purpose beyond creating a heavy chain dimer, the baculovirus/insect cell expression system was used to create truncated rod constructs. By expressing constructs consisting of 2, 7, or 15 heptads of rod sequence past the invariant proline that marks the beginning of the rod (Fig. 1A, constructs a–c), we demonstrate here that an inhibited state cannot be attained by a monomeric species of any length. Native gels showed that all these constructs were monomeric (Fig. 2A, lane 5). The observation that more than 100 amino acids of the myosin rod show no tendency to dimerize suggests that the rod sequence adjacent to the heads has evolved to favor a labile coiled-coil, either for regulatory or mechanical reasons. Phosphorylation of the RLC did not increase the actin-activated ATPase activity of the monomers (Fig. 3A) or the rate of actin movement in a motility assay (data not shown). A specific interaction of the RLCs with sequences present near the head–rod junction is therefore not sufficient to obtain regulation.
Figure 1.
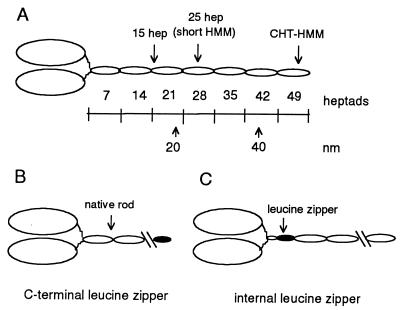
(A) Schematic diagram of double-headed heavy meromyosin. An invariant proline (amino acid 849 in the gizzard heavy chain) marks the end of the head and the beginning of the heptad repeat in the rod. Each oval-shaped segment of the rod represents 7 heptad repeats. One heptad repeat has a repeated pattern of seven amino acids (abcdefg), where the a and d positions have a preponderance of hydrophobic residues (11). The rise per residue is 0.15 nm. CHT-HMM, HMM prepared by chymotryptic digestion of myosin. (B) Illustration of the C-terminal leucine zipper constructs. The C-terminal leucine zipper constructs are named for the number of heptads of native sequence before the leucine zipper sequence, e.g., 2-heptad/zipper has 2 heptads of rod sequence followed by the 32-aa leucine zipper sequence. (C) Illustration of the internal leucine zipper constructs, in which native rod sequence follows the leucine zipper sequence.
Figure 2.
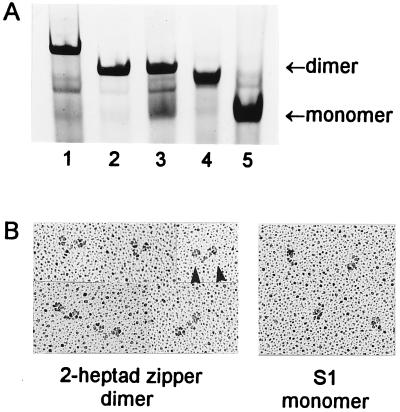
(A) Native gel showing the dimerization state of the expressed constructs. Lane 1, dimeric skeletal rod chimera (construct j); lane 2, dimeric 25-heptad short HMM with a C-terminal leucine zipper (similar to construct h); lane 3, 25-heptad short HMM without the zipper, showing an equilibrium between dimeric and monomeric species; lane 4, dimeric 2-heptad construct with a C-terminal leucine zipper (construct d); lane 5, monomeric 15-heptad construct (construct c). (B) Metal-shadowed images of the dimeric 2-heptad construct with a C-terminal leucine zipper (Left; construct d) show pairs of heads (arrowheads), in contrast to the single heads of monomeric S1 (Right).
Figure 3.
Actin-activated ATPase activity of monomers and C-terminal leucine zipper constructs. Phosphorylated species, open symbols; unphosphorylated species, filled symbols. (A) Monomeric 15-heptad S1 (construct c). (B) C-terminal 2-heptad/zipper (construct d). (C) C-terminal 7-heptad/zipper (construct e). (D) C-terminal 15-heptad/zipper (circles; construct h) and 25-heptad short HMM/zipper (squares).
An earlier study showed that even two heads do not ensure regulation: the activity of an expressed myosin with two heads and a short rod consisting of 25-heptad repeats (“short HMM”) was poorly regulated by phosphorylation and could not fully attain an off state. The loss of regulation is probably related to the labile nature of this dimeric construct, because an equilibrium with monomer was detected by native gels (7) (Fig. 2A, lane 3). In contrast, the activity of HMM prepared by chymotryptic cleavage of myosin, which contains approximately 46 heptads of rod sequence, is completely dependent on phosphorylation (12). An expressed stable dimer with 37 heptads of rod sequence (construct k) had properties like chymotryptic HMM, establishing that between 175 and 259 amino acids (i.e., 25–37 heptads) are required to achieve dimer stability and full regulatory properties.
C-Terminal Zippered Constructs.
The lack of regulation of the monomeric constructs, as well as the previously characterized short HMM, could be due either to the lack of dimerization or to the loss of a necessary rod segment. These alternative explanations could be distinguished by creating stable dimers with the addition of a 32-aa GCN4 leucine zipper sequence (4.5 heptads) to the C terminus of the smooth muscle myosin sequence (Fig. 1B). Despite the small number of residues, the leucine zipper forms a stable α-helical coiled-coil by virtue of the predominance of leucine residues in the “d” position of the heptad repeat (5). Metal-shadowed images of the expressed 2-heptad/zipper construct showed pairs of heads, as expected if the zipper mediates dimer formation (Fig. 2B, construct d). Native gels confirmed that stable dimers were formed by addition of the leucine zipper following varying lengths of native rod sequence (Fig. 2A, lanes 2 and 4).
Stable dimerization via the leucine zipper confers some regulation by light chain phosphorylation, unlike the complete lack of regulation observed with the monomers. The extreme case is the addition of the zipper sequence at the first heptad past the invariant proline (i.e., 0 heptads of native sequence, construct d). Actin-activated ATPase activity of this construct showed a 3- to 5-fold degree of regulation by phosphorylation. To measure motility, a small piece of the rod that contains the epitope for monoclonal antibody S2.2 was added after the leucine zipper, thus providing a uniform attachment site for motility. The 0-heptad/zipper construct moved actin 3.4-fold faster in the phosphorylated state (0.75 ± 0.09 μm/sec, n = 2) than in the unphosphorylated state (0.22 ± 0.06 μm/sec, n = 2), again showing partial regulation.
Similarly, with constructs containing 2 heptads or 7 heptads of native sequence before the zipper sequence, the activity of the unphosphorylated dimeric species was not fully inhibited, and thus there was a loss of the complete off state (Fig. 3 B and C; constructs d and e). A trend toward an increasing degree of regulation was observed as the length of the native rod sequence increased. The activity of the unphosphorylated species was completely inhibited once zipper sequences were added after 15 or 25 heptads of the rod (Fig. 3D, construct h). Full regulation is considered here to be those constructs that show a greater than 10-fold change in activity with phosphorylation by steady-state ATPase activity. (Constructs containing the zipper sequence after 2–25 heptads of native sequence did not contain an epitope tag for motility and thus only ATPase activity was measured.) Formation of a stable dimer also increased Vmax approximately 5-fold compared with its monomeric counterpart, suggesting that dimerization alters myosin’s kinetic properties (Fig. 3, compare A and D). Dimerization plus a length of rod that is approximately equal to the length of the myosin head is therefore necessary to fully restore phosphorylation-dependent regulation.
Internal Zippered Constructs.
The incomplete inhibition of the short dimeric zipper constructs in the unphosphorylated state could be caused by the lack of a critical piece of the rod or because the presence of the zipper prevents a favorable interaction that occurs in the native molecule. To distinguish between these possibilities, we determined whether regulation could be rescued by addition of rod sequence after the C-terminal 2-heptad/zipper construct. The resulting chimeric construct of 37 heptads contains an internal leucine zipper, with native rod residues 867–898 replaced with the leucine zipper sequence (Fig. 1C; construct i). Based on both ATPase and in vitro motility experiments, full regulation was restored by addition of native rod sequence (Table 1). If the unstable region of the rod adjacent to the heads needs to “melt” so that some part of the two heads can form a stereospecific interaction necessary for the completely inhibited state, then this lability is restricted to the first 2-heptad repeats. The presence of the zipper per se therefore does not prohibit formation of the off state.
Table 1.
Regulatory properties of wild-type and leucine zipper constructs
| Constructs* | RLC | Actin-activated ATPase, sec−1 | Motility, μm/s |
|---|---|---|---|
| C-terminal leucine zipper after 2 heptads (construct d) | Dephos | 0.91 | ND |
| Phos | 3.55 | ND | |
| Wild-type 37-heptad (construct k) | Dephos | 0.10 | None (2) |
| Phos | 1.9 | 0.96 ± 0.15 (2) | |
| 37 Heptad, but internal leucine zipper replaces native sequence from 3-7 heptads (amino acids 867-898) (construct i) | Dephos | 0.18 | None (3) |
| Phos | 2.9 | 0.69 ± 0.08 (3) | |
| 37 Heptad, but internal leucine zipper inserted after 2 heptads of rod sequence (between amino acids 866 and 867), thus translocating heptads 3-37 by 5 nm (construct f) | Dephos | 0.99 | 0.29 ± 0.05 (3) |
| Phos | 3.22 | 0.71 ± 0.08 (3) |
Actin-activated ATPase activities were obtained at 40 μM actin-tropomyosin under the conditions described in Materials and Methods. Values are averages from two independent preparations. Actin filament motility was measured at 30°C (13). Constructs were attached to the substratum via monoclonal antibody S2.1. The motility of the C-terminal 2 heptad was not determined (ND), because the epitope for Ab S2.1 was not present in this construct. Dephos, dephosphorylated RLC; phos, phosphorylated RLC.
Details of the constructs in this table are described in Materials and Mehtods. The 37-heptad construct has properties similar to chymotryptic HMM.
n = number of independent preparations.
To test whether there is specificity in the piece of rod that is required to restore regulation, two additional internal zipper chimeras with translocated rods were created. In the first, the leucine zipper sequence was inserted into the 37-heptad construct after 2 heptads of rod sequence (construct f). The resulting construct thus has rod heptads 3–37 displaced by the length of the leucine zipper (approximately 5 nm), relative to their position in the wild-type molecule. If the heads interact with the rod, they would now encounter a different sequence than that in the wild-type molecule. Full regulation was not obtained with this construct, either by ATPase activity or by motility (Table 1). A second chimera, with the leucine zipper inserted after 7 heptads of rod sequence (construct g), thus displacing rod heptads 7–37 by 5 nm relative to their position in the wild-type molecules, also showed an incomplete off state. Both internal zipper constructs suggest that regulation can be fully restored only if the native rod is present with correct spatial positioning relative to the heads.
Chimeric Rod Constructs.
Chimeras containing the smooth muscle myosin head attached to the rod from unregulated sarcomeric myosins (construct j) were expressed to determine if the rod derived from a regulated myosin contains unique features. Approximately 35% of the residues in the smooth muscle myosin subfragment-2 (S-2) region are identical to those found in the striated rods. The actin-activated ATPase activity of a smooth head/skeletal rod chimera was fully regulated (phosphorylated chimera, Vmax = 4.2 ± 0.6 sec−1; dephosphorylated chimera <0.2 sec−1 between 10 and 70 μM actin). Some element of the rod that is important for regulation has been conserved in the sarcomeric rods, whether it is a pattern of charge or a specific sequence, which may relate to the fact that the mechanical properties of sarcomeric myosins are modulated by light chain phosphorylation (14, 15).
Primitive myosin II molecules, such as that of Dictyostelium, are regulated approximately 4-fold by light chain phosphorylation but do not achieve the more complete off state seen in purified vertebrate smooth/non-muscle myosins (16, 17). Their degree of regulation is similar to that seen for the C-terminal 7-heptad/zipper constructs of smooth muscle myosin. This primitive myosin II regulation may simply require dimerization, without any contribution from head–rod interactions. One could speculate that a fully off state for myosin II molecules is not absolutely necessary but would certainly offer an evolutionary advantage in terms of energy savings for a cell.
Relevance to in Vivo Regulation.
Dimerization per se thus provides a degree of regulation by light chain phosphorylation greater than that obtained with any monomeric species (Fig. 4). Complete inhibition of the unphosphorylated state of vertebrate smooth/non-muscle myosin entails a mechanism wherein specific interactions mediated by the the rod play a key role. It is possible that the coiled-coil rod must be approximately the length of the myosin head (19 nm) to stabilize interactions not only between the neck regions, but also between the motor domains. When unphosphorylated smooth muscle myosin adopts the folded conformation, which is myosin’s most inhibited state, the heads are close to each other and bent back toward the rod, consistent with interactions mediated by the rod (9, 18). Such an inhibited folded monomer is likely to be present in vertebrate non-muscle cells.
Another implication from this study is that if cellular conditions prohibit the specific interaction of the heads with the rod, then unphosphorylated myosin could slowly cycle. Physiological studies with smooth muscle do in fact suggest that dephosphorylated crossbridges can slowly cycle and maintain force (19). A head–rod interaction could be prevented either because of constraints imposed by the native thick filament structure or because of accessory proteins, such as smooth muscle kinase-related protein (also called telokin) (20), that bind near the head–rod junction.
In conclusion, unlike myosin’s mechanical properties, which are retained to some extent by the motor domain alone, regulation of activity requires a more complex set of interactions involving multiple domains of the myosin molecule, including elements of both myosin heads and a portion of the myosin rod.
Acknowledgments
We thank Susan Lowey for helpful discussions during the course of this work, and Peter Kim for the gift of the GCN4 DNA. This work was supported by National Institutes of Health Grants HL38113 (K.M.T.) and AR35661 (H.L.S.).
Footnotes
Abbreviations: HMM, heavy meromyosin; RLC, regulatory light chain.
References
- 1.Harrington W F, Rodgers M E, Davis J S. In: Molecular Mechanisms in Muscular Contraction. Squire J M, editor. New York: Macmillan; 1990. pp. 241–263. [Google Scholar]
- 2.Itakura S, Yamakawa H, Toyoshima Y Y, Ishijima A, Kojima T, Harada Y, Yanagida T, Wakabayashi T, Sutoh S. Biochem Biophys Res Commun. 1993;196:1504–1510. doi: 10.1006/bbrc.1993.2422. [DOI] [PubMed] [Google Scholar]
- 3.Trybus K M. J Muscle Res Cell Motil. 1994;15:587–594. doi: 10.1007/BF00121066. [DOI] [PubMed] [Google Scholar]
- 4.Cremo C R, Sellers J R, Facemyer K C. J Biol Chem. 1995;270:2171–2175. doi: 10.1074/jbc.270.5.2171. [DOI] [PubMed] [Google Scholar]
- 5.O’Shea E K, Klemm J D, Kim P S, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 6.O’Reilly D R, Miller L K, Luckow V A. Baculovirus Expression Vectors: A Laboratory Manual. New York: Freeman; 1992. [Google Scholar]
- 7.Trybus K M. J Biol Chem. 1994;269:20819–20822. [PubMed] [Google Scholar]
- 8.Trybus K M, Lowey S. J Biol Chem. 1985;260:15988–15995. [PubMed] [Google Scholar]
- 9.Trybus K M, Lowey S. J Biol Chem. 1984;259:8564–8571. [PubMed] [Google Scholar]
- 10.White H D. Methods Enzymol. 1982;85:698–708. doi: 10.1016/0076-6879(82)85057-x. [DOI] [PubMed] [Google Scholar]
- 11.McLachlan A D, Karn J. Nature (London) 1982;299:226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- 12.Sellers J R. J Biol Chem. 1985;260:15815–15819. [PubMed] [Google Scholar]
- 13.Trybus K M, Chatman T A. J Biol Chem. 1993;268:4412–4419. [PubMed] [Google Scholar]
- 14.Sweeney H L, Bowman B F, Stull J T. Am J Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 15.Sweeney H L, Yang Z, Zhi G, Stull J T, Trybus K M. Proc Natl Acad Sci USA. 1994;91:1490–1494. doi: 10.1073/pnas.91.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostrow B D, Chen P, Chisholm R L. J Cell Biol. 1994;127:1945–1955. doi: 10.1083/jcb.127.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross R A, Cross K E, Sobieszek A. EMBO J. 1986;5:2637–2641. doi: 10.1002/j.1460-2075.1986.tb04545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Offer G, Knight P. J Mol Biol. 1996;256:407–416. doi: 10.1006/jmbi.1996.0096. [DOI] [PubMed] [Google Scholar]
- 19.Kamm K E, Stull J T. Annu Rev Pharmacol Toxicol. 1985;25:593–620. doi: 10.1146/annurev.pa.25.040185.003113. [DOI] [PubMed] [Google Scholar]
- 20.Shirinsky V P, Vorotnikov A V, Birukov K G, Nanaev A K, Collinge M, Lukas T J, Sellers J R, Watterson A M. J Biol Chem. 1993;268:16578–16583. [PubMed] [Google Scholar]