Abstract
The SELEX method and oligonucleotide combinatorial chemistry discovery process yields high-affinity/high-specificity ligands for virtually any molecular target. Typically, the enormous starting libraries used in the SELEX process contain 1014–1015 sequences. We now ask if the smaller sequences, complexity of extant organisms, and evolutionary history provide useful interactions between oligonucleotides and at least some unexpected targets. That is, do organisms contain a robust “linkage map” between their oligonucleotides and proteins and/or small molecules that enriches life?
We have accepted the challenge of writing an inaugural article, hoping to stimulate and provoke other scientists to wonder with us about a chance to discover surprising molecular interactions in biology between nucleic acids and other intracellular components. We will chart the conceptual triangle surrounding in vitro evolution [the SELEX (for systematic evolution of ligands by exponential enrichment) methodology]: insights into the prebiotic earth, applied medical research, and novel regulatory elements and circuits in cells (identified through the genomic SELEX process). We came to this set of ideas through the study of bacteriophage T4 development, with a focus on translational regulation. It was that work and the extraordinary environment for RNA studies that exists at the University of Colorado in Boulder that led us to our present interests.
Translational Regulation in Bacteriophage T4-Infected Escherichia coli
Among the first experiments on coupled transcription and translation in vitro were those done in Lipmann’s laboratory by one of us (1). For the first time, the synthesis of active, full-length enzymes from a DNA template was achieved. The templates used for most of those experiments were from T-even bacteriophage, which ultimately led to our interest in T4 translation and its regulation.
What a time to study a simple developing system—Peter Geiduschek’s and Dick Epstein’s laboratories (40, 41) had made clear the phage transcriptional pattern, while the invention of high-resolution SDS gels by Uli Laemmli (42) made it possible to detect a corresponding translational pattern. We thought that the development of this organism could be understood, given the power of T4 genetics (2) and the availability of mutations in most of the essential genes.
The early work we did at the University of Colorado was so simple. We received T4 mutant strains from people (who for the most part still shared things) and ran gels to see what happened to phage gene expression after infections of nonpermissive E. coli with those T4 mutants. We learned something interesting every day. We saw the developmental pattern (3), the global regulation, just as everyone knew we would, but we kept stumbling onto surprises. The first big surprise was that amber (the chain-terminating codon UAG) mutations in gene 32, which encodes the single-stranded DNA binding protein of T4, led to overexpression of the amber fragment (a fact easily observed because amber fragments in T4-infected cells are stable to proteolysis). Temperature-sensitive mutations in gene 32 led to overproduction of the full-size protein (4). After some minor experimentation, all straightforward, we proved that the gene 32 protein is an autogenous translational repressor (5–7). This set of experiments was done at the same time that many people were proclaiming that all gene-specific regulation was transcriptional. The T4 gene 32 experiments led to a rapid understanding of many of the translational regulatory loops for E. coli ribosomal protein expression. (We remember saying, with such pleasure, that the phrase “autogenous regulation of its own synthesis” was redundant, like the names Gail Storm and Rip Torn.)
Shortly after we discovered the gene 32 regulatory loop, we stumbled onto a similar story for gene 43. Gene 43 encodes the bacteriophage T4 DNA polymerase, and again we found that amber fragments and temperature-sensitive mutants of that protein were overexpressed. Once again, the regulatory loop was autogenous and translational (8). Now we had two examples of DNA-binding proteins that repressed their own translation, in each case by binding to the translational initiation domain of their respective mRNAs.
At this time, data were also accumulating that suggested that the regA gene product was a translational repressor. Because we were working hard on a region of the rIIB gene that was loaded with mutations, we were quickly able to identify the domain in the T4 rIIB mRNA that was the target for its translational repression by the regA protein (9–13); again, a translational repressor worked by binding to the initiation domain of a target message and blocking protein synthesis. The T4 regA protein, whose structure has been determined (14), blocks translation initiation on many target messages, using target sequences and structures that are different for individual messages. The mechanism of target selection by this protein that performs global translational regulation is still not clear though we have data that suggest that it interacts with single-stranded RNA in a sequence-specific manner (D.B., J. Brown, C. H. Kang, L.G., and P. Allen, unpublished work).
During these years, we were setting the stage, preparing our minds, for the discovery of the SELEX methodology and its applications. The thesis students and postdoctoral associates in our laboratory, the very bright undergraduates, and the brilliant faculty in Boulder (especially Gary Stormo, Mike Yarus, Tom Cech, and Olke Uhlenbeck, charter members of the first “RNA Club,” David Hirsh, and our non-Boulder colleagues in the phage world, David Shub and Ed Brody) spoke constantly about the powers of RNA. We always were thinking about fancy uses of RNA. One satisfying paper from our laboratory was about translational regulation of the E. coli infC gene (which encodes the translation initiation factor IF3); that paper is a soaring piece of rhetoric and logic about a fancy RNA that we had caught, we thought, showing off (15). The ideas were largely unsupported by evidence (at the time, and probably now), yet the thinking, the wondering, the adventurous academic environment were all helping to make single-stranded oligonucleotides bold creatures in our thoughts.
The First SELEX Experiment
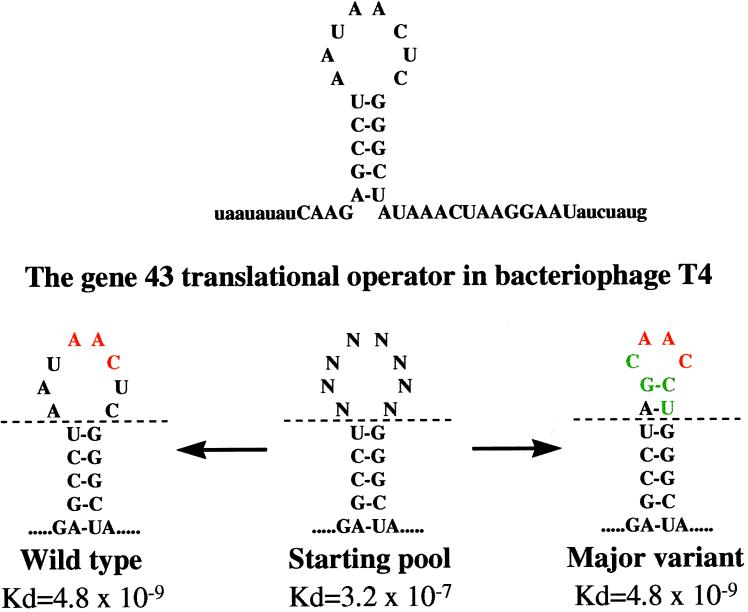
The experiments on autogenous translational repression by gene 43 protein continued in our laboratory, in part because specific mRNA binding by a DNA-binding protein had general interest. The mRNA target was known to be close to the initiating AUG and to include a hairpin 5′ to the Shine and Dalgarno sequence. Craig Tuerk, then a student about to wrap up his dissertation [his thesis work included the first observation that an abundant RNA tetraloop sequence provided stability to an RNA hairpin (16)], was considering the creation of 24 single-base mutations in the 8-nt loop of the mRNA target to understand the requirements for those positions in repressor binding. Yet a history existed, from our laboratory and elsewhere, of randomizing oligonucleotide sequences in preparation for genetic and/or biochemical analysis (17, 18), and we had realized that selection from a pool of oligonucleotides was possible. He randomized the entire loop to provide 48 (65,536) different sequences, and then selected the winners from that pool. [He understood, of course, that 65,536 sequences was a better pool to use to understand what nucleotides mattered than a mere 24; his advisor might have asked after the easy, straightforward experiment,“Well, what about the double mutants, and the triples, etc.?”] He used binding of the pool to the gene 43 protein as a way to partition the best binding sequences away from most of the pool. The pool contained fixed sequences on both ends to facilitate amplification of the better binding sequences so that the selection/partitioning could be reiterated. He named the process of in vitro evolution SELEX, for systematic evolution of ligands by exponential enrichment. The experiment worked better than we hoped, yielding two winning loop sequences (as shown in Fig. 1; ref. 19).
Figure 1.
The sequences and reasonable secondary structures of two RNAs that bind with identical affinities to bacteriophage T4 DNA polymerase are shown. The green nucleotides in the major variant are the positions of difference between the two RNAs.
The isolation of the wild-type T4 loop sequence did not surprise us (although later we learned that it is the rare nucleic acid binding protein whose natural target sequence is the highest-affinity sequence for that protein). The isolation of a “quadruple mutant” as a second high-affinity sequence astounded us. We could never have isolated such a mutant even after heavy mutagenesis with a powerful in vivo selection (that we didn’t have), and not even the most ornery thesis advisor would ever have suggested the creation of such a mutant using in vitro techniques. Many discussions about the major variant centered on the likelihood that the sequence was never tried in nature (in one extreme calculation, based on several assumptions, 100 million liters of a 1 M wild-type virus stock would be expected to yield a single virus with the desired mutations).
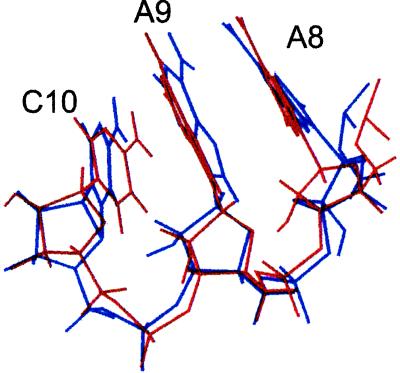
Amazingly, it turned out that the quadruple mutant RNA structure is isomorphic with the wild-type structure (Fig. 2) as shown by NMR studies (20, 21). We acknowledged that possibility at the time but we thought it unlikely. We were entranced with the existence of the isoenergetic quadruple mutant (of whatever structure—we didn’t care). We speculated that RNA libraries contain many solutions to the problem of high-affinity binding. We went crazy imagining the possibilities of using single-stranded oligonucleotides as ligands for virtually any molecular target. We realized we could identify drugs and diagnostic agents, and we said so, both in a patent application and the original publication (19, 22). Kevin Struhl’s laboratory had done a wonderful experiment with double-stranded DNA, using a transcription factor to partition from a pool those sequences that were winners for that protein (23); however, because double-stranded DNA binding is determined largely by simple sequence consensus, as opposed to more complex sequence/structure (shapes), these experiments did not lead to an appreciation of what might be done with in vitro evolution (the SELEX methodology) on single-stranded oligonucleotide pools.
Figure 2.
The conserved AACs of two RNAs that bind bacteriophage T4 DNA polymerase are shown. The AACs are in red in Fig. 1. The NMR structures and discussion are reproduced with permission from refs. 20 and 21 (Copyright 1996, Am. Chem. Soc.).
Aptamer Shapes and Pockets
Several aptamers (24), the ligands that emerge from the SELEX method, have been studied structurally. A recent review has been written that describes four aptamer structures (25). ATP, FMN, arginine, and citrulline have been used to select aptamers; those aptamers create pockets using noncanonical base pairs and backbone distortions and/or turns. Each structure exhibits two types of conservation: first, there are bases that covary and that provide conserved secondary structure domains that form structural scaffolds; and second, there are bases that are absolutely conserved. The latter nucleotides often are single-stranded or in noncanonical base pairs that directly interact with the target molecule. This is exactly what one might have predicted from similar attributes of proteins that bind small molecules. Ed Brody (personal communication) has argued that we will be unable to predict in detail the shapes of these pockets (or the larger surfaces that interact with protein targets), because the most idiosyncratic regions of the oligonucleotide will always be at the binding interface and will always use rule-bending noncanonical interactions. The scaffolding will be easy, but the most important domains will remain obscure. The most amazing ligand is the theophylline-binding aptamer identified by Jenison et al. (26); the structure has been obtained by Zimmerman et al. (G. Zimmerman, A. Pardi, R. Jenison, and J. P. Simorre, personal communication). In this structure, an extraordinary backbone turn, accompanied by a so-called 1-3-2 stack (in which stacking interactions occur between three adjacent nucleotides at a bend so severe that the third nucleotide stacks between the first and the second), begs the scientist to declare, “My goodness, an oligonucleotide can form any shape required to provide binding affinity and specificity!” Of course, this is exactly what drives the power of the SELEX process.
There is some need to understand the role of induced fit in aptamer binding. While everyone understands that induced fit extracts an entropic price, already we see that some aptamers without their targets are disordered according to NMR data. Induced fit is a phrase that demands careful language: if an aptamer alternates rapidly between two or three preferred structures, one of which binds the target, do we agree to say that upon binding the aptamer has undergone induced fit? Conversely, what fraction of the time must an aptamer exist in that final structure, but without its target, before we say that a preexisting state was sampled rather than created? Our point is simple—the scaffolding from covarying base pairs provides a very close approximation of the final structure, both before and after target binding, and thus the only unknown is how many states are occupied by the conserved and bulged or looped nonhelical nucleotides or noncanonical base pairs? If it’s a few, we suggest that this is not what people usually mean when they say “induced fit.”
Insights into the Prebiotic Earth
At around the same time that we were developing the SELEX protocol, Andrew Ellington and Jack Szostak were doing similar experiments aimed at finding RNAs that bound to organic dyes (24). Because the dyes were known intercalating agents, because many solutions to that problem were also evident, because no ligands were so striking as the pair of ligands for gene 43 protein, and because Jack Szostak was determined to study the early prebiotic catalytic potential of oligonucleotides, their work did not draw them toward drug discovery. In fact, the extraordinary work from Szostak’s laboratory (27) over the last 7 years has been aimed, almost without exception, at trying to understand the catalytic potential of RNA (or DNA).
Szostak has reviewed the ribozyme literature (27). Clearly, in vitro evolution is a good way to find nucleic acids with new catalytic activities. The attempts to find ribozymes using transition state analogues as targets have not been very successful (27), while direct selections have worked pretty well. The reported catalytic activities of these ribozymes have been low (as are the activities of natural ribozymes), but the central question from these experiments has been related to the prebiotic world (and/or the early biotic world). Thus slow catalytic rates are not discouraging for this enterprise; the issue is what an RNA or DNA can do. As we have all written repeatedly, proteins won and the reasons they won probably include better catalytic potential. Scientists in this field have an ethic about “cheating”: it is not fair, they assert, to use nonnatural nucleotides [say, one with a histidine hanging off the 5 position of a pyrimidine (28)] to provoke faster or different catalytic activities. Probably these rules were not articulated on the prebiotic earth, since at that early moment the earth itself didn’t know what chemicals we would call natural (as in “fair”) billions of years later. Intellectually this is an impossible, yet fascinating, question: can we imagine or deduce the relevant conditions on earth so that a meaningful experiment, an experiment that limits our theories for the chemical evolution that occurred, can be done in a modern laboratory? [We wonder what Trotsky thought in 1917 and can’t agree on even that. In the same way, we find the ribozyme/prebiotic earth research endeavor (27) a little disconnected from what may have existed long ago, and yet very thrilling; the experimental work in this area is wonderful.]
Applied Medical Research: Pharmaceuticals and Diagnostic Reagents
Most pharmaceuticals and many diagnostic reagents bind to proteins, and high-affinity/high-specificity binding aptamers are easy to obtain with the SELEX method. Very soon after the SELEX protocol was aimed at disease targets, nuclease-resistant aptamers were reported (29–31). Such aptamers are routinely obtained (using front-loaded SELEX, which incorporates stabilizing nucleotides into the library so that the winners found through SELEX are stable without post-SELEX manipulations), having (as above) high affinities and specificities for their intended targets. The diagnostic applications of these compounds is obvious; in fact, both ELISA-like assays and fluorescence-activated cell sorter (FACS) analyses have been done with aptamers (32, 33). Other issues remain for therapeutic uses of aptamers, yet all of those issues (pharmacokinetics, pharmacodynamics, synthesis at large scale and appropriate formulation chemistries, toxicity, immunogenicity, and efficacy in animals) have been addressed with success. The SELEX method may well be a fast, general method for producing efficacious drugs; we await only human trial data to discover if this dream is true.
More than 100 SELEX experiments have generated aptamers at either NeXstar or at laboratories at the University of Colorado. Perhaps another 100 SELEX experiments have been done away from Boulder. The data, still rather incomplete, for these ≈200 experiments are revealing.
About half the experiments have been aimed at nucleic acid binding proteins—that is, proteins that are said to interact with nucleic acids as part of their natural function. The other half have been aimed at proteins that are thought to do other things and to largely ignore nucleic acids naturally. The most striking fact is that the affinities of the winners from the first group are, if anything, weaker than the affinities of the second group; the affinities of SELEX-derived compounds aimed at regular proteins are beyond what we expect to see when single-chain antibodies or Fab fragments are interrogated in the same manner (34). It is not uncommon for a winning ligand from SELEX to have a nanomolar Kd when the target is a professional nucleic acid binding protein, while we have seen many picomolar Kd values for other protein targets. This is completely unexpected and serves to stimulate the kind of thinking we propose for a novel kind of regulatory loop.
Genomic SELEX and Biological Regulatory Loops
At first glance the SELEX method appears to be valuable for studies of the early earth (through the study of the catalytic powers of oligonucleotides), for studies of nucleic acid binding proteins (through the detailed study of high-affinity ligands), and for developing therapeutic and diagnostic compounds. The strongest statement we can make about the SELEX method is that it never really fails. All targets yield winning ligands with either modest affinities or with extraordinary affinities for their targets. These experiments have usually been performed with 1014–1015 unique oligonucleotides in the initial pool, the library used to find the winning ligand. No one has intentionally used a pool containing only 1010 sequences, and thus no one can be sure how good the winning ligands would be with respect to either affinity or specificity. The reason for not using the smaller library is simple: since large libraries can be used, and since the drive has been to find either great winning ligands or catalysts, why not use as many sequences as possible? But the number 1010 is an interesting number: it represents the approximate sequence complexity, the genome size, of a human. And thus a big question becomes “do natural sequences interact in novel ways, as either DNA or RNA, with unexpected target molecules in the organism?”
This question, as we have written earlier (35), is about the protein–nucleic acid linkage map; it is an exact analogue of the protein–protein linkage map articulated so beautifully by Fields and his colleagues (36). Think about E. coli; at ≈10−15 liters for its volume, a single molecule in E. coli is present at about 1 nM concentration. It is reasonable, even sensible, to wonder if under such conditions (small volumes packed with high concentrations of everything), specific oligonucleotides interact with specific proteins. We think that they do and that this is a general property of biology. This property is by no means restricted to professional nucleic acid binding proteins.
Imagine, then, a protein in any organism. Wouldn’t we like to know if that protein talks to any specific nucleic acid sequence in that organism (or to any nucleic acid sequence in an invasive pathogen of that organism, for good or for bad, for thwarting the pathogen or for increasing its growth potential)? We have taken this idea very seriously. We have prepared libraries of sized inserts of genomic nucleic acid sequences, ready for a SELEX protocol (called genomic SELEXs. These libraries have been made with E. coli, yeast, and human DNA (B.S.S., T.S., D.B., and L.G., unpublished work); soon they will be made for Drosophila melanogaster, Caenorhabditis elegans, mouse, and Arabidopsis. The idea is to investigate the interactions between a protein and the nucleic acids with which it comes in contact in vivo. Such an idea is more intriguing given the current and future availability of genome sequences. That is, we hope to understand the protein–nucleic acid linkage map in those organisms that are under the most thorough study, the organisms whose genetics, biochemistry, enzymology/metabolism, development, and genome sequencing might lead to some ideas about what constitutes an organism.
The genomic libraries (Fig. 3) were generated by random priming of sheared DNA from each of the organisms, followed by sizing to obtain overlapping fragments of any desired size (B.S.S., T.S., D.B., and L.G., unpublished work).
Figure 3.
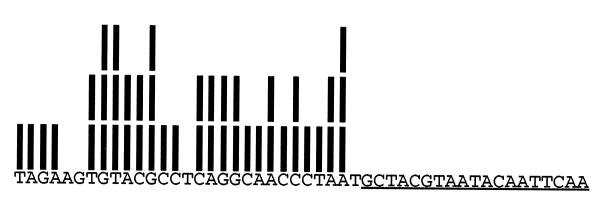
A perfect library generated by random priming should contain a complete set of genomic inserts starting (and ending) at each nucleotide of the genome. The diagram above shows a hypothetical set of library fragments with genomic inserts 36 bases long and a hypothetical binding site of 26 bases. These and only these fragments will survive to the final round of SELEX. The primer sequences (underlined) allow PCR amplification, while the T7 promoter (indicated with asterisks) is used when RNA is the target.
All sequences that we tested with PCR were represented in these libraries. Sequencing of individual library fragments suggests that genomic inserts nearly saturate the possible endpoints (Fig. 4).
Figure 4.
We developed a technique to examine the distribution of endpoints of genomic inserts that contain any short unique sequence (36). We isolated a set of library fragments that contain the Saccharomyces cerevisiae NDC1 gene sequence underlined above. Each bar above a nucleotide in the adjacent sequence means that nucleotide was adjacent to the T7 containing promoter in one of the 43 fragments examined. This limited analysis revealed 25 out of 29 possible endpoints! In our examination of five genes in four libraries, the largest “gap” that we found (i.e., stretch of nucleotides for which no endpoints were observed) is 9 nt. Assuming judicious choice of insert size, we are confident that every nucleic acid binding site for every protein is to be found in every library constructed by this method.
Using these libraries in the SELEX protocol should allow one to identify any sequence in any organism, whether DNA or RNA, that is the preferred site for any protein in that organism, as long as that site is present within a linear fragment of the size being screened. An enormous site, the catalytic center of ribosomal RNA (for example), would not be identified in our libraries. Needless to say, cDNA libraries (and cDNA libraries from specific tissues, times in development, or cellular locations) could be used as well.
The first set of experiments are revealing with respect to the simplest use of the genomic SELEX process. Genetic and biochemical approaches have revealed six sites in the E. coli genome at which the DNA-binding protein, metJ, interacts. The genomic SELEX process with double-stranded DNA from E. coli uncovered an additional five sites (Y.-y.H., D.B., and L.G., unpublished work). The bacteriophage MS2 coat protein binds tightly to a single sequence in the viral genome. When we asked if there are binding sites encoded by the E. coli genome, the answer was yes. (T.S., unpublished work). The U1A protein is known to interact with two primary sites in human RNA, and yet when the genomic SELEX process was performed with the U1A protein and human RNA, a large number of preferred targets were identified (B.S.S., T. Gibson, and L.G., unpublished work). We believe that the genomic SELEX process can expand the list of biological targets for any and every nucleic acid binding protein beyond what genetics or biochemistry can tell us. The genomic SELEX process is most obviously valuable in this area, since it extends the functions of known nucleic acid binding proteins to their entire set of biological targets. These first experiments also validate the libraries we have made as well as the methodologies. When known nucleic acid binding proteins are used, most of the known binding sites are found and, in addition, unknown sites are identified.
A second set of genomic SELEX experiments is aimed at proteins known to have an exquisite polyanion binding site but thought not to interact naturally with nucleic acids. Our favorite proteins in this class are secreted growth factors that contain heparin binding sites. We wonder if these proteins interact with a specific RNA or DNA intracellularly, before their secretion or when a fraction of the protein remains within the cell. We have made one guess (thus far) of a protein that we thought might talk to a site on RNA. We asked if basic fibroblast growth factor binds to a site in human RNA; we chose basic fibroblast growth factor because it has been used successfully in a variety of SELEX experiments (31, 37) and because a small fraction of the protein is synthesized with an amino-terminal extension that contains a nuclear localization signal (38). When challenged with basic fibroblast growth factor, the human genomic SELEX library yielded a single RNA winner with a nanomolar Kd; that RNA sequence is under study in our laboratory (Y.-y.H., B.S.S., Y.W., and L.G., unpublished work).
The third set of genomic SELEX experiments involves proteins that are neither known nucleic acid binding proteins nor polyanion binding proteins. Imagine that we are interested in β-galactosidase, for example, or any protein within any organism. Because the SELEX process yields winning ligands for virtually any protein, we must wonder if all proteins (or at least many) use some surface to interact with specific sites in the cognate genomic DNA or RNA. We believe, as in the two-hybrid protein paradigm, that winning natural ligands would suggest a biological regulatory loop that requires further investigation. People long have speculated on the large size of proteins; one element of large protein size that would be useful would be surfaces that promote regulatory loops that keep biological systems homeostatic (39). Moreover, since the SELEX process has worked on many small molecules, one could extend the idea of a protein–nucleic acid linkage map to include a metabolite–nucleic acid linkage map.
Conclusions and Perspectives
The success of in vitro evolution with large nucleic acid libraries has led us to wonder about biological regulatory loops in vivo. We have some of the same intuitions we had in the early 1970s when we were almost alone thinking that translational regulation was common. As noted above, we stumbled upon translational regulation in T4-infected E. coli because we were running SDS gels and looking at the patterns of protein synthesis—that is, we were simply taking a new technique (SDS gel electrophoresis was once a new technique!) and collecting data in an organism that interested us. The genomic SELEX process has the quality of being constructed from a technique (SELEX) and an idea—that the success of the SELEX process is predictive for interactions between many proteins and natural nucleic acids (and the additional idea that biology still has surprises for us). We have been talking about the broad implications of genomic SELEX for several years in our laboratory, yet it took us a long time to decide to test the idea. The problem with doing genomic SELEX (with basic fibroblast growth factor and β-galactosidase, representing applications two and three, above) is that we couldn’t decide how many negative experiments we would do before we discarded the idea; that is, if basic fibroblast growth factor had no high-affinity natural RNA or DNA sequence, is the idea of a broad protein–nucleic acid linkage map wrong? The genomic SELEX process is too difficult to expect all scientists to try every protein they study individually, and so we are developing methods that allow an approach in which many proteins are challenged simultaneously by all RNAs or DNAs from an organism.
While the emphasis in this paper seems aimed largely at the impact of any protein on expression of specific genes through preferred target sites, the reciprocal regulatory activities of natural nucleic acids on specific enzymes, cytokines, receptors, and other proteins are equally likely to be found through the genomic SELEX process. The selection of unnatural nucleic acids that inhibit the activity of a specific protein has proven to be trivial. We anticipate that nature will have observed and used the same property in specifically controlling the activity of proteins in vivo and that such a scheme can be uncovered using the genomic SELEX process.
Acknowledgments
We have had the good fortune to discuss the SELEX paradigm and its applications to applied and basic research with hundreds of people over the last decade. We thank them as well as the National Institutes of Health for supporting our research.
References
- 1.Gold L M, Schweiger M. Proc Natl Acad Sci USA. 1969;62:892–898. doi: 10.1073/pnas.62.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epstein R H, Bolle A, Steinberg C M, Kellenberger E, Boy de la Tour E, Chevalley R, Edgar R S, Sussman M, Denhardt G H, Lielausis A. Cold Spring Harbor Symp Quant Biol. 1963;28:375–392. [Google Scholar]
- 3.O’Farrell P Z, Gold L M. J Biol Chem. 1973;248:5502–5511. [PubMed] [Google Scholar]
- 4.Gold L, O’Farrell P Z, Russel M. J Biol Chem. 1976;251:7251–7262. [PubMed] [Google Scholar]
- 5.Russel M, Gold L, Morrissett H, O’Farrell P Z. J Biol Chem. 1976;251:7263–7270. [PubMed] [Google Scholar]
- 6.Lemaire G, Gold L, Yarus M. J Mol Biol. 1978;126:73–90. doi: 10.1016/0022-2836(78)90280-2. [DOI] [PubMed] [Google Scholar]
- 7.McPheeter D S, Stormo G D, Gold L. J Mol Biol. 1988;201:517–535. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- 8.Andrake M, Guild N, Hsu T, Gold L, Tuerk C, Karam J. Proc Natl Acad Sci USA. 1988;85:7942–7946. doi: 10.1073/pnas.85.21.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karam J, Gold L, Singer B S, Dawson M. Proc Natl Acad Sci USA. 1981;78:4669–4673. doi: 10.1073/pnas.78.8.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller E S, Winter R B, Campbell K M, Power S D, Gold L. J Biol Chem. 1985;260:13053–13059. [PubMed] [Google Scholar]
- 11.Miller E S, Karam J, Dawson M, Trojanowska M, Gauss P, Gold L. J Mol Biol. 1987;194:397–410. doi: 10.1016/0022-2836(87)90670-x. [DOI] [PubMed] [Google Scholar]
- 12.Shinedling S, Gayle M, Pribnow D, Gold L. Mol Gen Genet. 1987;207:224–232. doi: 10.1007/BF00331582. [DOI] [PubMed] [Google Scholar]
- 13.Winter R B, Morrissey L, Gauss P, Gold L, Hsu T, Karam J. Proc Natl Acad Sci USA. 1987;84:7822–7826. doi: 10.1073/pnas.84.22.7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang C, Chan R, Berger I, Lockshin C, Green L, Gold L, Rich A. Science. 1995;268:1170–1173. doi: 10.1126/science.7761833. [DOI] [PubMed] [Google Scholar]
- 15.Gold L, Stormo G, Saunders R. Proc Natl Acad Sci USA. 1984;81:7061–7065. doi: 10.1073/pnas.81.22.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuerk C, Gauss P, Thermes C, Groebe D R, Gayle M, Guild N, Stormo G, d’Aubenton-Carafa Y, Uhlenbeck O C, Tinoco I, Jr, Brody E N, Gold L. Proc Natl Acad Sci USA. 1988;85:1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Childs J, Villanueba K, Barrick D, Schneider T D, Stormo G D, Gold L, Leitner M, Caruthers M. UCLA Symp Mol Cell Biol. 1985;30:341–350. [Google Scholar]
- 18.Barrick D, Villanueba K, Childs J, Kalil R, Schneider T, Lawrence C, Gold L, Stormo G. Nucleic Acids Res. 1994;22:1287–1295. doi: 10.1093/nar/22.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 20.Mirmira S R, Tinoco I., Jr Biochemistry. 1996;35:7664–7674. doi: 10.1021/bi960414y. [DOI] [PubMed] [Google Scholar]
- 21.Mirmira S R, Tinoco I., Jr Biochemistry. 1996;35:7675–7683. doi: 10.1021/bi960415q. [DOI] [PubMed] [Google Scholar]
- 22.Gold, L. & Tuerk, C. (1993) U.S. Patent 5,270,163.
- 23.Oliphant A R, Brandl C J, Struhl K. Mol Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellington A, Szostak J. Nature (London) 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 25.Feigon J, Dieckmann T, Smith F W. Chem Biol. 1996;3:611–617. doi: 10.1016/s1074-5521(96)90127-1. [DOI] [PubMed] [Google Scholar]
- 26.Jenison R D, Gill S C, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 27.Lorsh J R, Szostak J. Acc Chem Res. 1994;29:103–110. doi: 10.1021/ar9501378. [DOI] [PubMed] [Google Scholar]
- 28.Eaton B E, Pieken W A. Annu Rev Biochem. 1995;64:837–863. doi: 10.1146/annurev.bi.64.070195.004201. [DOI] [PubMed] [Google Scholar]
- 29.Lin Y, Gill S C, Jayasena S D. Nucleic Acids Res. 1994;22:5229–5234. doi: 10.1093/nar/22.24.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green L, Jellinek D, Beebe L, Feistner B, Gill S, Jucker F, Janjic N. Chem Biol. 1995;2:683–695. doi: 10.1016/1074-5521(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 31.Jellinek D, Green L, Bell C, Lynott K, Gill N, Vargeese C, Kirschenheuter G, McGee D, Abesinghe P, Pieken W, Shapiro R, Rifkin D, Moscatelli D, Janjic N. Biochemistry. 1995;34:11363–11372. doi: 10.1021/bi00036a009. [DOI] [PubMed] [Google Scholar]
- 32.Drolet D W, Moon-McDermott L, Romig T S. Nat Biotechnol. 1996;14:1021–1025. doi: 10.1038/nbt0896-1021. [DOI] [PubMed] [Google Scholar]
- 33.Davis K A, Abrams B, Lin Y, Jayasena S D. Nucleic Acids Res. 1996;24:702–706. doi: 10.1093/nar/24.4.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.deKruif J, Van der Vuurst de Vries A-R, Cilenti L, Boel E, van Ewijk W, Logtenberg T. Immunol Today. 1996;17:453–455. doi: 10.1016/0167-5699(96)30057-y. [DOI] [PubMed] [Google Scholar]
- 35.Gold L. J Biol Chem. 1995;270:13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- 36.Evangelista C, Lockshon D, Fields S. Trends Cell Biol. 1996;6:196–199. doi: 10.1016/0962-8924(96)40002-2. [DOI] [PubMed] [Google Scholar]
- 37.Jellinek D, Lynott C K, Rifkin D B, Janjic N. Proc Natl Acad Sci USA. 1993;90:11227–11231. doi: 10.1073/pnas.90.23.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rifkin D B, Moscatelli D, Roghani M, Nagano Y, Quarto N, Klein S, Bikfalvi A. Mol Reprod Dev. 1994;39:102–105. doi: 10.1002/mrd.1080390115. [DOI] [PubMed] [Google Scholar]
- 39.Savageau M A. Biochemical Systems Analysis: A Study of Function and Design in Molecular Biology. Reading, MA: Addison–Wesley; 1976. [Google Scholar]
- 40.Bolle A, Epstein R H, Salser W, Geidushek E P. J Mol Biol. 1968;31:325–348. doi: 10.1016/0022-2836(68)90413-0. [DOI] [PubMed] [Google Scholar]
- 41.Bolle A, Epstein R H, Salser W, Geidushek E P. J Mol Biol. 1968;33:349–362. doi: 10.1016/0022-2836(68)90193-9. [DOI] [PubMed] [Google Scholar]
- 42.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]