Abstract
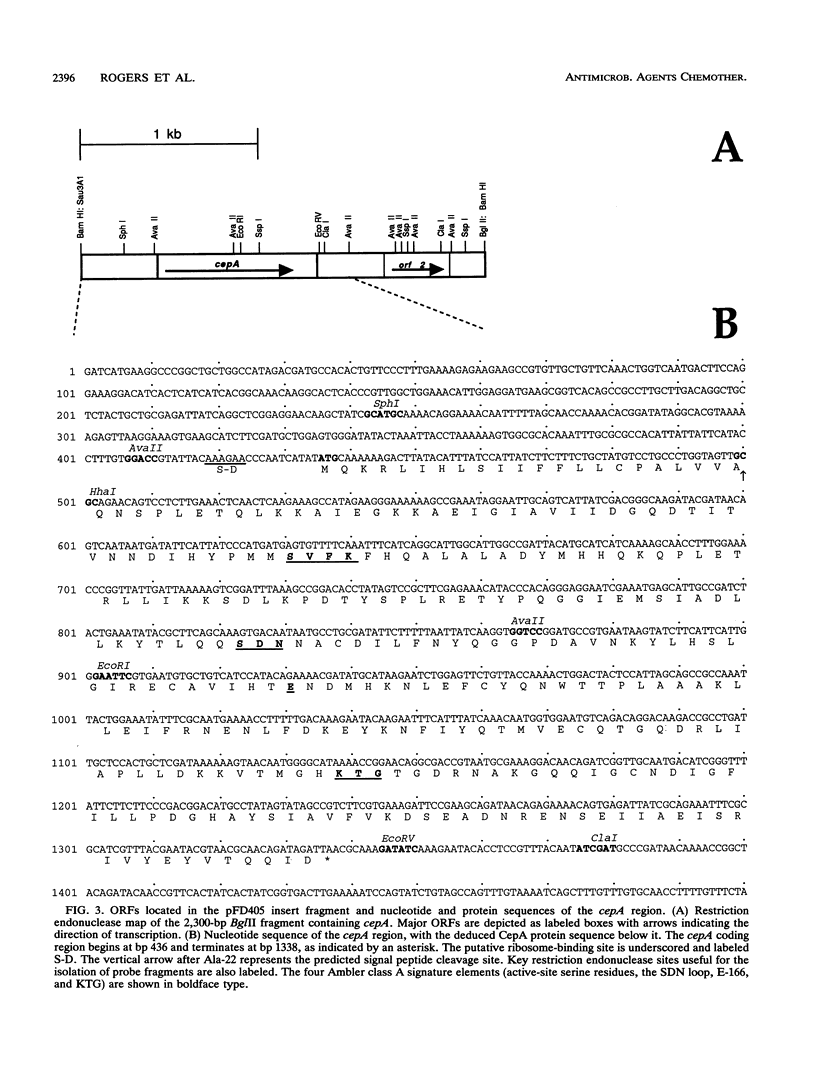
Bacteroides fragilis CS30 is a clinical isolate resistant to high concentrations of benzylpenicillin and cephaloridine but not to cephamycin or penem antibiotics. beta-Lactam resistance is mediated by a chromosomally encoded cephalosporinase produced at a high level. The gene encoding this beta-lactamase was cloned from genomic libraries constructed in Escherichia coli and then mated with B. fragilis 638 for identification of ampicillin-resistant (Apr) strains. Apr transconjugants contained a nitrocefin-reactive protein with the physical and enzymatic properties of the original CS30 isolate. The beta-lactamase gene (cepA) was localized by deletion analysis and subcloned, and its nucleotide sequence was determined. The 903-bp cepA open reading frame encoded a 300-amino-acid precursor protein (predicted molecular mass, 34,070 Da). A beta-lactamase-deficient mutant strain of B. fragilis 638 was constructed by insertional inactivation with the cepA gene of CS30, demonstrating strict functional homology between these chromosomal beta-lactamase genes. An extensive comparison of the CepA protein sequence by alignment with other beta-lactamases revealed the strict conservation of at least four elements common to Ambler class A. A further comparison of the CepA protein sequence with protein sequences of beta-lactamases from two other Bacteroides species indicated that they constitute their own distinct subgroup of class A beta-lactamases.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Coulson A. F., Frère J. M., Ghuysen J. M., Joris B., Forsman M., Levesque R. C., Tiraby G., Waley S. G. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991 May 15;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Arakawa Y., Ohta M., Kido N., Fujii Y., Komatsu T., Kato N. Close evolutionary relationship between the chromosomally encoded beta-lactamase gene of Klebsiella pneumoniae and the TEM beta-lactamase gene mediated by R plasmids. FEBS Lett. 1986 Oct 20;207(1):69–74. doi: 10.1016/0014-5793(86)80014-x. [DOI] [PubMed] [Google Scholar]
- Arakawa Y., Ohta M., Kido N., Mori M., Ito H., Komatsu T., Fujii Y., Kato N. Chromosomal beta-lactamase of Klebsiella oxytoca, a new class A enzyme that hydrolyzes broad-spectrum beta-lactam antibiotics. Antimicrob Agents Chemother. 1989 Jan;33(1):63–70. doi: 10.1128/aac.33.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Britz M. L., Wilkinson R. G. Purification and properties of beta-lactamase from Bacteroides fragilis. Antimicrob Agents Chemother. 1978 Mar;13(3):373–382. doi: 10.1128/aac.13.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 2c, 2d, 2e, 3, and 4. Antimicrob Agents Chemother. 1989 Mar;33(3):271–276. doi: 10.1128/aac.33.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. T. Nucleotide sequence of the Staphylococcus aureus PC1 beta-lactamase gene. Nucleic Acids Res. 1986 Jul 25;14(14):5940–5940. doi: 10.1093/nar/14.14.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornick N. A., Cuchural G. J., Jr, Snydman D. R., Jacobus N. V., Iannini P., Hill G., Cleary T., O'Keefe J. P., Pierson C., Finegold S. M. The antimicrobial susceptibility patterns of the Bacteroides fragilis group in the United States, 1987. J Antimicrob Chemother. 1990 Jun;25(6):1011–1019. doi: 10.1093/jac/25.6.1011. [DOI] [PubMed] [Google Scholar]
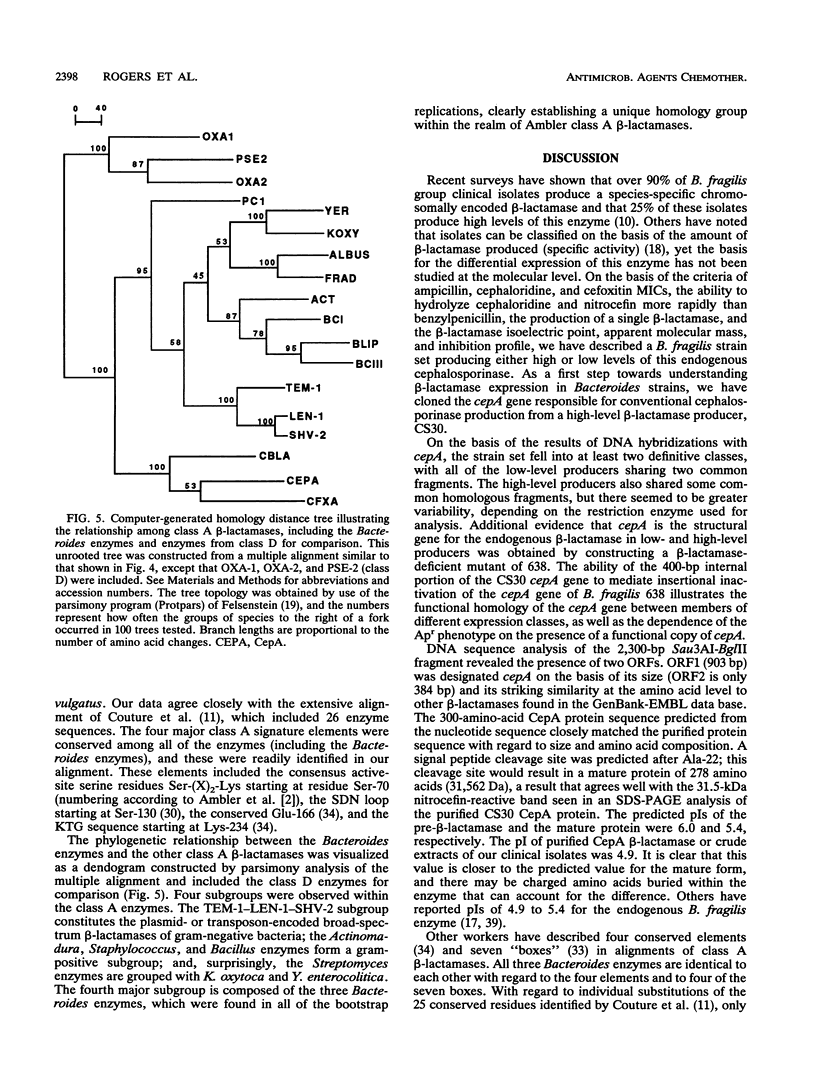
- Couture F., Lachapelle J., Levesque R. C. Phylogeny of LCR-1 and OXA-5 with class A and class D beta-lactamases. Mol Microbiol. 1992 Jun;6(12):1693–1705. doi: 10.1111/j.1365-2958.1992.tb00894.x. [DOI] [PubMed] [Google Scholar]
- Cuchural G. J., Jr, Tally F. P., Jacobus N. V., Aldridge K., Cleary T., Finegold S. M., Hill G., Iannini P., O'Keefe J. P., Pierson C. Susceptibility of the Bacteroides fragilis group in the United States: analysis by site of isolation. Antimicrob Agents Chemother. 1988 May;32(5):717–722. doi: 10.1128/aac.32.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey R. E. Leader peptidase. Mol Microbiol. 1991 Dec;5(12):2855–2860. doi: 10.1111/j.1365-2958.1991.tb01844.x. [DOI] [PubMed] [Google Scholar]
- Dale J. W., Godwin D., Mossakowska D., Stephenson P., Wall S. Sequence of the OXA2 beta-lactamase: comparison with other penicillin-reactive enzymes. FEBS Lett. 1985 Oct 21;191(1):39–44. doi: 10.1016/0014-5793(85)80989-3. [DOI] [PubMed] [Google Scholar]
- Dehottay P., Dusart J., De Meester F., Joris B., Van Beeumen J., Erpicum T., Frère J. M., Ghuysen J. M. Nucleotide sequence of the gene encoding the Streptomyces albus G beta-lactamase precursor. Eur J Biochem. 1987 Jul 15;166(2):345–350. doi: 10.1111/j.1432-1033.1987.tb13521.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R., Greenwood D. An investigation of beta-lactamases from clinical isolates of Bacteroides species. J Med Microbiol. 1992 Feb;36(2):89–95. doi: 10.1099/00222615-36-2-89. [DOI] [PubMed] [Google Scholar]
- Eley A., Greenwood D. Characterization of beta-lactamases in clinical isolates of Bacteroides. J Antimicrob Chemother. 1986 Sep;18(3):325–333. doi: 10.1093/jac/18.3.325. [DOI] [PubMed] [Google Scholar]
- Foweraker J. E., Hawkey P. M., Heritage J., Van Landuyt H. W. Novel beta-lactamase from Capnocytophaga sp. Antimicrob Agents Chemother. 1990 Aug;34(8):1501–1504. doi: 10.1128/aac.34.8.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Hasegawa P., Davis C. E. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie E. P., Salyers A. A. Use of targeted insertional mutagenesis to determine whether chondroitin lyase II is essential for chondroitin sulfate utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1986 Jun;166(3):966–971. doi: 10.1128/jb.166.3.966-971.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Knowles J. R. Directed selective pressure on a beta-lactamase to analyse molecular changes involved in development of enzyme function. Nature. 1976 Dec 23;264(5588):803–804. doi: 10.1038/264803a0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Houba S., Willem S., Duez C., Molitor C., Dusart J., Frère J. M., Ghuysen J. M. Nucleotide sequence of the gene encoding the active-site serine beta-lactamase from Actinomadura R39. FEMS Microbiol Lett. 1989 Dec;53(3):241–246. doi: 10.1016/0378-1097(89)90224-3. [DOI] [PubMed] [Google Scholar]
- Huovinen P., Huovinen S., Jacoby G. A. Sequence of PSE-2 beta-lactamase. Antimicrob Agents Chemother. 1988 Jan;32(1):134–136. doi: 10.1128/aac.32.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Carlino A., Madonna M. J., Lampen J. O. Cloning and sequencing of the metallothioprotein beta-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J Bacteriol. 1985 Oct;164(1):223–229. doi: 10.1128/jb.164.1.223-229.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Pastor F. I., Lampen J. O. Cloning and sequencing of the blaZ gene encoding beta-lactamase III, a lipoprotein of Bacillus cereus 569/H. J Bacteriol. 1987 Feb;169(2):579–586. doi: 10.1128/jb.169.2.579-586.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Joris B., Lepage S., Dusart J., Frère J. M. Role of the conserved amino acids of the 'SDN' loop (Ser130, Asp131 and Asn132) in a class A beta-lactamase studied by site-directed mutagenesis. Biochem J. 1990 Oct 15;271(2):399–406. doi: 10.1042/bj2710399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaurin B., Grundström T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ghuysen J. M., Dive G., Renard A., Dideberg O., Charlier P., Frère J. M., Kelly J. A., Boyington J. C., Moews P. C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988 Mar 1;250(2):313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ledent P., Dideberg O., Fonzé E., Lamotte-Brasseur J., Kelly J. A., Ghuysen J. M., Frère J. M. Comparison of the sequences of class A beta-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991 Nov;35(11):2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Neugebauer K., Sprengel R., Schaller H. Penicillinase from Bacillus licheniformis: nucleotide sequence of the gene and implications for the biosynthesis of a secretory protein in a Gram-positive bacterium. Nucleic Acids Res. 1981 Jun 11;9(11):2577–2588. doi: 10.1093/nar/9.11.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolich M. P., Shoemaker N. B., Salyers A. A. A Bacteroides tetracycline resistance gene represents a new class of ribosome protection tetracycline resistance. Antimicrob Agents Chemother. 1992 May;36(5):1005–1012. doi: 10.1128/aac.36.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord C. E., Lindqvist L., Olsson-Liljequist B., Tunér K. Beta-lactamases in anaerobic bacteria. Scand J Infect Dis Suppl. 1985;46:57–63. [PubMed] [Google Scholar]
- Nord C. E., Olsson-Liljequist B. Resistance to beta-lactam antibiotics in Bacteroides species. J Antimicrob Chemother. 1981 Dec;8 (Suppl 500):33–42. doi: 10.1093/jac/8.suppl_d.33. [DOI] [PubMed] [Google Scholar]
- O'Callaghan C. H., Morris A., Kirby S. M., Shingler A. H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972 Apr;1(4):283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson-Liljequist B., Dornbusch K., Nord C. E. Characterization of three different beta-lactamases from the Bacteroides fragilis group. Antimicrob Agents Chemother. 1980 Aug;18(2):220–225. doi: 10.1128/aac.18.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson B., Dornbusch K., Nord C. E. Susceptibility to beta-lactam antibiotics and production of beta-lactamase in Bacteroides fragilis. Med Microbiol Immunol. 1977 Oct 7;163(3):183–194. doi: 10.1007/BF02126677. [DOI] [PubMed] [Google Scholar]
- Ouellette M., Bissonnette L., Roy P. H. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A. C., Smith C. J. Genetic and biochemical analysis of a novel Ambler class A beta-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob Agents Chemother. 1993 May;37(5):1028–1036. doi: 10.1128/aac.37.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbielski A., Melzer B. Nucleotide sequence of the gene encoding the SHV-2 beta-lactamase (blaSHV-2) of Klebsiella ozaenae. Nucleic Acids Res. 1990 Aug 25;18(16):4916–4916. doi: 10.1093/nar/18.16.4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privitera G., Dublanchet A., Sebald M. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J Infect Dis. 1979 Jan;139(1):97–101. doi: 10.1093/infdis/139.1.97. [DOI] [PubMed] [Google Scholar]
- Sato K., Matsuura Y., Miyata K., Inoue M., Mitsuhashi S. Characterization of cephalosporinases from Bacteroides fragilis, Bacteroides thetaiotaomicron and Bacteroides vulgatus. J Antibiot (Tokyo) 1983 Jan;36(1):76–85. doi: 10.7164/antibiotics.36.76. [DOI] [PubMed] [Google Scholar]
- Seoane A., García Lobo J. M. Nucleotide sequence of a new class A beta-lactamase gene from the chromosome of Yersinia enterocolitica: implications for the evolution of class A beta-lactamases. Mol Gen Genet. 1991 Aug;228(1-2):215–220. doi: 10.1007/BF00282468. [DOI] [PubMed] [Google Scholar]
- Shoemaker N. B., Getty C., Gardner J. F., Salyers A. A. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J Bacteriol. 1986 Mar;165(3):929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Guthrie E. P., Salyers A. A., Gardner J. F. Evidence that the clindamycin-erythromycin resistance gene of Bacteroides plasmid pBF4 is on a transposable element. J Bacteriol. 1985 May;162(2):626–632. doi: 10.1128/jb.162.2.626-632.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloma A., Gross M. Molecular cloning and nucleotide sequence of the type I beta-lactamase gene from Bacillus cereus. Nucleic Acids Res. 1983 Jul 25;11(14):4997–5004. doi: 10.1093/nar/11.14.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Callihan D. R. Analysis of rRNA restriction fragment length polymorphisms from Bacteroides spp. and Bacteroides fragilis isolates associated with diarrhea in humans and animals. J Clin Microbiol. 1992 Apr;30(4):806–812. doi: 10.1128/jcm.30.4.806-812.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J. Characterization of Bacteroides ovatus plasmid pBI136 and structure of its clindamycin resistance region. J Bacteriol. 1985 Mar;161(3):1069–1073. doi: 10.1128/jb.161.3.1069-1073.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Parker A. C. Identification of a circular intermediate in the transfer and transposition of Tn4555, a mobilizable transposon from Bacteroides spp. J Bacteriol. 1993 May;175(9):2682–2691. doi: 10.1128/jb.175.9.2682-2691.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Malamy M. H. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J Bacteriol. 1990 May;172(5):2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene. 1983 Sep;23(3):331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Oyaizu Y., Oyaizu H., Woese C. R. Natural relationship between bacteroides and flavobacteria. J Bacteriol. 1985 Oct;164(1):230–236. doi: 10.1128/jb.164.1.230-236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R. A., Macrina F. L. Physical characterization of Bacteroides fragilis R plasmid pBF4. J Bacteriol. 1981 Feb;145(2):867–872. doi: 10.1128/jb.145.2.867-872.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]