Abstract
The interferon-induced double-stranded RNA-activated protein kinase, PKR, likely contributes to both the antiviral and the antiproliferative effects of interferon. We previously found that influenza virus avoids the translational inhibitory effects of activated PKR by activating a cellular inhibitory protein, termed P58IPK, based on its Mr of 58,000. P58IPK is a member of the tetratricopeptide family of proteins and possesses significant homology to the conserved J region of the DnaJ family of heat shock proteins. We earlier hypothesized that P58IPK was kept in an inactive state with its own inhibitor (termed I-P58IPK) in uninfected cells. We therefore attempted the purification and characterization of I-P58IPK. The following data suggest that we have identified the molecular chaperone, hsp40, as I-P58IPK. (i) The MonoP-purified I-P58IPK protein reacted with hsp40 antibody. (ii) This preparation demonstrated high specific activity in an in vitro functional assay containing only purified recombinant and native components. (iii) Purified, recombinant hsp40 protein inhibited P58IPK in an identical in vitro assay. (iv) Finally, we demonstrate that hsp40 directly complexes with P58IPK, in vitro, suggesting the inhibition occurs through a direct interaction. Our data, taken together, provide evidence for a novel intersection between the heat shock and interferon pathways, and suggest that influenza virus regulates PKR activity through the recruitment of a cellular stress pathway.
The interferon (IFN)-induced, serine–threonine protein kinase, PKR, is a critical component of the host defense response against virus infection (1, 2). PKR becomes autophosphorylated and activated in response to double-stranded RNA (dsRNA) and functions to block protein synthesis by phosphorylating the α subunit of eukaryotic initiation factor 2 (eIF-2α) (3). Translational regulation by PKR may play a role in growth regulation, as expression of PKR has been found to be growth-suppressive in yeast (4). Furthermore, expression of a dominant negative PKR mutant has been shown to transform NIH 3T3 cells, suggesting that PKR may function as a tumor suppressor protein (5–7). The kinase has also been hypothesized to play a role in inducing apoptosis (8) and regulating adipocyte differentiation (9). The platelet-derived growth factor signal transduction pathway has been found to require the activity of PKR in the induction of immediate early gene transcription (10). Finally, PKR has been suggested to be a regulator of the NF-kB signal transduction pathway (11).
Several cellular regulators of PKR have been previously described (12), but the best characterized is P58IPK (IPK, inhibitor of protein kinase) (13). P58IPK was first identified as a PKR inhibitor that is activated during influenza virus infection. P58IPK has since been purified, cloned, and extensively characterized (13, 14). P58IPK is a member of the tetratricopeptide repeat family of proteins, containing 9 tandemly arranged 34 amino acid repeats. Tetratricopeptide repeat domains are predicted to form amphipathic helices which may play a role in protein–protein interaction (15). Several tetratricopeptide repeat proteins have been identified thus far, but few have been assigned functional roles (for a review, see ref. 16). In addition to the tetratricopeptide repeat motifs, the C-terminal portion of P58IPK shares homology with the J domain of the DnaJ heat shock protein of Escherichia coli, placing P58IPK within the heat shock family of proteins (17). Finally, we found P58IPK to display oncogenic properties, possibly through its ability to down-regulate PKR (18).
The P58IPK–PKR regulatory pathway is highly complex. Evidence that P58IPK is regulated by its own inhibitor, earlier defined as I-P58IPK, includes the following: (i) P58IPK activity was undetectable in uninfected cells; (ii) P58IPK activity, but not physical protein levels, increased upon influenza virus infection; (iii) P58IPK activity was unmasked in uninfected cell extracts after biochemical fractionation with ammonium sulfate (P58IPK activity was found in the 40–60% ammonium sulfate fraction); and (iv) P58IPK activity was repressed by adding back the 60–80% ammonium sulfate fraction (which contains I-P58IPK activity) (19). Given the above observations, we hypothesized that infection by influenza virus released P58IPK from I-P58IPK, thus allowing P58IPK to directly act on PKR and inhibit the protein kinase. The goal of the present study was to identify I-P58IPK using biochemical purification and in vitro functional assays for I-P58IPK function. We can now report that the molecular chaperone, hsp40, also known as hdj-1, both inhibits and interacts with P58IPK, strongly suggesting that the cellular stress pathway participates in PKR regulation during influenza virus infection.
MATERIALS AND METHODS
Cells and Bacterial Strains.
Madin–Darby bovine kidney (MDBK) cells were grown in monolayer cultures in 850-cm2 roller bottles at 37°C in DMEM supplemented with 10% calf serum/2 mM l-glutamine/10 mM Hepes buffer. IFN-treated extracts were prepared as described (20). Glutathione S-transferase (GST)-tagged P58IPK (GST–P58IPK) was prepared as described (13). For the binding experiments, full-length human hsp40 was expressed from the T3 promoter of pBluescript (Stratagene) harbored in E. coli strain XL-1.
Purification of I-P58IPK.
Monolayer MDBK cells (2 × 1010) were harvested at ≈90% confluency as described (21). Cytoplasmic extracts were centrifuged at 100,000 × g for 1 h in a Beckman Ti 70.1 rotor. The supernatant (S-100) was fractionated by ammonium sulfate precipitation as described (21) and assayed for I-P58IPK activity. The active fraction was applied to a MonoQ HR 10/10 fast protein liquid chromatography (FPLC) anion-exchange column (Pharmacia). Proteins were eluted batch-wise with 300 mM and 500 mM KCl. Fractions were dialyzed into buffer B and assayed for I-P58IPK activity. The active fraction was resuspended into buffer A (75 mM Tris·CH3COOH, pH 9.3) and dialyzed against the identical buffer. The dialysate was applied to a MonoP HR 5/20 FPLC chromatofocusing column (Pharmacia). Proteins were eluted on a linear pH gradient of 9.3–6.0 with 10% Polybuffer-96 (Pharmacia) adjusted to pH 6.0. Fractions were collected, and pooled protein peaks were dialyzed into buffer B [20 mM Tris·HCl/100 mM KCl/0.1 mM EDTA/1 mM DTT/0.1 mM phenylmethylsulfonyl fluoride (PMSF)/5% glycerol] and assayed for the ability to inhibit P58IPK activity.
In Vitro Assays for I-P58IPK Activity.
Two assays were developed to detect I-P58IPK activity, one using crude and partially purified components and a second using highly purified components. The assay using crude components was used to screen for I-P58IPK activity in the ammonium sulfate and MonoQ fractions and is described in ref. 19.
The assay using purified components was used to test for I-P58IPK activity in the MonoP fractions and in the purified mitochondrial aspartate aminotransferase (mAAT) and hsp40 preparations. Fractions were mixed with GST–P58IPK purified from E. coli for 10 min at 30°C. PKR, purified from IFN-treated Daudi cells, was added, and the reaction was incubated at 30°C for 10 min. The total reaction volume was 30 μl in buffer with final concentrations as follows: 2 mM Hepes, pH 7.5/16.7 mM Tris·HCl/56.7 mM KCl/40 mM NaCl/2 mM MgCl2/2 mM MnCl2/1 mg/ml BSA/1.4 mM DTT/6 μg/ml aprotinin/0.02 mM PMSF/0.05 mM EDTA/6.6% glycerol/2 mM ATP. Poly I:C dsRNA was added at a final concentration of 0.1 μg/ml, in addition to 5 μCi (1 Ci = 37 GBq) of [γ-32P]ATP, and the reaction was incubated at 30°C for 10 min. Calf thymus histone protein IIA (10 μg) was added in the presence of 10 μCi [γ-32P]ATP, and the reaction was incubated at 30°C for 20 min. We earlier found a perfect correlation between the ability of PKR to phosphorylate histones and its natural substrate, eIF-2α (20). The reaction was stopped with the addition of 30 μl of stop buffer [2× protein disruption buffer (150 mM Tris·HCl, pH 6.8/2.6M 2-mercaptoethanol/3.7% SDS/18.5% glycerol/17 mM EDTA/20 μg/ml RNase A) at 60°C], boiled for 5 min, and analyzed by SDS/PAGE (14% gel). The degree of substrate phosphorylation was visualized by autoradiography and quantified by PhosphorImager analysis.
mAAT and hsp40.
Recombinant purified hsp40 was kindly provided by R. Morimoto (Northwestern University, Evanston, IL) and was prepared as described (22). Polyclonal antiserum to hsp40 was prepared using recombinant protein as immunogen (unpublished data). Recombinant mAAT protein was kindly provided by Marino Martinez-Carrion (University of Missouri, Kansas City, KS) and purified as described (23). The mAAT antibody was provided by P. D. Berk, prepared as described (24).
Western Blot Analysis of P58IPK.
In vitro kinase reactions were performed as described above (without [γ-32P]ATP). Proteins were separated by SDS/PAGE and transferred to a nitrocellulose filter, according to the procedure of Towbin and coworkers (25). Filters were blocked in PBS containing 5% nonfat dry milk and subsequently probed with monoclonal antibodies specific for P58IPK (2F8 and 9F10) at 1:25 each (26). Blots were washed 4 times with 0.1% Tween 20 in PBS and hybridized with secondary antibody, goat anti-mouse IgG conjugated to horseradish peroxidase (GIBCO/BRL). Blots were washed repeatedly with 0.3% and 0.1% Tween 20 in PBS, and the signal was visualized by enhanced chemiluminescence (Amersham).
Two-Dimensional Non-Equilibrium pH Gel Electrophoresis (NEpHGE).
Analyses were carried out essentially as described by O’Farrell and coworkers (27). Protein samples were prepared as follows. Lammeli sample buffer (6×) was added to a final concentration of 0.5×, and samples were boiled for 5 min. RNase A (1 μg) was added, and samples were incubated at room temperature for 15 min and then boiled for 2 min. RNase A (10 μg) and 10 μg BSA were then added as pH markers, and 3 μl of 3 M 2-mercaptoethanol was added. Samples were lyophilized in a Speed Vac (Savant). Lyophilized samples were resolubilized with 30 μl of NEpHGE sample buffer [9.5 M urea/2.0% Nonidet P-40/5% 2-mercaptoethanol/2% Servalyte ampholines, pH 2–11 (Serva)] and incubated for 10 min at 30°C. Samples were loaded onto 1.5 mm × 15 cm tube gels (5% acrylamide/0.2% bis-acrylamide/2% Servalytes, pH 2–11/2% Nonidet P-40/8 M urea), overlaid with 0.5× NEpHGE sample buffer, and electrophoresed in a Hoefer DE-102 Tube Gel Electrophoresis Apparatus for 2400 volt-hours. After the first dimension run, the tube gels were extruded and layered onto a 1.5 mm × 14 cm slab gel, overlaid with 0.5% SeaKem agarose (FMC) containing 0.2× Lammeli sample buffer, and analyzed by SDS/PAGE (12.5% gel).
In Vitro Transcription and Translation.
hsp40 plasmid DNA was prepared using a Wizard Mini-Prep Kit (Promega) as per the manufacturer’s protocol. Full-length capped hsp40 transcript was prepared in a mMessage mMachine Large Scale In vitro Transcription Kit (Ambion, Austin, TX) using the T3 promoter of pBluescript. A linear transcript was generated by cleavage at a HindIII site immediately downstream of the hsp40 coding sequence. Transcript length and purity were examined by use of PAGE on a 5% acrylamide 7.5 M urea denaturing gel. Message-dependent rabbit reticulocyte lysate was prepared as described (28). The in vitro transcribed product (140 ng) was used to program the recitulocyte lysate in the presence of [35S]methionine as described (28). The amount of translated product was determined by scintillation counting of trichloroacetic acid-precipitable protein. The translated product was also visualized by SDS/PAGE and autoradiography.
GST–P58IPK Binding Assays.
P58IPK–hsp40 binding assays were performed using GST-tagged P58IPK as a substrate for binding in vitro translated hsp40. Binding experiments were performed as described in ref. 26.
RESULTS
Purification of the Cellular Inhibitor of P58IPK.
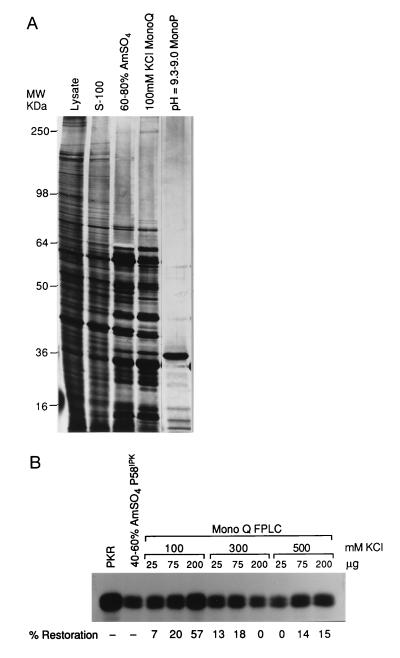
As mentioned earlier, we accumulated preliminary data that suggested a protein inhibitor of P58IPK existed in uninfected cells (19). We attempted to purify this regulator using standard biochemical purification and two independent in vitro functional assays. The procedure for the purification is described in Materials and Methods. Briefly, cytoplasmic extracts, prepared from approximately 2 × 1010 cells, were subjected to centrifugation at 100,000 × g. The S-100 supernatant was further fractionated by ammonium sulfate precipitation. The 60–80% ammonium sulfate fraction was found to inhibit P58IPK (data not shown; Table 1) and was further purified using a MonoQ FPLC anion-exchange column. The MonoQ fractions were then tested for their ability to inhibit P58IPK. Fractions were mixed with partially purified P58IPK (present in the 40–60% ammonium sulfate fraction). After a brief incubation, these components were mixed with extracts prepared from IFN-treated 293 cells (as a PKR source). PKR was immunoprecipitated from the mix and assayed for activity. An inhibitor of P58IPK would be expected to reverse the P58IPK-mediated inhibition of PKR, resulting in a stimulation of PKR activity and enhanced histone phosphorylation. The 100 mM KCl MonoQ flow-through fraction contained the highest I-P58IPK activity, restoring nearly 60% of PKR function (Fig. 1B).
Table 1.
Summary of the purification of the inhibitor of P58
| Purification step | Volume, ml | Protein,* mg | Total activity,† units | Specific activity,‡ units/mg | Purification, -fold | Yield, % |
|---|---|---|---|---|---|---|
| Cytoplasmic extract | 265.00 | 1439.00 | — | — | — | — |
| 100,000 × g | 255.00 | 1201.00 | — | — | — | — |
| Ammonium sulfate | 16.50 | 110.00 | 4939 | 44.9 | 1.0 | 100 |
| MonoQ FPLC | 28.00 | 55.00 | 3212 | 58.4 | 1.3 | 65 |
| MonoP FPLC | 9.46 | 0.036 | 445 | 12364 | 275 | 9 |
Protein was measured by the Micro BCA protein assay (Pierce) except for the MonoP fractions in which protein was estimated from silver-stained gels.
One unit of activity is defined as the amount of protein required to cause 1% restoration of PKR activity in the presence of P58IPK.
Specific activity for the MonoP fraction was corrected for the activity of GST–P58IPK relative to the 40–60% ammonium sulfate P58IPK used in previous fractions.
Figure 1.
Purification of the I-P58IPK inhibitor from uninfected MDBK cells. (A) Polypeptide analysis of the fractions containing I-P58IPK activity. From left to right, the crude cytoplasmic cell lysate, 100,000 × g supernatant (S-100), 60–80% ammonium sulfate, 100 mM KCl MonoQ FPLC flow through, and pH = 9.3–9.0 MonoP FPLC eluate fractions were analyzed by SDS/PAGE (12% gel) with the proteins visualized by silver staining. Positions of molecular weight markers are shown on the left. (B) In vitro histone phosphorylation assay examining I-P58IPK activity in the MonoQ FPLC fractions. To demonstrate P58IPK activity, IFN-treated extracts were treated with buffer alone (PKR) or with the 40–60% ammonium sulfate fraction (source of P58IPK), followed by the PKR immunoprecipitation and kinase assay as described in Materials and Methods. To test for I-P58IPK activity, the 40–60% ammonium sulfate fraction was prereacted with the indicated amounts of the flow through (100 mM) MonoQ or high salt batch eluted fractions (300 mM or 500 mM). IFN-treated extracts were then added, and PKR was subsequently immunoprecipitated and assayed. The 100% PKR activity was defined by incubation with buffer alone (PKR), and 0% activity was defined as PKR activity remaining after incubation with P58IPK alone (40–60% ammonium sulfate). The percentage of restoration of PKR activity (due to inhibition of P58IPK activity) was quantified by PhosphorImager analysis of histone phosphorylation, as shown at the bottom of B.
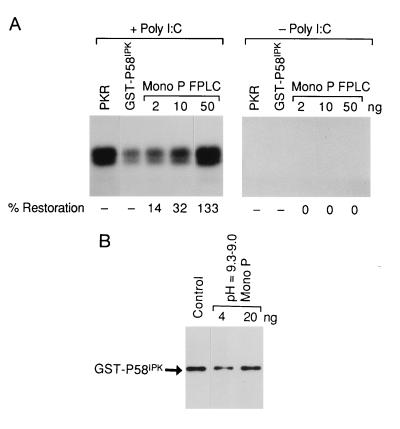
The active fraction was then applied to a MonoP FPLC chromatofocusing column. The eluted fractions were then tested for I-P58IPK inhibitory activity using the alternative in vitro assay, composed of purified proteins. The second assay was developed as the crude component assay was unsuitable for screening the highly purified MonoP fractions. Conversely, the assay using purified components could not be used with the ammonium sulfate or MonoQ fractions because of contaminating kinases and phosphoproteins. Again we were testing for a reversal of P58IPK-mediated inhibition of PKR and thus a restoration of PKR activity. The dose-dependent activity was found to reside exclusively in the pH = 9.3–9.0 fraction (Fig. 2A and data not shown). Only 50 ng of the MonoP fraction was required to completely inhibit P58IPK and restore essentially all the PKR activity. Restoration of PKR activity by the MonoP fraction required preincubation with recombinant P58IPK, suggesting that the protein was not acting on PKR itself (data not shown). Control experiments, performed in the absence of dsRNA, showed that this MonoP fraction did not activate PKR (Fig. 2A). Furthermore, Western blot analysis, using P58IPK monoclonal antibodies, showed that the reduction of P58IPK activity was not due to proteolysis (Fig. 2B). The active fractions throughout the purification were analyzed both for their specific activity (Table 1) and for their polypeptide content (Fig. 1A). The specific activity of the inhibitor of P58IPK increased from approximately 45 units per mg of protein in the 60–80% ammonium sulfate fraction to 12,364 units per mg in the MonoP fraction, reflecting a 275-fold increase during the purification procedure. Significantly, one major protein (Mr = 38,000) present in the highly active MonoP fraction was detected by silver staining.
Figure 2.
Analysis of I-P58IPK activity in the MonoP purified fraction. (A) I-P58IPK activity using an in vitro assay and purified components. (Left) Restoration of PKR activity in an in vitro assay using dsRNA, purified native PKR (0.1 μg), GST–P58IPK (0.075 μg), and the pH = 9.3–9.0 MonoP fraction (protein amounts are indicated on top of the panel). The MonoP fraction was preincubated with P58IPK before PKR addition, as described in detail in Materials and Methods. PKR activity, as measured by histone phosphorylation, was quantified by PhosphorImager analysis. The 100% activity is defined as the activity of PKR alone (PKR), and 0% activity was defined as PKR activity after mixing with GST–P58IPK (GST–P58IPK). The percentage of PKR restoration is shown at the bottom. (Right) The same experiment minus dsRNA. (B) Decreased P58IPK activity is not due to P58IPK proteolysis. GST–P58IPK was incubated with buffer alone (Control) or with the indicated amounts of the pH = 9.3–9.0 MonoP fraction under reaction conditions identical to those in the in vitro functional assay described A. The reaction was analyzed by SDS/PAGE and Western blot analysis using P58IPK-specific monoclonal antibody.
Identification of I-P58IPK.
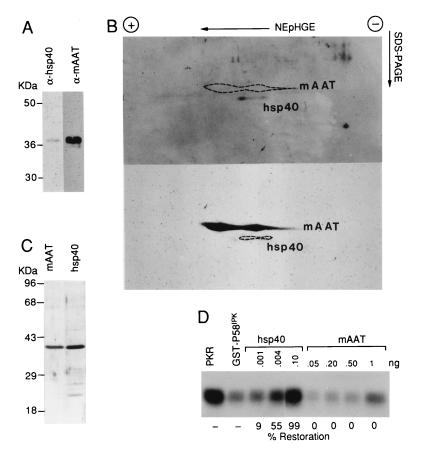
To determine its identity, the MonoP purified protein was digested with the protease LysC, and the peptide fragments were subjected to microsequence analysis. Three of three peptide fragments were 100% identical to the bovine mAAT (data not shown). In accordance with our MonoP analysis, mAAT is known to have an isoelectric point of ≈9.0 and Mr of ≈40,000 (29). However, when we tested purified mAAT in our in vitro assay, we were unable to inhibit P58IPK and restore PKR activity (Fig. 3D). Given these results, and the well documented enzymatic functions and subcellular localization of mAAT (30), we explored the possibility that we had copurified mAAT with the bona fide inhibitor of P58IPK. Several observations led us to consider a member of the heat shock family of proteins, specifically the eukaryotic DnaJ homolog, hsp40. First, the homology of P58IPK to the DnaJ protein family suggests that P58IPK, and thus any coregulator, may be involved in the stress response. Second, heat shock proteins interact primarily through direct binding, which is consistent with our evidence of how P58IPK acts and how it is presumably regulated (31). Finally and most importantly, the hsp40 protein has a Mr of 38,000 and pI of ≈9.0, essentially identical to the protein identified in our MonoP purified fraction (32).
Figure 3.
The MonoP purified preparation contains both hsp40 and mAAT. (A) Western blot analysis of the active MonoP fraction. The MonoP fraction (0.5 μg) was analyzed by one-dimensional SDS/PAGE, followed by Western blot analysis. The blot was first probed with hsp40 -pecific (α-hsp40) antibody and then stripped and reprobed with antisera specific for mAAT (α-mAAT). (B) Analysis of pH = 9.3–9.0 MonoP fraction by NEpHGE and Western blotting. The MonoP fraction (0.5 μg) was electrophoresed in two dimensions by NEpHGE as described in Materials and Methods. The second-dimension gel was transferred to a nitrocellulose membrane and probed with antisera to hsp40 for Western blot analysis (Upper). The same blot was then stripped and reprobed with antisera to mAAT (Lower). To demonstrate the differential migration, the position of the reciprocal protein is indicated by an area marked by dotted lines. (C) Characterization of purified hsp40 and mAAT. Purified recombinant hsp40 or mAAT was electrophoresed on a 12% polyacrylamide gel and subjected to silver staining. Positions of molecular weight markers are indicated at the left. (D) In vitro assay measuring the ability of purified hsp40 or mAAT to inhibit P58IPK function. The in vitro assay was performed as described in Fig. 2A. The amounts of purified hsp40 and mAAT, which were both preincubated with GST–P58IPK, are indicated at the top of the figure and the percentage of PKR restoration is indicated at the bottom of the figure, with 100% and 0% PKR activity defined as in Fig. 2A.
Initially, we reacted hsp40 antibody with the MonoP purified fraction in a Western blot analysis. We found that the 38-kDa protein band reacted with the hsp40 antisera. As expected, antisera to mAAT also reacted with the polypeptide, and quantitative Western blot analysis showed mAAT present in a 50-fold higher amount (Fig. 3A and data not shown). This is consistent with the observation that peptide sequence information was obtained exclusively from mAAT, as the copurified protein would represent a significantly smaller proportion of the purified fraction. Because the proteins were indistinguishable by one-dimensional electrophoresis, we attempted to identify the two proteins by two-dimensional NEpHGE (NEpHGE), an established technique for hsp40 analysis (Fig. 3B) (33). Using Western blot analysis, we were able to distinguish the more abundant and basic mAAT from hsp40, unequivocally demonstrating the presence of the two proteins in our purified preparation. The two-dimensional NEpHGE positions of the proteins were confirmed using purified hsp40 and mAAT (data not shown).
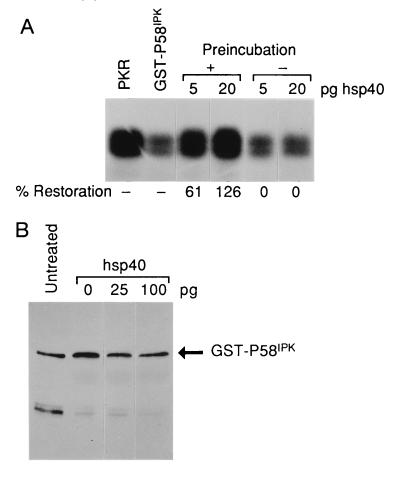
Definitive proof that hsp40, not mAAT, was I-P58IPK required comparison of their functional activity in our in vitro assay. Therefore, we obtained highly purified recombinant hsp40 and mAAT. The purity of the two protein preparations was confirmed by silver staining of the SDS/polyacrylamide gel (Fig. 3C). Using our in vitro assay, which included native PKR and recombinant GST–P58IPK, we found that highly purified, recombinant hsp40 reversed P58IPK inhibition of PKR in a dose-dependent manner (Fig. 3D). Importantly, purified mAAT (tested at up to 10-fold higher amounts than hsp40) failed to inhibit P58IPK activity. The lack of mAAT function also suggests that the I-P58IPK specific activity in Table 1 is severely underestimated due to the relatively low concentrations of hsp40 in our purified preparation. However, this apparent low yield is not surprising considering published reports that suggest hsp40 is a relatively non-abundant protein (34, 35). Similar to our native purified preparation, recombinant hsp40 failed to activate PKR in the absence of dsRNA (data not shown) and was unable to block P58IPK activity unless preincubated with P58IPK before addition of PKR (Fig. 4A). Finally, it was critical to determine whether decreased P58IPK function was due to hsp40-induced P58IPK proteolysis. hsp40 did not induce the degradation of recombinant P58IPK as determined by Western blot analysis (Fig. 4B). The stoichiometry of the hsp40-mediated inhibition of P58IPK deserves comment. While virtually equimolar amounts of P58IPK are required for PKR inhibition, we found that picogram quantities of hsp40 were sufficient to inhibit nanogram amounts of P58IPK. We feel this may indicate that hsp40 is able to disrupt secondary or tertiary structures within P58IPK, consistent with the role of hsp40 as a molecular chaperone. Furthermore, the recent observation that P58IPK is able to homooligermize would suggest that even disruptions of quaternary structure could have a drastic effect on P58IPK activity (36).
Figure 4.
Characterization of I-P58IPK hsp40 activity. (A) Preincubation of hsp40 with P58IPK is required for I-P58IPK activity. The in vitro assay was performed with purified components as described in the legend to Fig. 2A. In the experiments performed without preincubation, GST–P58IPK and PKR were mixed before the addition of hsp40 to the in vitro reaction. (B) Hsp40 does not induce the degradation of GST–P58IPK. GST–P58IPK was either untreated or incubated under the precise in vitro assay conditions with the indicated amounts of hsp40. The reaction was analyzed by SDS/PAGE and Western blot analysis using P58IPK specific monoclonal antibody, as described in Materials and Methods. The faster-migrating protein is a breakdown product of the GST–P58IPK fusion protein and does not appear to contribute to the reaction.
Although hsp40 antibody recognized our purified MonoP fraction and recombinant highly purified hsp40 inhibited P58IPK activity in vitro, we cannot rule out that I-P58IPK is not hsp40 itself, but a highly related protein, which is therefore reactive with the antibody and possesses inhibitory activity due to conserved hsp40 functional domains. Unequivocal demonstration that I-P58IPK is hsp40 requires microsequencing the homogeneously purified protein, a goal currently not attainable due to the mAAT contamination and the relatively low hsp40 yields.
P58IPK Interacts with hsp40 in Vitro.
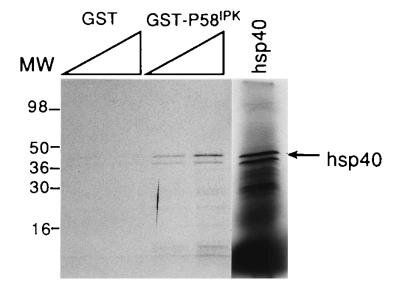
Since we originally hypothesized that the inhibition of P58IPK occurred by a direct interaction, we carried out an in vitro analysis of P58IPK–hsp40 binding using a GST pull-down experiment. hsp40 was translated in vitro, in the presence of [35S]methionine and reacted with crude extracts from E. coli expressing GST (as a control) or GST–P58IPK. Analysis of the bound proteins, which remained after glutathione-agarose selection and washing steps, revealed that the full-length hsp40 translation product formed a stable, titratable complex with GST–P58IPK, but not significantly with GST alone (Fig. 5). It should be noted that this assay was also used to map the PKR–P58IPK interactive domains, and that these experiments show that GST–P58IPK displays specificity in binding (26). These data indicate that the binding of hsp40 to P58IPK is specific and are consistent with the idea that complex formation is required for the hsp40 inhibition of P58IPK. It remains to be determined whether a stable interaction between hsp40 and P58IPK can be detected inside the cell.
Figure 5.
hsp40 complexes with P58IPK in vitro. Increasing amounts of E. coli extracts containing either GST or GST–P58IPK were incubated with equal counts of [35S]methionine labeled, in vitro translated hsp40 product. After addition of glutathione-agarose beads and appropriate washings, the bound material was subjected to SDS/PAGE on 14% acrylamide gels. The in vitro translated starting material is shown in the right lane (hsp40), and the full-length product is indicated by the arrow. It should be noted that the two protein bands visible in the input lane react specifically with hsp40 antisera, indicating the smaller product is a result of either internal initiation or partial degradation (data not shown). Molecular weight markers are indicated on left.
DISCUSSION
Translational regulation in influenza virus-infected cells may be analogous to translational control during conditions of heat shock or stress. In influenza virus-infected cells, there is a dramatic shutoff of cellular protein synthesis leading to the selective translation of viral mRNAs (1). In heat shocked or stressed cells, there similarly is often a complete cessation of “normal” cellular protein synthesis and a subsequent redirection of the translation of exclusively heat shock mRNAs (37). One might speculate that, in both virus-infected and stressed cells, protein synthesis is initially inhibited through the transient activation of PKR and phosphorylation of eIF-2α. This report suggests that in the case of influenza virus infection, P58IPK, normally bound to hsp40, dissociates from hsp40 and interacts with PKR to block eIF-2α phosphorylation and restimulate protein synthesis. Whether a specific influenza viral gene product causes the dissociation of hsp40 and P58IPK remains to be determined. The restart of mRNA translation in virus-infected cells during “recovery” is restricted to viral mRNAs, likely due to the cis-acting sequences present in the 5′ untranslated region which recruit cellular and/or viral trans-acting factors (38, 39). We currently do not know whether the PKR/P58IPK/hsp40 pathway plays a role in the regulation of the translation of heat shock mRNAs, which also have unique elements within the 5′ untranslated region hypothesized to play a role in their selective translation (39). It is clear, however, that heat shock induces an transient increase in eIF-2α phosphorylation, at least in certain cell lines (40). The regulatory pathways involving PKR and P58IPK are likely more intertwined with stress control given the recent discovery of a novel hsp90-related protein which was identified as an additional P58IPK interactive and inhibitory protein by a yeast two-hybrid library screen (M. Gale and M. Katze, unpublished work).
In addition to PKR, there are two other protein kinases which phosphorylate eIF-2α on serine 51, HRI (heme regulated inhibitor of translation), present predominantly in reticulocytes, and GCN2 of Saccharomyces cerevisiae. Like PKR, there is evidence that both of these kinases are regulated by stress (41). The yeast GCN2 kinase is stimulated by deprivation of amino acids or purines. In addition to inhibiting general protein synthesis, GCN2 plays a role in selective translation and specifically stimulates translation of GCN4, a transcriptional activator of amino acid biosynthetic genes. HRI is an eIF-2α kinase that is activated in rabbit reticulocytes by heme deprivation and stress conditions that elicit a heat shock response. Furthermore, both hsp90 and the EC1 antigen (possibly in conjunction with hsp70) bind to HRI, although the molecular details regarding these interactions and their roles in HRI regulation remain unclear (42).
This report is not the first example of viruses regulating or using components of the stress pathway to aid in their replication. Perhaps the most elegant example has been documented in the bacteriophage system. Infection leads to an increased synthesis of stress proteins, such as GroEL and DnaK, in spite of a general decrease in total protein synthesis (43). These stress proteins interact with viral components and aid in the correct assembly and replication of a variety of bacteriophages such as lambda, T4, and T5 (43). In regard to eukaryotic viruses, there is substantial evidence that many viruses induce a stress response in host cells as detected by increased levels of particular stress proteins or their cognate mRNAs. In most cases, the induced stress proteins have been shown to be members of the hsp60, hsp70, and hsp90 families (31). In some cases, a direct association between stress proteins and viral components has been demonstrated in virus-infected cells. Seeger and colleagues observed that hepatitis B virus requires hsp90 as an essential host factor for virus replication (44). Other work has shown that an hsp60 related protein, but not other heat shock proteins, was associated with purified HIV and SIV virions (45). It is also notable that the HIV gag protein binds to cyclophilins, the expression of which is induced by heat shock and which appears to play a role in the stress response (46). Finally, our data suggest that influenza virus recruits hsp40 as a mechanism for down-regulating the potential lethal effects of PKR on viral replication.
In closing, we found that hsp40 functions as a P58IPK inhibitor and in this way modulates the activity of PKR and mRNA translation. hsp40 is a eukaryotic homologue of the bacterial DnaJ heat shock protein. Morimoto and colleagues have directly shown that hsp40 can serve as a molecular chaperone, participating in the proper folding and assembly of selected polypeptides (47). Evidence also has been presented which shows that hsp40 interacts with the translational machinery and may play an important role in the biogenesis of proteins, inducing the proper folding of polypeptides as they emerge from ribosomes (48). Work on a yeast hsp40 homologue, ydj-1, now suggests another novel role for hsp40; hsp40 may influence the signal transduction and steroid signaling properties of hsp90 (49). Early related work by Edwards et al. also implicated heat shock as a mechanism for modulating steroid receptor activity (50). Indeed, it has been revealed that steroid receptor may require hsp40, in addition to hsp90 and hsp70, for assembly and maintenance of the aporeceptor in the absence of ligand and for proper folding of the activated receptor after ligand binding (51, 52). It is intriguing that hsp40, like PKR, has therefore been implicated in two areas of gene regulation, translational control and signal transduction. The common player in these pathways is P58IPK, which has been found to interact with both proteins. The evidence presented in the current report therefore points to potential roles for P58IPK not only in signal transduction, but also in the regulation of protein synthesis under conditions of cellular stress.
Acknowledgments
We gratefully acknowledge Marino Martinez-Carrion for the purified mAAT, Billy Burgess for microsequence analysis, Ara Hovanessian for the PKR monoclonal antibody, and Paul Berk for the mAAT antisera. We also thank Marlene Wambach for technical assistance, Marjorie Domenowske for figure preparation, and Dagmar Daniel for manuscript preparation. This investigation was supported by National Institutes of Health Grants AI 22646 and RR 00166 to M.G.K. M.W.M. was supported by Public Health Service, National Research Service Award T32 GM07270, from the National Institute of General Medical Sciences. W.J.H. and W.J.W. were supported by National Science Foundation Grant MCB9421946 and National Institutes of Health Grant R01 GM33551. B.C.F. was supported by a predoctoral National Research Service Award fellowship.
Footnotes
Abbreviations: IFN, interferon; dsRNA, double-stranded RNA; eIF-2α, α subunit of eukaryotic initiation factor 2; MDBK cells, Madin–Darby bovine kidney cells; GST, glutathione S-transferase; FPLC, fast protein liquid chromatography; mAAT, mitochondrial aspartate aminotransferase; NEpHGE, non-equilibrium pH gel electrophoresis; S-100, 100,000 × g supernatant.
References
- 1.Katze M G. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 607–630. [Google Scholar]
- 2.Meurs E, Chong K L, Galabru J, Thomas N, Kerr I, Williams B R G, Hovanessian A G. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 3.Galabru J, Hovanessian A G. J Biol Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 4.Dever T E, Chen J J, Barber G N, Cigan A M, Feng L, Donahue T F, London I M, Katze M G, Hinnebusch A G. Proc Natl Acad Sci USA. 1993;90:4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barber G N, Wambach M, Thompson S, Jagus R, Katze M G. Mol Cell Biol. 1995;15:3138–3146. doi: 10.1128/mcb.15.6.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 7.Meurs E, Galabru J, Barber G N, Katze M G, Hovanessian A G. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S B, Esteban M. Virology. 1994;199:491–496. doi: 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Petryshyn R A. Eur J Biochem. 1991;195:41–48. doi: 10.1111/j.1432-1033.1991.tb15673.x. [DOI] [PubMed] [Google Scholar]
- 10.Mundschau L J, Faller D V. J Biol Chem. 1995;270:3100–3106. doi: 10.1074/jbc.270.7.3100. [DOI] [PubMed] [Google Scholar]
- 11.Maran A, Maitra R K, Kumar A, Dong B, Xiao W, Li G, Williams B R G, Torrence P F, Silverman R H. Science. 1994;265:789–792. doi: 10.1126/science.7914032. [DOI] [PubMed] [Google Scholar]
- 12.Lee T G, Katze M G. Prog Mol Subcell Biol. 1994;14:48–65. doi: 10.1007/978-3-642-78549-8_4. [DOI] [PubMed] [Google Scholar]
- 13.Lee T G, Tang N, Thompson S, Miller J, Katze M G. Mol Cell Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korth M J, Lyons C N, Wambach M, Katze M G. Gene. 1996;170:181–188. doi: 10.1016/0378-1119(95)00883-7. [DOI] [PubMed] [Google Scholar]
- 15.Lamb J R, Tugendreich S, Hieter P. Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 16.Goebl M, Yanagida M. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 17.Cyr D M, Langer T, Douglas M G. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 18.Barber G N, Thompson S, Lee T G, Strom T, Darveau A, Katze M G. Proc Natl Acad Sci USA. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee T G, Tomita J, Hovanessian A G, Katze M G. J Biol Chem. 1992;267:14238–14243. [PubMed] [Google Scholar]
- 20.Katze M G, Tomita J, Black T, Krug R M, Safer B, Hovanessian A G. J Virol. 1988;62:3710–3717. doi: 10.1128/jvi.62.10.3710-3717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee T G, Tomita J, Hovanessian A G, Katze M G. Proc Natl Acad Sci USA. 1990;87:6208–6212. doi: 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zylicz M, Yamamoto T, McKittrick N, Sell S, Georgopoulos C. J Biol Chem. 1985;260:7591–7598. [PubMed] [Google Scholar]
- 23.Altieri F, Mattingly J R, Jr, Rodriguez-Berrocal F J, Youssef J, Iriarte A, Wu T, Martinez-Carrion M. J Biol Chem. 1989;264:4782–4786. [PubMed] [Google Scholar]
- 24.Zhou S L, Stump D, Surrentino D, Potter B J, Berk P D. J Biol Chem. 1992;267:14456–14461. [PubMed] [Google Scholar]
- 25.Towbin H, Staehlin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polyak S J, Tang N, Wambach M, Barber G N, Katze M G. J Biol Chem. 1996;271:1702–1707. doi: 10.1074/jbc.271.3.1702. [DOI] [PubMed] [Google Scholar]
- 27.O’Farrell P Z, Goodman H M, O’Farrell P H. Cell. 1977;12:1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 28.Katze M G, Wambach M, Wong M-L, Garfinkel M S, Meurs E, Chong K L, Williams B R G, Hovanessian A G, Barber G N. Mol Cell Biol. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonderegger P, Jaussi R, Christen P, Gehring H. J Biol Chem. 1982;257:3339–3345. [PubMed] [Google Scholar]
- 30.Isola L M, Zhou S L, Kiang C L, Stump D D, Bradbury M W, Berk P D. Proc Natl Acad Sci USA. 1995;92:9866–9870. doi: 10.1073/pnas.92.21.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindquist S. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 32.Ohtsuka K. Biochem Biophys Res Commun. 1993;197:235–240. doi: 10.1006/bbrc.1993.2466. [DOI] [PubMed] [Google Scholar]
- 33.Hattori H, Kaneda T, Lokeshwar B, Laszlo A, Ohtsuka K. J Cell Sci. 1993;104:629–638. doi: 10.1242/jcs.104.3.629. [DOI] [PubMed] [Google Scholar]
- 34.Sugito K, Yamane M, Hattori H, Hayashi Y, Tohnai I, Ueda M, Nobuo T, Ohtsuka K. FEBS Lett. 1995;358:161–164. doi: 10.1016/0014-5793(94)01417-y. [DOI] [PubMed] [Google Scholar]
- 35.Yamane M, Hattori H, Sugito K, Hayashi Y, Tohnai I, Ueda M, Nishazawa K, Ohtsuka K. Cell Struct Funct. 1995;20:157–166. doi: 10.1247/csf.20.157. [DOI] [PubMed] [Google Scholar]
- 36.Gale M, Jr, Tan S L, Wambach M, Katze M G. Mol Cell Biol. 1996;16:4172–4181. doi: 10.1128/mcb.16.8.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duncan R F. In: Translational Control. Hershey J, Mathews M, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 271–294. [Google Scholar]
- 38.Garfinkel M S, Katze M G. J Biol Chem. 1993;268:22223–22226. [PubMed] [Google Scholar]
- 39.Park Y W, Katze M G. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 40.Duncan R, Hershey J W B. J Biol Chem. 1984;259:11882–11889. [PubMed] [Google Scholar]
- 41.Hinnebusch A G. Semin Cell Biol. 1994;5:417–426. doi: 10.1006/scel.1994.1049. [DOI] [PubMed] [Google Scholar]
- 42.Matts R L, Hurst R. J Biol Chem. 1989;264:15542–15547. [PubMed] [Google Scholar]
- 43.Jindal S, Malkovsky M. Trends Microbiol. 1996;2:89–91. doi: 10.1016/0966-842x(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 44.Jianming H, Seeger C. Proc Natl Acad Sci USA. 1996;93:1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartz S R, Pauza C D, Ivanyi J, Jindal S, Welch W J, Malkovsky M. J Med Primatol. 1996;23:151–154. doi: 10.1111/j.1600-0684.1994.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 46.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 47.Freeman B C, Morimoto R I. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 48.Frydman J, Nimmesgern E, Ohtsuka K, Hartl F U. Nature (London) 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 49.Kimura Y, Yahara I, Lindquist S. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 50.Edwards D P, Estes P A, Fadok V A, Bona B J, Onate S, Nordeen S K, Welch W J. Biochemistry. 1992;31:2482–2491. doi: 10.1021/bi00124a007. [DOI] [PubMed] [Google Scholar]
- 51.Caplan A J, Langley E, Wilson E M, Vidal J. J Biol Chem. 1995;270:5251–5257. doi: 10.1074/jbc.270.10.5251. [DOI] [PubMed] [Google Scholar]
- 52.Pratt W B, Welsh M J. Semin Cell Biol. 1994;5:83–93. doi: 10.1006/scel.1994.1012. [DOI] [PubMed] [Google Scholar]