Abstract
The cytokine interleukin-2 (IL-2) is produced by T cells when they recognize a foreign antigen. Transcription of the IL-2 gene is tightly controlled by the combined actions of multiple transcriptional activators. However, the contribution of sequences in the IL-2 core promoter and the architecture of the IL-2 regulatory region to setting levels of IL-2 transcription are not understood. We have probed these properties of the human IL-2 promoter to understand how the regulatory and core promoter regions cooperate in response to T cell stimulation, thereby setting high levels of inducible transcription. We found that the IL-2 core promoter contains a TATA box that is critical for inducible expression. Moreover, the spacing and orientation between the IL-2 regulatory and core promoter regions is important for setting the level of transcription. The regulatory region of the IL-2 promoter is capable of mediating high levels of expression even when the helical phasing between transcription factor binding sites is perturbed. Although long considered an enhancer, our studies indicate that the regulatory region in the IL-2 promoter is better considered as a proximal regulatory element, since it lacks multiple properties associated with enhancer elements.
Keywords: Interleukin-2, enhancer, promoter, transcription
1. Introduction
Interleukin-2 (IL-2) is a cytokine that is expressed by activated T lymphocytes immediately following exposure to an antigen (Ullman et al., 1990). IL-2 expression is regulated at the levels of transcription and RNA stability (Chen et al., 1998; Crabtree and Clipstone, 1994; Goodbourn, 1994; Jain et al., 1995). Two independent signals are necessary to activate IL-2 transcription in a T cell: antigen presentation by an antigen presenting cell to a T cell receptor and a costimulatory signal often mediated by CD28 (Crabtree and Clipstone, 1994). IL-2 transcriptional activation has served as a model for understanding how input from multiple complex signaling pathways can be integrated at a single promoter to turn a dormant gene on.
The promoter of the IL-2 gene is rather compact, consisting of a core region (from approximately −40 to +40) and an upstream regulatory region (from approximately −300 to −40) (Figure 1A). The IL-2 core promoter contains a near consensus TATA box from −32 to −25, but the function of this element and other sequences in the core promoter have not been determined. In addition to the TATA box, a number of elements have been identified and characterized in core promoters in higher eukaryotes, including the initiator element (Inr), the downstream promoter element (DPE), and the motif ten element (MTE), which all bind subunits of the general transcription factor TFIID. Core promoters can also contain TFIIB recognition elements, BREu and BREd, which bind the general transcription factor TFIIB (Juven-Gershon et al., 2006). Each of these elements has a consensus sequence, which aids in their identification in core promoters; however, mutational analysis must be performed to understand the contribution of a putative core promoter element to setting the level of transcription.
Figure 1.
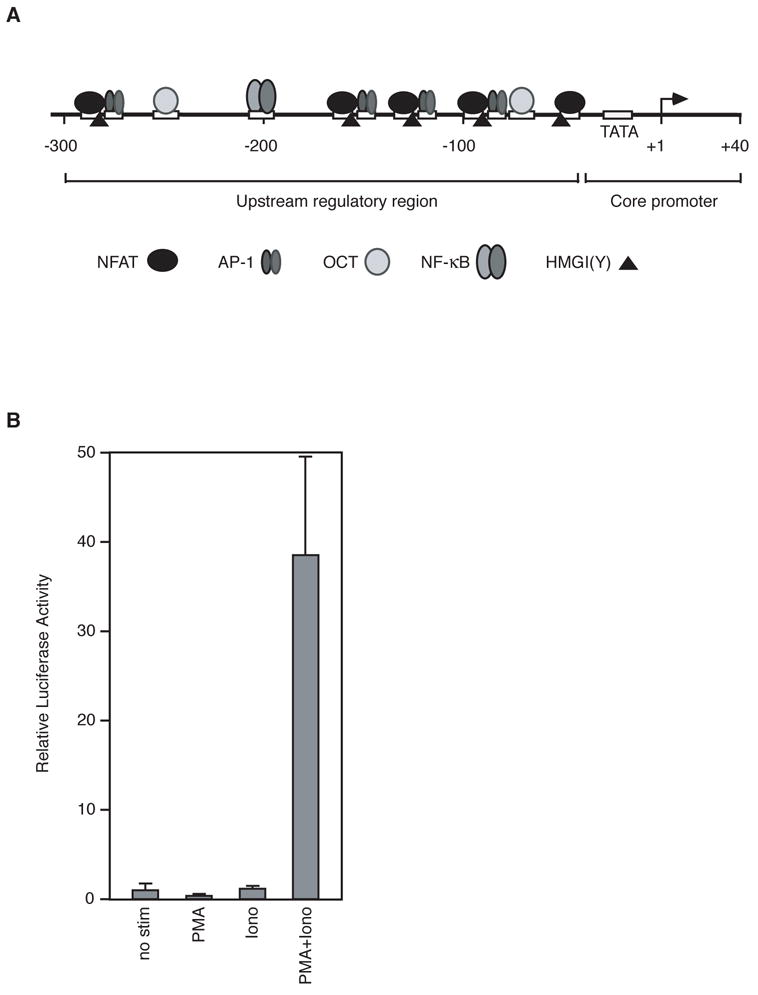
The human IL-2 promoter. (A) Schematic depicting the core region, regulatory region, and locations of transcription factor binding sites in the human IL-2 promoter. (B) Luciferase expression from an IL-2 reporter construct transfected into Jurkat cells requires co-stimulation with ionomycin and PMA. Data are plotted normalized to the level of expression observed in unstimulated cells. Each bar was obtained from a minimum of three independent transfections. Errors represent one standard deviation.
The IL-2 regulatory region is required for inducible IL-2 transcription (Durand et al., 1987; Fujita et al., 1986). Many transcriptional activators bind to the IL-2 regulatory region (Figure 1A) including members of the NFAT, AP-1, Oct-1, and NF-κB families (Rooney et al., 1995; Rothenberg and Ward, 1996). There are four composite sites in the IL-2 regulatory region to which NFAT and AP-1 bind cooperatively, and an additional site for binding NFAT is present at position −45 (Durand et al., 1988; Rooney et al., 1995). There are two Oct sites (Kamps et al., 1990; Ullman et al., 1991) and one NF-κB site in the regulatory region (Hoyos et al., 1989; Serfling et al., 1989). Maximal induction of IL-2 only occurs when the regulatory region is present in its intact form; mutation of any one of the activator binding sites significantly reduces expression from transient reporter constructs (Hentsch et al., 1992; Rooney et al., 1995).
The regulatory region of the IL-2 promoter is considered an enhancer (Durand et al., 1987; Fujita et al., 1986). Enhancers are a class regulatory regions required for high-level transcription from an associated core promoter (Khoury and Gruss, 1983; Maniatis et al., 1987). Enhancers contain sequences that bind transcriptional activators, often cooperatively (Tjian and Maniatis, 1994). In natural promoters, enhancers can be found upstream or downstream of the core promoter, and in many cases thousands of base pairs away. Using molecular techniques, enhancers have been found to activate transcription when: 1) placed near an unrelated core promoter, 2) moved far away from a core promoter, 3) placed upstream or downstream of a core promoter, and 4) their orientation is reversed.
Specific nucleoprotein complexes form at some enhancers, and are referred to as enhanceosomes. An enhanceosome is composed of the enhancer DNA and all proteins required for proper activation of transcription from the native core promoter, arranged in a precise higher-order structure (Merika and Thanos, 2001; Thanos and Maniatis, 1995). The function of such a complex was elegantly demonstrated by studies performed in the Maniatis, Thanos, and Grosschedl laboratories on the enhancers of the interferon-β (IFNβ) (Falvo et al., 1995; Thanos and Maniatis, 1992; Thanos and Maniatis, 1995; Yie et al., 1999), T cell receptor-α (Giese et al., 1992; Giese and Grosschedl, 1993; Giese et al., 1995), and tumor necrosis factor-α (Falvo et al., 2000) promoters. The precise architectural arrangement of an enhanceosome is critical for transcriptional activation. Disrupting even a single protein-DNA interaction, or changing the spacing between elements in the DNA, profoundly affects the assembly of the enhanceosome and ultimately the regulation of transcription. For instance, changing the helical phasing of the IFNβ enhancer by the addition of a half turn of DNA drastically reduced transcriptional activation (Thanos and Maniatis, 1995). Architectural proteins, such as HMGI(Y), have been shown to facilitate the assembly and function of enhanceosomes (Giese et al., 1992; Thanos and Maniatis, 1992). The presence of HMGI(Y) binding sites in enhancers is considered to be an indicator of the formation of an enhanceosome.
Several features of the IL-2 upstream regulatory region indicate that it is an enhancer and perhaps transcriptional activation of this gene is governed by an enhanceosome. Placing the IL-2 regulatory region upstream of a heterologous promoter and reversing its orientation provided evidence that it functions like a classic enhancer (Durand et al., 1987; Fujita et al., 1986). Moreover, the regulatory region of the IL-2 promoter contains multiple binding sites for the architectural protein HMGI(Y) (Himes et al., 2000).
Here we have characterized core promoter elements, the regulatory region, and architectural features of the human IL-2 promoter. Systematic mutagenesis of the core promoter showed that the TATA box is critical for function. No other sequences in the region from −44 to +8 were found to be important for high levels of inducible transcription. We tested whether the helical phasing of transcription factor binding sites in the regulatory region is important for inducible IL-2 transcription, with the goal of revealing whether the IL-2 promoter is activated by formation of an enhanceosome. Insertions of a half helical turn of DNA at three different positions did not decrease the function of the IL-2 regulatory region. Moreover, reversing the orientation of a single NFAT/AP-1 composite site in the IL-2 promoter did not decrease the function of the regulatory region. When the properties of the IL-2 regulatory region were further tested we found that it did not efficiently function when its orientation was reversed or it was placed upstream of a heterologous core promoter, and inserting 500 bp of DNA between the regulatory region and core promoter abolished function. These experiments lead us to propose that the IL-2 promoter is composed of a tightly coupled core promoter and proximal regulatory region that does not behave as a classical enhancer.
2. Materials and methods
2.1. Preparation of mutated promoters
The pBS-IL-2-luc plasmid, which has been described previously (Ferguson et al., 2001), was used as a template for making all mutant IL-2 promoters. pBS-IL-2-luc contains the region from −326 to +45 of the human IL-2 promoter (cloned between Xho I and Hind III sites) upstream of the firefly luciferase gene in the vector pBluescript-KS(+). Unless indicated otherwise, mutant plasmids were constructed using PCR methodology and verified by DNA sequencing. For each mutant promoter, PCR was used to generate two DNA fragments both of which contained the designated mutations in overlapping regions. The two DNA fragments were then used together as a PCR template to generate a third DNA fragment. This final DNA fragment was digested using Xho I and Hind III and ligated into the pBS-IL-2-luc plasmid, which was also cut with Xho I and Hind III. To generate the helical phasing mutants, 5 bp of DNA were inserted to generate a Pvu II site on the upstream side of position −191, −107, or −60. For the ARRE-2 flip and ARRE-2 knock out mutants the region between −285 and −265 was either reversed or changed to an unrelated sequence. The −41/−290 flip mutant was engineered using 70 to 90 long oligonucleotides. The mutants containing insertions between the regulatory region and core promoter were made by engineering an Nde 1 site on the upstream side of −37 (to make the 6 bp insertion) and subsequently inserting a 500 bp TATA-less DNA fragment.
2.2. Mammalian Cell Culture and Transfections
Jurkat T cells (human lymphoma T cells) were maintained in RPMI media, supplemented with 10% fetal bovine serum and antibiotics (1% Pen/Strep) in a humidified incubator containing 5% CO2. Jurkat cells were split to 2.5 × 105 cells/ml 24 hr prior to transfection, so there would be about 5 × 105 cells/ml at the time of transfection. XtremeGene Q2 transfection reagent (Roche) was used for the transient transfection of DNA constructs into Jurkat cells. In general, 80 μl of TE (pH 8.0), 100 ng of pRL-Null-Renilla luciferase reporter, and 500–1000 ng of pBS-IL-2-luc were incubated at room temperature for 10 minutes. After 10 minutes, a mixture of 80 μl of Optimem (GIBCO) and 3.2 μl XtremeGene Q2 transfection reagent (Roche) was added. The mixture was incubated for an additional 10 minutes at room temperature. The total reaction mixture was added to 5 × 105 Jurkat cells suspended in 1 ml of RPMI media. Cells incubated for 5 hours at 37°C, 5% CO2. After this time, 1 ml of RPMI containing 20% fetal bovine serum was added to the transfected cells for a final concentration of 10% fetal bovine serum. After 19 hours, cells were stimulated using 20 ng/ml of phorbol-12-myristate 13-acetate (PMA), 1 μM ionomycin (Calbiochem), and 10 mM CaCl2. After six hours of stimulation, cells were lysed in 0.5 ml 1X Passive Lysis Buffer (Promega). A dual-luciferase reporter assay (Promega) was used to measure reporter gene expression by monitoring levels of firefly luciferase and Renilla luciferase within a sample. Luciferase units were read on a luminometer. Inducible gene expression was quantitated by calculating the ratio of firefly to Renilla luciferase. Results from the luciferase assays are represented as the mean of at least three independent transfections. Errors represent one standard deviation.
3. Results
3.1. The IL-2 promoter contains a TATA box that is critical for inducible expression in T cells
The IL-2 promoter contains a regulatory region and core promoter region as diagrammed in Figure 1A. We utilized a reporter plasmid containing this promoter upstream of the firefly luciferase gene. As shown in Figure 1B, when transfected into Jurkat T cells this reporter plasmid exhibits a robust response that is dependent on the addition of both the calcium ionophore ionomycin and phorbol 12-myristate 13 acetate (PMA), two chemicals that together can be used to mimic the costimulatory signal required to activate T cells (Flanagan and Crabtree, 1992). All promoter mutations were made in the context of this reporter plasmid.
We began our characterization of the IL-2 promoter by performing mutagenesis of the core region. The sequence of the nontemplate strand of the −44 to +8 region of the human IL-2 promoter is shown in the schematic at the top of Figure 2. The three regions of this promoter that have similarity to known core promoter elements are indicated below the sequence, with consensus positions indicated in bold. The putative TATA box is a 7 out of 8 match with the consensus sequence (T-A-T-A-A/T-A-A/T-A/G) (Bucher, 1990). There is a 4 out of 7 match with the BREd consensus sequence (G/A-T-T/G/A-T/G-G/T-T/G-T/G) (Deng and Roberts, 2005), and a 4 out of 6 match with the consensus Inr sequence (T/C-T/C-A+1-N-T/A-T/C-T/C) (Javahery et al., 1994; Lo and Smale, 1996). No significant similarities to other known core promoter elements such as the BREu, DPE, and MTE are found in the IL-2 promoter.
Figure 2.
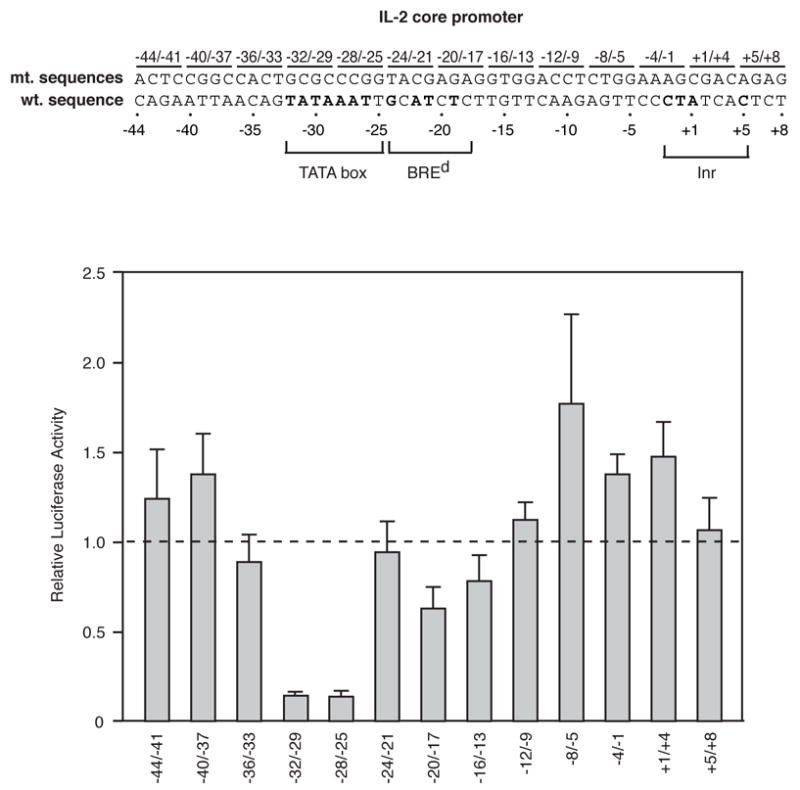
Systematic mutagenesis of the human IL-2 core promoter. The sequence of the nontemplate strand of the −44 to +8 region of the IL-2 promoter is shown. Positions that match the consensus sequences for the TATA box, BREd, and Inr are indicated in bold. Transversion mutations were made in four basepair blocks as indicated. Mutants were transfected into Jurkat cells and stimulated luciferase activities were normalized to that observed with the wild type construct (indicated by the dashed line).
To begin to determine which sequences in the IL-2 core promoter influence inducible expression in cells, the core promoter from −44 to +8 was systematically mutated in blocks of 4 basepairs, thereby creating 13 mutant promoters. Transversion mutations were generated, and the mutant sequences are shown above the wild type sequence in the schematic (Figure 2). Each mutant promoter construct was transfected into Jurkat T cells, which were simulated with ionomycin and PMA. The bar plot displays the firefly luciferase to Renilla luciferase ratios of the mutant promoters normalized to wild-type IL-2 (indicated by the dashed line) after stimulation of transfected Jurkat cells. The two mutations in the TATA box (−32/−29 and −28/−25) substantially reduced inducible IL-2 expression. Two other mutations subtly decreased inducible expression (−20/−17 and −16/−13). The −20/−17 mutant is within the putative BREd. Some mutants increased levels of expression, with the −8/−5 mutant showing the greatest increase. These results show that the TATA box is the primary core sequence critical for IL-2 expression.
To further explore the contributions of sequence in the BREd and Inr regions to inducible IL-2 expression we directly mutated these elements alone and in combination with the TATA box. The sequences of these mutations are shown at the top of Figure 3. The bar plot shows the effect of each of these mutations on expression, normalized to that from the wild type IL-2 construct, indicated by the dashed line. Mutating neither the BREd nor the Inr region reduced expression alone or when combined with the TATA mutation. Curiously, in the context of the TATA mutation, mutating the Inr region increased expression. The reason for this is unknown.
Figure 3.
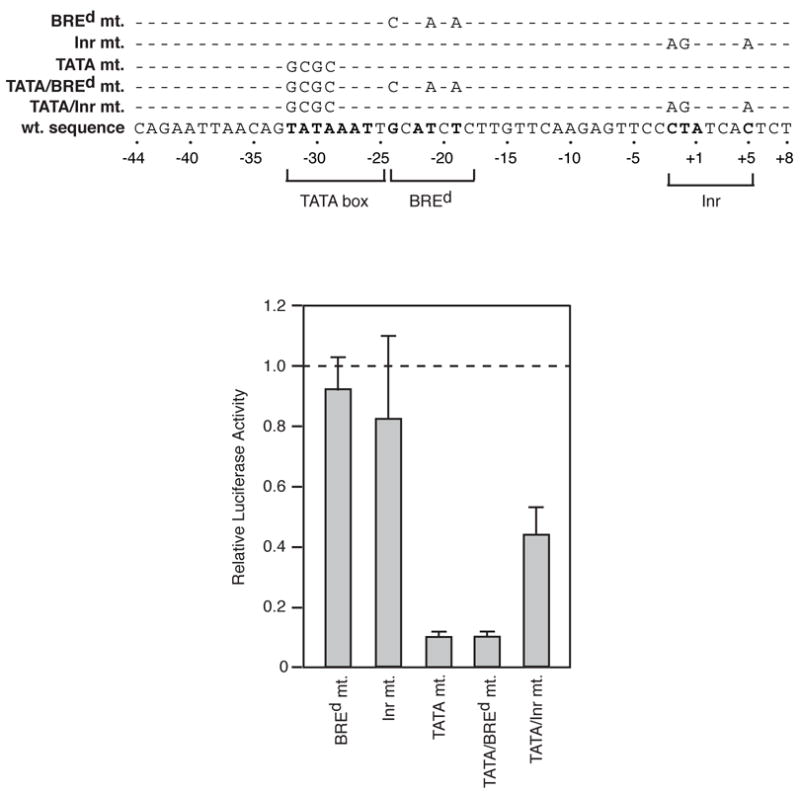
Mutation of putative IL-2 core promoter elements. The sequence of the nontemplate strand of the −44 to +8 region of the IL-2 promoter is shown. Positions that match the consensus sequences for the TATA box, BREd, and Inr are indicated in bold. Mutations generated in these elements are indicated above the sequence. Mutants were transfected into Jurkat cells and stimulated luciferase activities were normalized to that observed with the wild type construct (indicated by the dashed line).
3.2. The IL-2 regulatory region does not have characteristic properties of an enhanceosome
To test the potential that an enhanceosome forms in the IL-2 regulatory region we changed the helical phasing by inserting 5 bp of DNA at three different positions, all of which were between known regulatory protein binding sites (Figure 4A). Previous studies with the IFNβ promoter showed that changing the helical phasing decreased transcriptional activation, although all regulatory elements remained occupied (Thanos and Maniatis, 1992). The three IL- 2 mutants with 5 bp insertions at −60, −107, and −191 did not decrease inducible expression compared to the wild type promoter (Figure 4B). Insertions at −60 and −191 showed expression levels comparable to wild type, whereas insertion at −107 caused approximately a two-fold increase in expression. Hence, the overall architecture of the regulatory region is not critical to its function in transcriptional activation.
Figure 4.
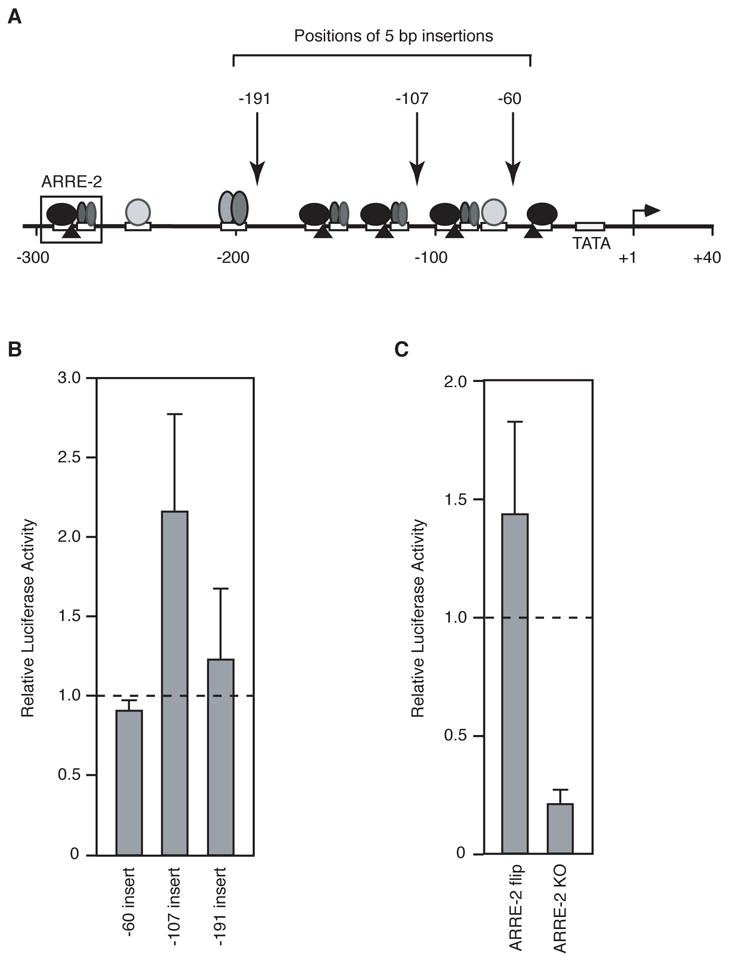
Tests of the enhanceosome properties of the IL-2 regulatory region. (A) Diagram of the IL-2 promoter and mutations made within the regulatory region. (B) Five basepair insertions do not disrupt the function of the IL-2 regulatory region. Five basepairs of DNA were inserted immediately upstream of the three positions indicated in panel A. Mutants were transfected into Jurkat cells and stimulated luciferase activities were normalized by that observed with the wild type construct (indicated by the dashed line). (C) Reversing the orientation of the ARRE-2 element does not disrupt IL-2 activation. The ARRE-2 element (indicated in panel A) was flipped (ARRE-2 flip) or replaced with unrelated sequence (ARRE-2 KO). Mutants were transfected into Jurkat cells and stimulated luciferase activities were normalized to that observed with the wild type construct (indicated by the dashed line).
As an additional test of the potential of the IL-2 regulatory region to form an enhanceosome we asked whether the orientation of a critical composite element is important for inducible transcription. NFAT and AP-1 proteins bind the well characterized ARRE-2 element cooperatively (Figure 4A). We reversed the orientation of this element such that the NFAT and AP-1 sites were switched (i.e. the NFAT site was downstream of the AP-1 site), but the sequence of the composite site was maintained. We reasoned that switching the positions of the NFAT and AP-1 sites could disrupt higher-order (long range) interactions between proteins involved in activating IL-2 transcription. In other words, if the IL-2 promoter formed an enhanceosome, then changing the orientation of the ARRE-2 element should decrease inducible expression from the reporter construct. When the orientation of ARRE-2 element was reversed (ARRE-2 flip) the activity was slightly higher than wild type (Figure 4C). For comparison, we mutated the ARRE-2 element, destroying the NFAT and AP-1 sites (ARRE-2 KO). The ARRE-2 knock-out construct exhibited significantly lower activity than the wild type construct. We conclude that the sequence of the ARRE-2 element, but not its orientation is critical for activation. Together the data in Figure 4 indicate that the IL-2 regulatory region does not form an enhanceosome with classical properties.
3.3. The orientation, spacing, and pairing between the IL-2 regulatory region and core promoter are important for setting the level of inducible expression
Since the IL-2 regulatory region did not display characteristics of an enhanceosome, we decided to determine which of the properties of an enhancer it possessed. Previously, two portions of the regulatory region (but not the complete regulatory region) had been shown to function when placed upstream of a heterologous core promoter in either orientation (Durand et al., 1987; Fujita et al., 1986). We placed the complete IL-2 regulatory region upstream of the strong adenovirus major late core promoter (AdMLP). The level of inducible expression from this chimeric promoter was approximately three-fold lower than that of the native IL-2 promoter (Figure 5). We next reversed to the orientation of the regulatory region upstream of its native core promoter (−41/−290 flip). Flipping the entire regulatory region caused a two-fold decrease in activation compared to the wild type promoter. It appears that both the native orientation and pairing with the core promoter are important for maximal levels of activation by the IL-2 regulatory region.
Figure 5.
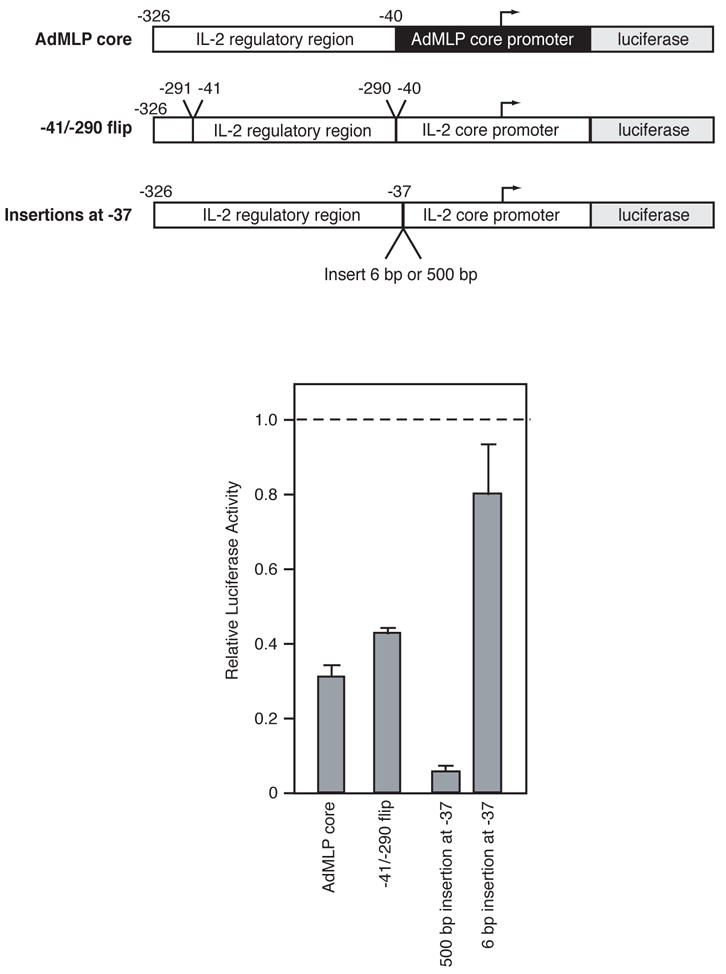
Tests of the enhancer properties of the IL-2 regulatory region. Diagrams of the four constructs used to test the enhancer properties of the IL-2 promoter are shown. Mutants were transfected into Jurkat cells and stimulated luciferase activities were normalized by that observed with the wild type construct (indicated by the dashed line).
Enhancers can also be located great distances away from core promoters, and when they are moved away from a core promoter their function is not reduced. To determine the effect of separating the IL-2 regulatory region from the core promoter we inserted 500 bp of DNA at position −37. This insertion abolished inducible expression (Figure 5). In fact, the level of expression was lower than any other mutation that we have tested. To ensure that the effect of this insertion was not simply due to disruption of an unknown regulatory element, we inserted 6 bp of DNA at −37 and found that this mutant promoter exhibited levels of inducible expression only subtly lower than that of the wild type promoter. Ironically, the 6 bp insertion between the regulatory and core regions caused expression to decrease more than any of the three 5 bp insertions within the regulatory region. Hence, the helical phasing or spacing between the regulatory and core regions of the IL-2 promoter is more important than the helical phasing between elements within the regulatory region. We conclude that the IL-2 regulatory region is not an enhancer, and suggest that instead it is a proximal regulatory region.
4. Discussion
Here we have characterized the sequence and architectural properties of the human IL-2 promoter. We found that the core promoter contains a TATA box that is critical for expression whereas the cryptic BREd and Inr elements do not contribute to inducible expression. Systematic mutation of the entire core promoter from −44 to +8 did not uncover elements other than the TATA box that contribute strongly to inducible expression. Our experiments with the regulatory region of the IL-2 promoter indicate that it is best thought of as a proximal regulatory region instead of an enhancer. The regulatory region loses almost all function when moved 500 bp from the core promoter. Moreover, it is only partially active when fused upstream of a heterologous core promoter and when its orientation is reversed. Finally, the function of the regulatory region of the IL-2 promoter does not require a highly defined architecture; neither altering the helical phasing between important promoter elements nor flipping the orientation of a critical composite element reduced the function of the promoter. Hence, the IL-2 promoter does not have some of the classical properties associated with enhanceosomes, although it remains possible that higher order structure is important for the function of the regulatory region.
The importance of the TATA box in IL-2 expression was expected. The sequence of the IL-2 TATA box is very close to the consensus sequence and the spacing from the transcriptional start site is reasonable (Bucher, 1990). We were, however, surprised to find that neither the BREd nor the Inr is important for high levels of inducible expression. Although neither of these sequences perfectly matches the consensus sequence for the element, each shares reasonable similarity with the ideal sequence. When present in other promoters, both the Inr and the BREd can function along with a TATA box to set levels of transcription (Juven-Gershon et al., 2006). This does not appear to be the case in the human IL-2 promoter.
We initially set out to determine whether the IL-2 regulatory region forms anenhanceosome. Previous studies of the IFNβ promoter showed that activation required specific regulatory proteins to bind regulatory elements that are precisely located around the DNA helix (Thanos and Maniatis, 1995). Changing the helical phasing by inserting a half turn of DNA between critical binding sites substantially reduced IFNβ transcriptional activation by disrupting the architecture of the enhanceosome, even though factor occupancy remained unchanged (Thanos and Maniatis, 1995). Thus, helical phasing and its influence on the arrangement of regulatory protein binding sites in the enhancer proved to be an important aspect of enhanceosome function. Here we probed the existence and function of an enhanceosome at the IL-2 promoter by changing the helical phasing between critical protein binding sites in the IL-2 regulatory region. If the crosstalk between regulatory proteins such as NFAT, AP-1, Oct, NF-κB, and HMGI(Y) along the regulatory region of the IL-2 promoter is dependent on helical phasing, then 5 bp insertions within the regulatory region should significantly reduce inducible expression.
We found that 5 bp insertions at three positions throughout the regulatory region did not decrease inducible expression from the IL-2 reporter. Moreover, reversing the orientation of the ARRE-2 element did not impair promoter function. These results lead us to conclude that the IL-2 regulatory region does not have the documented characteristics of an enhanceosome, in spite of the fact that this region contains multiple binding sites for transcriptional activators and the architectural protein HMGI(Y). It remains possible that the nucleoprotein complex that forms in the IL-2 regulatory region requires long range interactions between bound factors, but the exact helical phasing of these factors is not critical to their function.
Additional experiments were performed in order to determine whether the IL-2 regulatory region has properties of an enhancer. An enhancer is defined as a regulatory region that can activate a heterologous promoter, as well as its cognate promoter, and does so in a distance and orientation-independent manner. The IL-2 regulatory region has been referred to as an enhancer in publications for many years. This was based on work that showed portions of the regulatory region imparted inducible expression when fused upstream of heterologous core promoters in either orientation (Durand et al., 1987; Fujita et al., 1986). In both of these studies, large pieces of the regulatory region (either −319 to −127 or −326 to −52) were placed upstream of a heterologous TATA box, with the spacing being similar to or less than that found in the natural IL-2 promoter. In both cases, the chimeric promoters were not induced to the same level as the natural IL-2 promoter (approximately 5 fold lower) and the reverse orientation resulted in even lower levels of induced expression. Additional tests of the enhancer properties of the IL-2 regulatory region had not been performed.
Here we tested the regulatory region of the human IL-2 promoter for three general properties of an enhancer. First, we created a chimeric promoter in which the IL-2 regulatory region was fused upstream of a very strong core promoter (from the AdMLP) with a spacing identical to that of the natural IL-2 promoter. Although expression from this chimeric promoter was inducible, the level of expression was approximately one third of that observed with the natural IL-2 promoter. Second, we reversed the orientation of the IL-2 regulatory region upstream of its native core promoter. This mutant promoter was also inducible, but the level of expression was approximately half of that observed with the natural IL-2 promoter. Finally, we inserted 500 bp of DNA between the regulatory region and the core promoter, and found that this insertion decreased levels of expression 20-fold. Our results lead us to conclude that the IL-2 regulatory region lacks critical properties of an enhancer and is best referred to as a proximal regulatory region.
Acknowledgments
This research was supported by a Public Health Service grant GM55235 from the National Institutes of Health. J.R.W. and K.G. were supported in part by NIH Predoctoral Training Grant T32 GM07135.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- Chen CY, Del Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Clipstone NA. Signal transmission between the plasma membrane and nucleus of T lymphocytes. Annu Rev Biochem. 1994;63:1045–1083. doi: 10.1146/annurev.bi.63.070194.005145. [DOI] [PubMed] [Google Scholar]
- Deng W, Roberts SG. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand DB, Bush MR, Morgan JG, Weiss A, Crabtree GR. A 275-basepair fragment at the 5′ end of the interleukin 2 gene enhances expression from a heterologous promoter in response to signals from the T-cell antigen receptor. J Exp Med. 1987;165:395–407. doi: 10.1084/jem.165.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand DB, Shaw JP, Bush MR, Replogle RE, Belagaje R, Crabtree GR. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol Cell Biol. 1988;8:1715–1724. doi: 10.1128/mcb.8.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvo JV, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFNβ gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- Falvo JV, Uglialoro AM, Brinkman BM, Merika M, Parekh BS, Tsai EY, King HC, Morielli AD, Peralta EG, Maniatis T, Thanos D, Goldfeld AE. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol Cell Biol. 2000;20:2239–2247. doi: 10.1128/mcb.20.6.2239-2247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson HA, Kugel JF, Goodrich JA. Kinetic and mechanistic analysis of the RNA polymerase II transcription reaction at the human interleukin-2 promoter. J Mol Biol. 2001;314:993–1006. doi: 10.1006/jmbi.2000.5215. [DOI] [PubMed] [Google Scholar]
- Flanagan WM, Crabtree GR. In vitro transcription faithfully reflecting T-cell activation requirements. J Biol Chem. 1992;267:399–406. [PubMed] [Google Scholar]
- Fujita T, Shibuya H, Ohashi T, Yamanishi K, Taniguchi T. Regulation of human interleukin-2 gene: functional DNA sequences in the 5′ flanking region for the gene expression in activated T lymphocytes. Cell. 1986;46:401–405. doi: 10.1016/0092-8674(86)90660-4. [DOI] [PubMed] [Google Scholar]
- Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Giese K, Grosschedl R. LEF-1 contains an activation domain that stimulates transcription only in a specific context of factor-binding sites. EMBO J. 1993;12:4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- Goodbourn S. T-cell activation. Transcriptional regulation in activated T cells. Curr Biol. 1994;4:930–932. doi: 10.1016/s0960-9822(00)00209-8. [DOI] [PubMed] [Google Scholar]
- Hentsch B, Mouzaki A, Pfeuffer I, Rungger D, Serfling E. The weak, fine-tuned binding of ubiquitous transcription factors to the Il-2 enhancer contributes to its T cell-restricted activity. Nucl Acids Res. 1992;20:2657–2665. doi: 10.1093/nar/20.11.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes SR, Reeves R, Attema J, Nissen M, Li Y, Shannon MF. The role of high-mobility group I(Y) proteins in expression of IL-2 and T cell proliferation. J Immunol. 2000;164:3157–3168. doi: 10.4049/jimmunol.164.6.3157. [DOI] [PubMed] [Google Scholar]
- Hoyos B, Ballard DW, Bohnlein E, Siekevitz M, Greene WC. Kappa B-specific DNA binding proteins: role in the regulation of human interleukin-2 gene expression. Science. 1989;244:457–460. doi: 10.1126/science.2497518. [DOI] [PubMed] [Google Scholar]
- Jain J, Loh C, Rao A. Transcriptional regulation of the IL-2 gene. Curr Opin Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Biochem Soc Trans. 2006;34:1047–1050. doi: 10.1042/BST0341047. [DOI] [PubMed] [Google Scholar]
- Kamps MP, Corcoran L, LeBowitz JH, Baltimore D. The promoter of the human interleukin-2 gene contains two octamer-binding sites and is partially activated by the expression of Oct-2. Mol Cell Biol. 1990;10:5464–5472. doi: 10.1128/mcb.10.10.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G, Gruss P. Enhancer elements. Cell. 1983;33:313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Lo K, Smale ST. Generality of a functional initiator consensus sequence. Gene. 1996;182:13–22. doi: 10.1016/s0378-1119(96)00438-6. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Science. 1987;236:1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- Rooney JW, Sun YL, Glimcher LH, Hoey T. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol Cell Biol. 1995;15:6299–6310. doi: 10.1128/mcb.15.11.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Ward SB. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin 2 gene regulation. Proc Natl Acad Sci USA. 1996;93:9358–9365. doi: 10.1073/pnas.93.18.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfling E, Barthelmas R, Pfeuffer I, Schenk B, Zarius S, Swoboda R, Mercurio F, Karin M. Ubiquitous and lymphocyte-specific factors are involved in the induction of the mouse interleukin 2 gene in T lymphocytes. EMBO J. 1989;8:465–473. doi: 10.1002/j.1460-2075.1989.tb03399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Ullman KS, Flanagan WM, Edwards CA, Crabtree GR. Activation of early gene expression in T lymphocytes by Oct-1 and an inducible protein, OAP40. Science. 1991;254:558–562. doi: 10.1126/science.1683003. [DOI] [PubMed] [Google Scholar]
- Ullman KS, Northrop JP, Verweij CL, Crabtree GR. Transmission of signals from the T lymphocyte antigen receptor to the genes responsible for cell proliferation and immune function: the missing link. Ann Rev Immunol. 1990;8:421–452. doi: 10.1146/annurev.iy.08.040190.002225. [DOI] [PubMed] [Google Scholar]
- Yie J, Senger K, Thanos D. Mechanism by which the IFN-beta enhanceosome activates transcription. Proc Natl Acad Sci USA. 1999;96:13108–13113. doi: 10.1073/pnas.96.23.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]


