Abstract
Arsenic, a contaminant of water supplies worldwide, is one of the most toxic inorganic ions. Despite arsenic's health impact, there is relatively little structural detail known about its interactions with proteins. Bacteria such as Escherichia coli have evolved arsenic resistance using the Ars operon that is regulated by ArsR, a repressor protein that dissociates from DNA when As(III) binds. This protein undergoes a critical conformational change upon binding As(III) with three cysteine residues. Unfortunately, structures of ArsR with or without As(III) have not been reported. Alternatively, de novo designed peptides can bind As(III) in an endo configuration within a thiolate-rich environment consistent with that proposed for both ArsR and ArsD. We report the structure of the As(III) complex of Coil Ser L9C to a 1.8-Å resolution, providing x-ray characterization of As(III) in a Tris thiolate protein environment and allowing a structural basis by which to understand arsenated ArsR.
Keywords: arsenic-binding proteins, coiled coil peptides, crystallography, heavy metal toxicity, protein design
Arsenic toxicity is a worldwide problem as a natural contaminant of water supplies. Despite the fact that it is a human toxin and carcinogen, few structural reports on its interaction with biological ligands have appeared. Escherichia coli and other bacteria have evolved a detoxification mechanism that employs the arsRDABC operon (1). Arsenic removal by these encoded proteins is initiated when As(III) binds to ArsR, resulting in the dissociation of the repressor protein from the promoter DNA. The structure of the ArsA component of the ATP-dependent extrusion pump ArsAB with antimonite bound in close proximity to the nucleotide-binding site was able to provide some insight into the active transport of As(III) out of the cell (2). Additionally, structural characterization of substrate and product complexes of the arsenate reductase ArsC, which reduces arsenate (AsO43−) to arsenite (AsO2−), has helped elucidate this step of the arsenic detoxification pathway (3, 4). Recent studies of E. coli ArsD indicate that it is a metallochaperone, transporting As(III) to ArsA for extrusion (5). Extended x-ray absorption fine structure and mutagenesis studies have shown that ArsR coordinates As(III) with three cysteine thiolates at a distance of 2.25 Å, a coordination mode that ArsD is also proposed to employ (1, 5). However, no x-ray or NMR structures of ArsR, the repressor protein, or ArsD have been reported to date. Furthermore, AsS3 structures have not been reported for any biologically relevant small-molecule thiolates, such as glutathione, and only a handful of related AsS3 complexes with aromatic ligands or chelated alkyl dithiolate coordination are reported (6).
We set out to model the putative As(III) coordination environments of ArsR and ArsD in a designed peptide system. Previously, we have shown that the three-stranded coiled coil TRI peptides [Ac-G(LaKbAcLdEeEfKg)4G-NH2] designed with the heptad repeat strategy, such as TRI L16C (sequences of all peptides are in Table 1), were able to model the unusual trigonal thiolate Hg(II) coordination geometry seen in the MerR metalloregulatory protein (7). We have also examined the binding of the heavy metals Cd(II), Pb(II), and Bi(III) to the TRI peptides to better understand their modes of toxicity (8, 9). Based on this success, we examined whether the trigonal thiolate metal-binding cavities of TRI derivatives would be good models of the active site of ArsR. As(III) was determined by extended x-ray absorption fine structure to bind to the peptides TRI L12C and TRI L16C with As–S bond lengths of 2.24 and 2.25 Å, respectively, as compared with the 2.25-Å As–S distance reported for ArsR (10). Armed with this structural similarity, we felt that an x-ray structure of an arsenated form of the TRI peptides could reveal important structural insights for the binding of As(III) to ArsR.
Table 1.
Peptide sequences
| Peptide | gabcdefg abcdefg abcdefg abcdefg |
|---|---|
| CSL9C | Ac-E WEALEKK CAALESK LQALEKK LEALEHG-NH2 |
| Coil Ser | Ac-E WEALEKK LAALESK LQALEKK LEALEHG-NH2 |
| Coil VaLd | Ac-G VEALEKK VAALESK VQALESK VEALEHG-NH2 |
| TRI | Ac-G LKALEEK LKALEEK LKALEEK LKALEEK G-NH2 |
| TRI L16C | Ac-G LKALEEK LKALEEK CKALEEK LKALEEK G-NH2 |
| TRI L12C | Ac-G LKALEEK LKACEEK LKALEEK LKALEEK G-NH2 |
C and N termini are capped by Ac and NH2 groups, respectively.
The design of coiled coils and other secondary protein folds has been an active area of research with many structurally characterized examples (11, 12). The substantial research incorporating metal cofactors into designed peptide frameworks has been described in several recent reviews (13–15). Helical scaffolds have been the predominant structural motif used for the incorporation of many types of metal-binding sites, including hemes (16), iron–sulfur clusters (17), dinuclear metal centers (18), copper centers (19, 20), and heavy metal-binding trigonal thiolate sites (7, 21). β-Hairpins and other globular structures that bind As(III), Zn(II), and other metal cofactors have also been designed (22–25).
Despite the increasing number of reports on the design of metallopeptides, detailed structural characterization of many of these systems remains elusive. The class of designed metallopeptides that has been structurally characterized most thoroughly is four-helix bundles comprised of dimers of helix–turn–helix peptides. A 2.8-Å resolution structure of a heme-binding four-helix bundle maquette has been reported, but by far the most detailed structural information has been obtained for the due-ferri (DF) family of peptides designed by DeGrado et al., which contain carboxylate-bridged dinuclear metal centers in the center of a four-helix bundle (26, 27). NMR structures of both the apo and the di-Zn(II) forms of DF1 reveal slight changes in the protein fold upon metal binding (28). Additionally, a 1.9-Å structure of a di-Mn(II)-DF2 derivative contains four crystallographically independent bundles that unveil subtle differences in ligand coordination to the dinuclear centers (29). The analysis of these structures and those of other due-ferri-family derivatives provides insight into how changes in the hydrophobic core affect ligand coordination to these dinuclear sites and has allowed for the design of catalytic functionality into this type of simple designed framework (27, 31).
One of the first designed peptides to be structurally characterized was Coil Ser, which was crystallized as an antiparallel three-stranded coiled coil (32). It was suggested that the antiparallel orientation was obtained because of the steric bulk of the N-terminal tryptophans. We have been able to show that the replacement of the interior residue Leu 9 for a cysteine in Coil Ser, denoted CSL9C, creates a trigonal thiolate metal-binding site within a parallel coiled coil similar to that of TRI L9C (33). CSL9C was shown spectroscopically to have similar behavior to TRI L9C with regard to affinity, pH-dependence, and geometry of metal binding. These results corroborated previous solution studies of Coil Ser derivatives suggesting that parallel aggregates were formed at higher pH values (34). In this study, we are using CSL9C for structural studies of the arsenic-bound peptide because it has proven more facile to form diffraction-quality crystals in comparison to the TRI peptides.
The structure presented here of As(CSL9C)3 provides an examination for As(III) in a homoleptic tristhiolate environment in a protein. It is precisely this structure that has been proposed for As(III) binding to ArsR and ArsD. As shown below, there are subtle, but potentially significant, conformational restrictions placed on the As(III) thiolate environment. An analysis of this structure not only enhances our understanding of As detoxification by microbes but, in addition, provides surprising insights for the next generation of designed metallopeptides.
Results
Overall Structure.
The crystal structure of the arsenated three-stranded coiled coil Coil Ser L9C As(CSL9C)3 has been determined to a 1.8-Å resolution. Although the sequence of CSL9C differs by only one amino acid from the parent peptide, Coil Ser, the prototypical de novo designed homomeric antiparallel coiled coil, the structure of As(CSL9C)3 is a well folded, parallel three-stranded coiled coil similar to the related peptide, Coil VaLd, which has alternating interior hydrophobic layers of valine and leucine residues (35). The final model of As(CSL9C)3 contains 762 protein atoms, 50 solvent molecules, 1 As(III) ion, and 4 Zn(II) ions. The rmsd between the search model, which is comprised of the first 26 amino acids of Coil VaLd, and the final refined solution was 1.2 Å. All nonglycine residues fall in the most-favored α-helical region of a Ramachandran plot. The N-terminal capping acetyl groups have their carbonyl oxygens hydrogen-bonded to alanine 4 of their respective chains. Side chain residues, with the exception of Glu 24 A, Glu 3 B, and Glu 20 B, coordinated to Zn(II) and Glu 6 B, which are involved in hydrogen bonding to a solvent molecule, are in preferred conformations as analyzed by the rotamer check utility in Coot.
The Arsenic-Binding Site.
The center of the coiled coil structure contains an As(III) ion coordinated to the three cysteine residues in a trigonal pyramidal geometry, as seen in Fig. 1. Refinement of structure with the As(III)–S distance to 2.25 Å restrained to 2.25 ± 0.05 Å resulted in a mean As–S bond distance of 2.28 Å and S–As–S angles of 91°, 92°, and 88°, which are similar to small molecule As–S3 complexes in the literature (6, 36, 37). The orientation of the leucine residues in the layer above (Leu 5) and below (Leu 12) the metal-binding site differ, as seen in Fig. 2, with those in the Leu 5 layer, directed more toward the center of the coiled coil than those in the Leu 12 layer.
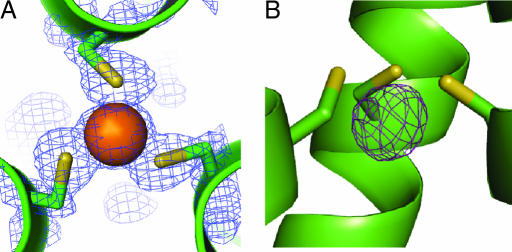
Fig. 1.
A top-down view from the N terminus of the coiled coil and a side view of the trigonal thiolate As(III)-binding site illustrate its pyramidal coordination in the interior of the three-stranded coiled coil. (A) Top-down view of the As(III) ion coordinated to the three cysteine residues in position 9 of the heptad repeat with 2Fo − Fc electron density contoured at 1.5 σ overlaid. (B) Side view of the metal-binding site with an Fo− Fc omit map contoured at 5 σ demonstrating the endo coordination of the As(III) ion 1.3 Å below the plane of the Cys Sγ-atoms and on an equal level with the β-methylene protons.
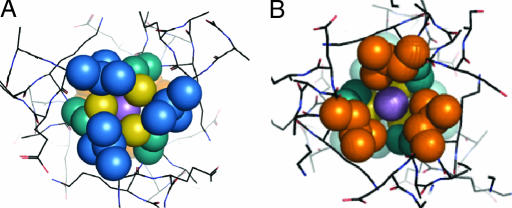
Fig. 2.
The packing of the leucine layers above and below the As(III) ion, shown as a purple sphere, is illustrated. (A) Top-down view of the leucine 5 layer (blue). (B) Leu 12 layer (orange) looking up the helical axis from the C terminus; this shows the orientation of the d residues in this layer toward the helical interface. This illustrates that if a metal that prefers tetrahedral coordination such as Cd(II) or Zn(II) were coordinated to the cysteines in the same manner as As(III), the fourth exogenous ligand (e.g., water) would coordinate on the C-terminal side of the metal ion.
The four Zn(II) ions per asymmetric unit present in the structure lie at crystal-packing interfaces linking the trimers together, as seen in Fig. 3. One dinuclear site that is coordinated by side chains from three separate coiled coils contains a μ-(η1,η2)-bridging carboxylate from Glu 24 C that links two Zn(II) ions separated by 4.97 Å. Glu 27 C, His 28 C, Glu 3 C, Glu 24 A, His 28 A, and a water molecule complete the coordination sphere, which is shown in detail in supporting information (SI) Fig. 6A. A second set of Zn(II) ions 4.73 Å apart connects two trimers and is coordinated by Glu 1 B, Glu 24 B, Glu 27 B, His 28 B, Glu 6 A, and two water molecules, as can be seen in SI Fig. 6B.
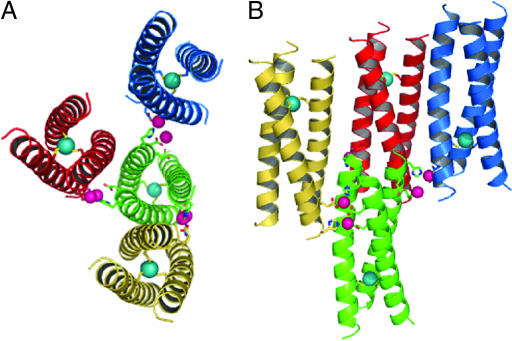
Fig. 3.
The zinc-mediated packing of the coiled coils with Zn(II) and As(III) ions is shown in pink and cyan, respectively. (A) Bottom-up view of a central coiled coil (green) with the six Zn(II) ions that are coordinated by side chains at the C-terminal end. (B) Side view of the trimeric structure of As(CSL9C)3 with a different perspective of the Zn(II) coordination to the exterior residues of the coiled coils. In both panels, only two of eight symmetry related Zn(II) ions are included for clarity.
Side Chain Electrostatic Interactions.
There are interhelical electrostatic interactions between e and g glutamate and lysine side chains of the heptad repeat on all three interhelical interfaces, which can be observed in detail in SI Fig. 7. There are additional electrostatic interactions not typical for a three-stranded coiled coil between the b and g residues Glu 24 A and Lys 22 B and between the indole nitrogens of Trp 2 C and Glu 1 A and Trp 2 B and Glu 1 C. Several interhelical interactions are mediated by water molecules, for example, between Glu 1 C and Glu 6 B. In addition, there are several interactions between the backbone and side chains, such as between the hydroxyl group of Ser 14 C and the carboxyl of the main chain of the same helix at Ala 10.
Discussion
Implications for Arsenic Toxicity.
Although arsenic is highly toxic and a carcinogen, its interactions with biomolecules have not been thoroughly investigated. Whereas As(V) in the form of arsenate is thought to bind to phosphate-binding sites (38), As(III) is known to have high affinity for thiolate-rich sites (39). An association constant of 1 × 107 was reported for formation of the trigonal thiolate AsL3 complex with glutathione as measured by calorimetric and spectroscopic methods (39). We expect that the affinity of As(III) for CSL9C would be greater than or equal to that of glutathione based on our previous studies with the TRI peptides that demonstrate the effect of peptide self-association on the affinity of Cd(II) and Hg(II) (40). Because slow kinetics (3-h incubation at 37°C for glutathione at millimolar concentration) and low extinction coefficient hinder direct determination of the affinity of CSL9C for As(III), spectroscopic measurement of the displacement of As(III) from As(CSL9C)3 by equimolar Cd(II) at pH 8.0 was attempted. This experiment revealed that As(III) was not replaced by Cd(II) after 5 h, indicating that the Ka of As(III) binding to CSL9C is either greater than the value of 2.7 × 107 that was measured for Cd(II) or that the kinetics of As(III) release are too slow to observe displacement on this time scale (33).
The structure of As(CSL9C)3 reveals a structural example of As(III) coordination in a designed peptide environment. The best crystallographic model for the As(III) site yields an average As–S bond distance of 2.28 Å, which is within the error of the crystallographic coordinates of our previously determined extended x-ray absorption fine structure values, and an average S–As–S angle of 90° that are in good agreement with small molecule As(III) complexes with trigonal pyramidal Tris-arylthiolate coordination, in which As(III)–S bond distances ranging from 2.23 to 2.29 Å and S–As–S angles with a mean value of 98° were reported (6, 36, 37). The compression of these angles in our structure compared with the small molecule may be a result of peptide aggregation and side chain rotamer effects. It is very likely that there are similar angles when As(III) binds to the ArsR and ArsD proteins, with a schematic model for ArsR being proposed with As(III)–S distances of 2.25 Å and mean S–As–S 92° angles (1).
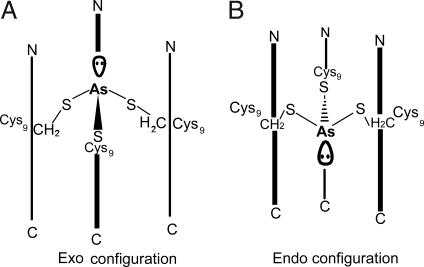
This analysis does not address an important conformational aspect of As(III) binding to a planar three-sulfur environment. The As(III) ion is predicted to have a stereochemically active lone pair. Examination of small-molecule trigonal thiolate As(III) structures in the literature shows that, in two cases, the As(III) has a pyramidal geometry with the arsenic located above (exo conformer) the plane of the sulfur atoms of the ligands (based on the As-S-Cβ atoms of the complexes). In one case where the As(III) is oriented below the ring (endo conformer); however, it was thought that this geometry was due to long-range oxygen interactions that stabilized the structure (37). Fig. 4 shows the endo and exo conformations as they would exist inside a three-stranded coiled coil. Surprisingly, the endo conformation of the As(III) is observed in CSL9C despite the fact that there are no long-range interactions stabilizing this orientation. Although several factors, such as packing of the adjacent hydrophobes, may be responsible for the observed endo orientation, we believe that it results because the cysteine rotamers are optimized in this configuration to accommodate the relatively short As–S bonds. Previous models for As(III) binding to ArsR have proposed the arsenic binding to Cys 32, Cys 34, and Cys 37 in an α-helical region of the protein. The simultaneous interaction of arsenic with Cys 32 and Cys 34 is particularly interesting because this is a Cys-X-Cys motif not normally associated with metal binding and likely requiring significant perturbation of the structure. Models assessing this perturbation have uniformly used an As(III)-bound exo to the three cysteines; however, this study demonstrates that future models should consider the possibility that As(III) adopts an endo conformation. Thus, it may be useful to reconsider predictions for As(III) coordination to ArsR and possibly ArsD detoxification proteins.
Fig. 4.
Scheme showing the two possible orientations of As(III) within a three-stranded coiled coil. (A) In the exo configuration, the As(III) ion would be located above both Sγ atoms and β-methylene protons of the cysteine side chains, and the lone pair would point toward the N terminus. (B) In the endo configuration, which is observed in the structure of As(CSL9C)3, the As(III) ion would be located below the Sγ atoms and approximately level with the β-methylene protons of the cysteine side chains. The lone pair would then be directed toward the C terminus.
Influence on Protein Design Strategy.
Design of coiled coils has been an active area of research since the report of a parallel two-stranded coiled coil by Hodges et al. (41). Since then, design of two-, three-, and four-stranded aggregates has been accomplished (11). Through the design of coiled coils, it has been discovered that the energetics of multiple conformations can be quite similar; this is underscored by the reports of crystal structures of homomeric assemblies with different aggregation states (42) and helix orientations (43).
The design of coiled coils that associate specifically into parallel or antiparallel aggregates has been made possible by manipulating electrostatic interactions between e and g positions of the heptad repeat (44, 45). The parallel heteromeric two-stranded coiled coil Velcro peptide and the three-stranded coiled coil ABC peptide were designed by incorporating repulsive electrostatic interactions for homomeric species and favorable electrostatic interactions for heteromeric aggregates (46, 47). Additionally, a polar amino acid was incorporated into a hydrophobic core position of the heptad repeat to discourage an antiparallel orientation that would require an unfavorable interaction between hydrophobic and polar side chains, which would be alleviated by formation of a parallel aggregate. Heteromeric antiparallel three-stranded coiled coils were designed by using the steric matching of hydrophobic residues to optimize packing in addition to the incorporation of favorable electrostatic interactions (48). In light of this knowledge of the requirements of electrostatic interactions and hydrophobic packing for the formation of stable aggregates, it is somewhat surprising that Coil Ser was found to adopt an antiparallel orientation, in which two of the e–g interhelical interfaces in the trimeric structure contain pairs of lysine-lysine and glutamate-glutamate residues. Coil Ser has few interhelical interactions in comparison to As(CSL9C)3. In Coil Ser, there are only four interhelical electrostatic interactions between lysine and glutamate residues, between the two helices that are parallel to one another, whereas As(CSL9C)3 has 10, at all three interfaces, which are illustrated in SI Fig. 7. The pH conditions under which crystals of Coil Ser and As(CSL9C)3 were obtained may have an influence on the orientation. Coil Ser was previously crystallized at pH 5.0, where the hydrogen bonding capabilities of glutamate residues are partially minimized by protonation, whereas As(CSL9C)3 was crystallized at pH 8, where glutamates would be fully deprotonated. There are also fewer water molecules involved in interactions with the peptide Coil Ser, which, in the structure presented here, are instrumental in interhelical interactions. The orientation of As(CSL9C)3 as a parallel three-stranded coiled coil is not surprising, and 2D NMR studies at pH 7.7 indicate a parallel orientation is obtained in solution (O. Iranzo and V.L.P., unpublished data), even without metal ion coordination templating the orientation (33).
In addition, the cysteine residues, which are in an a position, have their side chains pointed toward the center of the helix. The chi 1 dihedral angles (N-Cα-C-β-Sγ) for the three cysteine residues are −60° ± 1°, very close to the value of −65° for the most common cysteine rotamer (49). The average crystallographic B-factor for the Sγ atoms of the cysteine residues is 16, slightly lower than the average B-factor for the entire structure, which is 18. There is no evidence of alternate rotamers for the cysteine side chains. It is possibile that the presence of cysteine residues at high pH in the center of the coiled coil acts to direct the coiled coil to have a parallel orientation in solution, as was observed with polar glutamine and arginine residues in the design of parallel heteromeric GCN4 derivatives (50).
In several parallel coiled coil structures of GCN4-derivative peptides and in Coil VaLd, differences in hydrophobic packing in a and d hydrophobic layers were observed (35, 51, 52). In these structures and the one presented here, the hydrophobic layers are composed of either all a or all d residues, and the a leucine residues tend to orient more toward the center of the coiled coil than the d residues (35). In the antiparallel Coil Ser structure, the leucine layers are composed of two a and one d′ residues or one a and two d′ residues, which influences the packing (32). Because hydrophobic residues in a and d positions are known to orient differently with respect to the coiled coil axis, it is likely that, for a corresponding peptide with cysteines in a d position, the orientation of the cysteine side chains would be different from those in this case.
One of the original hypotheses as to why Coil Ser adopted an antiparallel orientation was that it did so to avoid steric clashes between the three bulky tryptophan residues at position 2 of the sequence (32). Although a reasonable possibility, this structure demonstrates that there is sufficient room for packing the tryptophan residues at the N terminus, suggesting that this is not the only influence on the orientation. In addition, in the B and C chains, the tryptophan residues are involved in interhelical electrostatic interactions with glutamate side chains. Although the coordination of As(III) to the cysteine residues likely stabilizes a parallel trimeric structure, other factors such as electrostatic and hydrophobic packing interactions certainly play a role in determining parallel orientation in solution.
We have successfully designed an As(III) coordination site with three cysteines in an endo trigonal pyramidal geometry. Although the observed As–S distances and S–As–S angles are within the expected range, the endo configuration was quite surprising. Our original design incorporated an exo As(III) with a coordination geometry similar to Pb(II) in the protein δ-aminolevulinic acid dehydratase (53). This observation raises the interesting question of whether there are generally structural differences between Pb(II) and As(III) coordination to three-stranded coiled coil peptides and to native proteins. Although both metal ions possess stereochemically active lone pairs and prefer trigonal pyramidal soft ligand-binding sites, As(III) is much smaller than Pb(II) (with a typical Pb(II)–S distance of 2.64 Å) (9). This difference in size and subsequent S–Pb–S angles may not allow Pb(II) to accept coordination as an endo conformer. In addition, the larger size of Pb(II) may perturb the helix, causing it to pucker out to be accommodated. Another key distinction between As(III) and Pb(II), other than size, is the difference in charge between the two ions. It is possible that nature uses these factors in discriminating between divalent and trivalent heavy metals in the metalloregulatory systems of the ArsR/SmtB family of transcriptional regulator proteins, in which ArsR is used ArsR for detection of As(III) and Sb(III), and CadC is used for detection of Cd(II) and Pb(II) (54).
Analysis of the packing of the Leu residues in the layers above and below the As(III)-binding site shows that there is room for a water molecule to bind between the Cys layer at position 9 and Leu 12, as shown in Fig. 2. Using Cd(II), our first designs with these peptides yielded equilibrium mixtures of three- and four-coordinate Cd(II) (55). By changing the leucine layer above the metal to alanine to allow better water access above the metal-binding site, we designed a peptide (TRI L12AL16C) that binds Cd(II) solely in a four-coordinate geometry, with three cysteinyl thiolates and one exogenous water. This observation would suggest that Cd(II) binds to these peptides as the exo conformer, consistent with the longer Cd(II)–S distances (2.54 Å), which are more reminiscent of Pb(II) than As(III) (8). These observations make one consider whether metalloregulatory proteins may be able to differentiate different toxic metals in part by their preference for endo [As(III)] versus exo [Pb(II)/Cd(II)] binding. It is known that Sb(III) stimulates the ArsR regulator; thus, it will be extremely interesting to see whether this ion, which is larger than As(III), will bind in an endo or exo configuration. There is one known structure of ArsA, the membrane translocating ATPase, with Sb(III) bound, but it is difficult to interpret its geometrical preferences because three Sb(III) ions are bound in close proximity with bridging cysteine thiolates and chloride ions (2). One Sb(III) ion in this structure has a trigonal cysteine coordination with two of the cysteines in an endo conformation, an indication that Sb(III) has the capability of binding in this conformation as well.
Crystallization of this peptide was not successful in the absence of Zn(II), and the resulting structure shows why it is essential. Zn(II) links the coiled coils together at crystal-packing interfaces on the exterior of the peptide by coordination to glutamate and histidine side chains. The binding of Zn(II) by the side chains of the C-terminal residues results in the C terminus of CSL9C being better folded in this region in comparison to Coil VaLd. A least-squares overlay of the Cα from residues 3 through 18 has an average rmsd of 0.2 Å from that of VaLd; however, if the entire helix is included, the rmsd is 1.7 Å, demonstrating the greater differences at the N and C termini, as can be seen in Fig. 5. It is likely that Zn(II) may facilitate the crystallization of other Coil Ser derivative peptides (e.g., CSL19C) that have been shown to have different metal-binding properties than CSL9C (33).
Fig. 5.
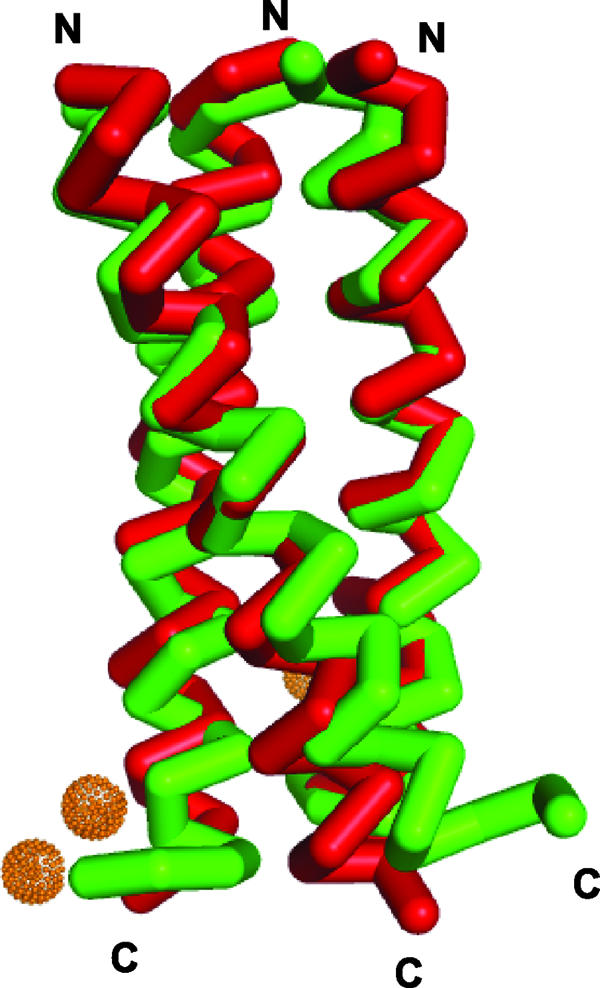
The overlay of Coil VaLd, shown in green (PDB entry 1COI), a related parallel three-stranded coiled coil with As(CSL9C)3 (red) demonstrates their structural similarity and highlights their divergence at the C and N termini where the Zn(II) ions hold CSL9C in a more helical conformation than VaLd.
Conclusions
We present here a unique structure of a designed three-stranded coiled coil peptide with As(III) bound to the hydrophobic interior in a trigonal thiolate coordination geometry. It is bound slightly below the plane of the cysteine residues in an endo trigonal pyramidal geometry. This structure provides a good model of the active site of the ArsR repressor protein when it is bound to As(III), which has not yet been structurally characterized. In addition, the discovery of Zn(II) ions linking the coiled coils together in the solid state is an interesting and potentially useful observation for the crystallization of other designed coiled coil peptides.
Experimental Procedures
Peptide Synthesis and Purification.
CSL9C was synthesized on an Applied Biosystems (Foster City, CA) 433A peptide synthesizer by standard protocols and purified and characterized as previously reported (33). As(CSL9C)3 was prepared by incubating 10-fold excess NaAsO2 with peptide in 50 mM potassium phosphate buffer (pH 7.0). The solution was allowed to react at room temperature for 36–48 h and then purified by RP-HPLC on a linear water to acetonitrile gradient. The lyophilized off-white powder was characterized by MALDI-MS in a sinapinic acid matrix to have the expected mass of 10,076 Da for the As(III)–peptide complex.
Crystallization.
As(CSL9C)3 was dissolved in distilled Milli-Q water to a concentration of 15 mg/ml. Crystals were grown by vapor diffusion at 20°C in a sitting drop with equal volumes of peptide and precipitant containing 100 mM imidazole buffer (pH 8.0), 200 mM Zn(OAc)2, and 40% PEG 400. Crystals were frozen in their mother liquor for data collection.
Data Collection and Refinement.
Data were measured at the Advanced Photon Source (APS) (COM-CAT Beamline 32-ID and GMCA-CAT Beamline 23-ID) at the Argonne National Laboratory and were collected on a MarCCD (Mar USA, Evanston, IL) at a wavelength of 1.00 Å at −180°C. Three hundred sixty frames of data were collected with a 1° rotation and 1-s exposure. Data were processed and scaled with the program HKL-2000 (56). The crystals were of the space group C2 with unit cell parameters a = 77.28, b = 44.20, c = 29.41, α = γ = 90°, and β = 119.5°.
The structure was solved by molecular replacement, using as a model the trimeric coiled coil Coil VaLd (Protein Data Bank entry 1COI) with the chain truncated to 26 residues. The side chains of the model were included. The Patterson-based program used was an updated version of GENPAT (35, 57). This model with side chains truncated to alanine was refined by rigid body refinement and restrained refinement in Refmac 5 in the CCP4 suite of programs (58, 59). 2Fo − Fc and Fo − Fc electron density maps generated with the CCP4 map utilities were used for building in side chains, metal ions, and C-terminal residues in Coot. Coot was used for finding water molecules (60). The refinement to 1.8 Å resulted in Rworking = 19.8% and Rfree = 25.4%. Data collection and refinement statistics are given in Table 2. The validity of the model was verified by a composite omit map generated in the Crystallography and NMR System (30). Figures were generated in Pymol and MIFit. The coordinates and structure factors have been deposited in the Protein Data Bank with the ID code 2JGO.
Table 2.
Data collection and refinement statistics
| Data collection statistics | |
| Data set | Native |
| Space group | C2 |
| Unit cell | a = 77.28; b = 29.41; c = 44.19; α = 90; β = 119.5; γ = 90 |
| Wavelength, Å | 1.00 |
| Resolution, Å | 1.8 (1.8–1.86) |
| Rsym, % | 6.5 (20.3) |
| 〈I/σI〉 | 10 (3) |
| Completeness, % | 98.4 (90.6) |
| Redundancy | 7 (5) |
| Refinement statistics | |
| Resolution, Å | 1.8 (13.7–1.8) |
| R-factor, % | 19.8 |
| Rfree, % | 25.4 |
| Protein atoms | 762 |
| Water molecules | 50 |
| Unique reflections | 8,137 |
| rmsd | |
| Bonds | 0.018 |
| Angles | 1.448 |
Parenthesized numbers are the values for the highest resolution shell.
Supplementary Material
Acknowledgments
We thank Joseph Brunzelle for data collection. This research was supported by National Institutes of Health Grant ES012236 (to V.L.P.), the Chemical Biology Interface Training Grant (to D.S.T.), and funds from the Michigan Economic Development Corporation Life Sciences Corridor (to J.A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2JGO).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701979104/DC1.
References
- 1.Shi W, Dong J, Scott RA, Ksenzenko MY, Rosen BP. J Biol Chem. 1996;271:9291–9297. doi: 10.1074/jbc.271.16.9291. [DOI] [PubMed] [Google Scholar]
- 2.Zhou T, Radaev S, Rosen BP, Gatti DL. EMBO J. 2000;19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin P, DeMel S, Shi J, Gladysheva T, Gatti DL, Rosen BP, Edwards BFP. Structure (London) 2001;9:1071–1081. doi: 10.1016/s0969-2126(01)00672-4. [DOI] [PubMed] [Google Scholar]
- 4.Messens J, Martins JC, Van Belle K, Brosens E, Desmyter A, De Gieter M, Wieruszeski J-M, Willem R, Wyns L, Zegers I. Proc Natl Acad Sci USA. 2002;99:8506–8511. doi: 10.1073/pnas.132142799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y-F, Walmsley AR, Rosen BP. Proc Natl Acad Sci USA. 2006;103:15617–15622. doi: 10.1073/pnas.0603974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaikh TA, Bakus RC, II, Parkin S, Atwood DA. J Organomet Chem. 2006;691:1825–1833. [Google Scholar]
- 7.Dieckmann GR, McRorie DK, Tierney DL, Utschig LM, Singer CP, O'Halloran TV, Penner-Hahn JE, DeGrado WF, Pecoraro VL. J Am Chem Soc. 1997;119:6195–6196. [Google Scholar]
- 8.Matzapetakis M, Farrer BT, Weng T-C, Hemmingsen L, Penner-Hahn JE, Pecoraro VL. J Am Chem Soc. 2002;124:8042–8054. doi: 10.1021/ja017520u. [DOI] [PubMed] [Google Scholar]
- 9.Matzapetakis M, Ghosh D, Weng T-C, Penner-Hahn JE, Pecoraro VL. J Biol Inorg Chem. 2006;11:876–890. doi: 10.1007/s00775-006-0140-7. [DOI] [PubMed] [Google Scholar]
- 10.Farrer BT, McClure CP, Penner-Hahn JE, Pecoraro VL. Inorg Chem. 2000;39:5422–5423. doi: 10.1021/ic0010149. [DOI] [PubMed] [Google Scholar]
- 11.Woolfson DN. In: Fibrous Proteins: Coiled-Coils, Collagen and Elastomers. Squire JM, editor. San Diego: Academic; 2005. pp. 79–112. [Google Scholar]
- 12.Pantoja-Uceda D, Santiveri CM, Jiménez MA. In: Protein Design—Methods and Applications, Methods in Molecular Biology. Guerois R, de la Paz ML, editors. Vol 340. Totowa, NJ: Humana; 2006. pp. 27–52. [Google Scholar]
- 13.Doerr AJ, McLendon GL. Inorg Chem. 2004;43:7916–7925. doi: 10.1021/ic0490573. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh D, Pecoraro VL. Curr Opin Chem Biol. 2005;9:97–103. doi: 10.1016/j.cbpa.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Barker PD. Curr Opin Struct Biol. 2003;13:490–499. doi: 10.1016/s0959-440x(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 16.Reedy CJ, Kennedy ML, Gibney BR. Chem Commun. 2003:570–571. doi: 10.1039/b211488g. [DOI] [PubMed] [Google Scholar]
- 17.Gibney BR, Mulholland SE, Rabanal F, Dutton PL. Proc Natl Acad Sci USA. 1996;93:15041–15046. doi: 10.1073/pnas.93.26.15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsh ENG, DeGrado WF. Proc Natl Acad Sci USA. 2002;99:5150–5154. doi: 10.1073/pnas.052023199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnepf R, Haehnel W, Wieghardt K, Hildebrandt P. J Am Chem Soc. 2004;126:14389–14399. doi: 10.1021/ja0484294. [DOI] [PubMed] [Google Scholar]
- 20.Kharenko OA, Kennedy DC, Demeler B, Maroney MJ, Ogawa MY. J Am Chem Soc. 2005;127:7678–7679. doi: 10.1021/ja042757m. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Suzuki K, Kanaori K, Tajima K, Kashiwada A, Hiroaki H, Kohda D, Tanaka T. Protein Sci. 2000;9:1327–1333. doi: 10.1110/ps.9.7.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petros AK, Reddi AR, Kennedy ML, Hyslop AG, Gibney BR. Inorg Chem. 2006;45:9941–9958. doi: 10.1021/ic052190q. [DOI] [PubMed] [Google Scholar]
- 23.Ramadan D, Cline DJ, Bai S, Thorpe C, Schneider JP. J Am Chem Soc. 2007;129:2981–2988. doi: 10.1021/ja067068k. [DOI] [PubMed] [Google Scholar]
- 24.Daugherty RG, Wasowicz T, Gibney BR, DeRose VJ. Inorg Chem. 2002;41:2623–2632. doi: 10.1021/ic010555a. [DOI] [PubMed] [Google Scholar]
- 25.Mulholland SE, Gibney BR, Rabanal F, Dutton PL. J Am Chem Soc. 1998;120:10296–10302. [Google Scholar]
- 26.Huang SS, Gibney BR, Stayrook SE, Leslie Dutton P, Lewis M. J Mol Biol. 2003;326:1219–1225. doi: 10.1016/s0022-2836(02)01441-9. [DOI] [PubMed] [Google Scholar]
- 27.Lombardi A, Summa CM, Geremia S, Randaccio L, Pavone V, DeGrado WF. Proc Natl Acad Sci USA. 2000;97:6298–6305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maglio O, Nastri F, Pavone V, Lombardi A, DeGrado WF. Proc Natl Acad Sci USA. 2003;100:3772–3777. doi: 10.1073/pnas.0730771100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeGrado WF, Costanzo LD, Geremia S, Lombardi A, Pavone V, Randaccio L. Angew Chem Int Ed. 2003;42:417–420. doi: 10.1002/anie.200390127. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan J, DeGrado WF. Proc Natl Acad Sci USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovejoy B, Choe S, Cascio D, McRorie D, DeGrado W, Eisenberg D. Science. 1993;259:1288–1293. doi: 10.1126/science.8446897. [DOI] [PubMed] [Google Scholar]
- 33.Iranzo O, Ghosh D, Pecoraro VL. Inorg Chem. 2006;45:9959–9973. doi: 10.1021/ic061183e. [DOI] [PubMed] [Google Scholar]
- 34.Wendt H, Berger C, Baici A, Thomas RM, Bosshard HR. Biochemistry. 1995;34:4097–4107. doi: 10.1021/bi00012a028. [DOI] [PubMed] [Google Scholar]
- 35.Ogihara NL, Weiss MS, DeGrado WF, Eisenberg D. Protein Sci. 1997;6:80–88. doi: 10.1002/pro.5560060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaikh TA, Parkin S, Atwood DA. J Organomet Chem. 2006;691:4167–4171. [Google Scholar]
- 37.Carter TG, Healey ER, Pitt MA, Johnson DW. Inorg Chem. 2005;44:9634–9636. doi: 10.1021/ic051522o. [DOI] [PubMed] [Google Scholar]
- 38.Willsky GR, Malamy MH. J Bacteriol. 1980;144:356–365. doi: 10.1128/jb.144.1.356-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spuches AM, Kruszyna HG, Rich AM, Wilcox DE. Inorg Chem. 2005;44:2964–2972. doi: 10.1021/ic048694q. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh D, Lee K-H, Demeler B, Pecoraro VL. Biochemistry. 2005;44:10732–10740. doi: 10.1021/bi0506674. [DOI] [PubMed] [Google Scholar]
- 41.Hodges R, Saund A, Chong P, St.-Pierre S, Reid R. J Biol Chem. 1981;256:1214–1224. [PubMed] [Google Scholar]
- 42.Gonzalez L, Brown RA, Richardson D, Alber T. Nat Struct Biol. 1996;3:1002–1010. doi: 10.1038/nsb1296-1002. [DOI] [PubMed] [Google Scholar]
- 43.Yadav MK, Leman LJ, Price DJ, Brooks CL, Stout CD, Ghadiri MR. Biochemistry. 2006;45:4463–4473. doi: 10.1021/bi060092q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monera OD, Kay CM, Hodges RS. Biochemistry. 1994;33:3862–3871. doi: 10.1021/bi00179a010. [DOI] [PubMed] [Google Scholar]
- 45.Schnarr NA, Kennan AJ. Org Lett. 2005;7:395–398. doi: 10.1021/ol047720l. [DOI] [PubMed] [Google Scholar]
- 46.O'Shea EK, Lumb KJ, Kim PS. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 47.Nautiyal S, Woolfson DN, King DS, Alber T. Biochemistry. 1995;34:11645–11651. doi: 10.1021/bi00037a001. [DOI] [PubMed] [Google Scholar]
- 48.Schnarr NA, Kennan AJ. J Am Chem Soc. 2004;126:14447–14451. doi: 10.1021/ja047496v. [DOI] [PubMed] [Google Scholar]
- 49.Ponder JW, Richards FM. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 50.Oakley MG, Hollenbeck JJ. Curr Opin Struct Biol. 2001;11:450–457. doi: 10.1016/s0959-440x(00)00232-3. [DOI] [PubMed] [Google Scholar]
- 51.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 52.Harbury PB, Kim PS, Alber T. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 53.Erskine PT, Duke EMH, Tickle IJ, Senior NM, Warren MJ, Cooper JB. Acta Crystallogr D. 2000;56:421–430. doi: 10.1107/s0907444900000597. [DOI] [PubMed] [Google Scholar]
- 54.Pennella MA, Giedroc DP. BioMetals. 2005;18:413–428. doi: 10.1007/s10534-005-3716-8. [DOI] [PubMed] [Google Scholar]
- 55.Lee K-H, Cabello C, Hemmingsen L, Marsh ENG, Pecoraro VL. Angew Chem Int Ed. 2006;45:2864–2868. doi: 10.1002/anie.200504548. [DOI] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 57.Nordman C. Acta Crystallogr A. 1994;50:68–72. [Google Scholar]
- 58.Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 59.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 60.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.