Abstract
Poxviruses (Poxviridae) are a family of double-stranded DNA viruses with no RNA stage. Members of the genus Orthopoxvirus (OPV) are highly invasive and virulent. It was recently shown that the taterapox virus (TATV) from a West African rodent is the sister of camelpox virus and therefore belongs to the clade closest to the variola virus (VARV), the etiological agent of smallpox. Although these OPVs are among the most dreaded pathogens on Earth, our current knowledge of their genomes, their origins, and their possible hosts is still very limited. Here, we report the horizontal transfer of a retroposon (known only from reptilian genomes) to the TATV genome. After isolating and analyzing different subfamilies of short interspersed elements (SINEs) from lizards and snakes, we identified a highly poisonous snake (Echis ocellatus) from West Africa as the closest species from which the SINE sequence discovered in the TATV genome (TATV-SINE) was transferred to the virus. We discovered direct repeats derived from the virus flanking the TATV-SINE, and the absence of any snake-derived DNA flanking the SINE. These data provide strong evidence that the TATV-SINE was actually transferred within the snake to the viral genome by retrotransposition and not by any horizontal transfer at the DNA level. We propose that the snake is another host for TATV, suggesting that VARV-related epidemiologically relevant viruses may have derived from our cold-blooded ancestors and that poxviruses are possible vectors for horizontal transfer of retroposons from reptiles to mammals.
Keywords: Bov-B long interspersed element, lateral transfer, Orthopoxvirus, retrotransposition, Sauria short interspersed element
Short interspersed elements (SINEs) are a type of retroposon (70–500 bp) that invade new genomic sites by a copy-and-paste mechanism through integration of a reverse-transcribed copy of RNA (1, 2). The retrotransposition of nonautonomous SINEs depends on the enzymatic retrotranspositional machinery of their autonomous partner long interspersed elements (LINEs), which encode a reverse transcriptase (RT) and endonuclease (EN). The mechanism of LINE retrotransposition (3) is termed “target-primed reverse transcription” (TPRT) and was adopted for partner SINE retrotransposition because the corresponding identical 3′ tail sequence of a SINE is recognized by the RT of its partner LINE (4–6). In TPRT, the EN initiates a nick on one strand of the targeted DNA. Subsequently, the 3′ OH introduced by this nick is used to prime cDNA synthesis, using an RNA transcript of a SINE or LINE as a template. TPRT usually results in a pair of direct repeats flanking the inserted retroposon, a hallmark of retrotransposition.
SINE families and subfamilies, which are defined by the presence of diagnostic nucleotides, namely a set of specific nucleotides that differ from those of a consensus sequence, can be found specifically within members of a particular phylogenetic lineage. Their role as effective markers for elucidating evolutionary history has been demonstrated in many studies (7–14). Apart from their vertical transfer, in rare cases SINEs can be transferred horizontally, such as in salmonid fishes (15). The horizontal transfer of SINE members of a particular SmaI subfamily was supposedly from coregonid salmonid fishes to two distantly related salmonid fishes. It also has been shown that salmonid SINEs were acquired by schistosomes by parasitic infection of salmonid fish (16), results which were recently expanded by our group (17).
The phenomenon of horizontal transfer has been particularly studied in bacteria (18) and viruses. Many well supported cases for horizontal transfer between viruses and their hosts are known. For example, it has been suggested that up to 50% of the poxvirus gene families show some evidence of horizontal gene transfer from their hosts (19). Two subfamilies of poxviruses can be distinguished, namely entomopoxvirinae (insects hosts) and chordopoxvirinae (vertebrate hosts). Their genomes range in size from 130 to 380 kbp and harbor a considerable number of genes (>200). All sequences of the viral DNA ligase family of poxviruses were proposed to have been acquired by the common ancestor of chordopoxvirinae from a mammalian host because all viral members show high similarity to mouse or human DNA ligase III sequences (20). Strong evidence for horizontal transfer events also comes from poxvirus-encoded dUTPase sequences, which in phylogenetic analyses cluster with mammalian host dUTPase sequences (21). However, there are no reports of retroposon sequences from eukaryotic hosts in poxvirus genomes. More common is the integration of sequences from viral sources into poxvirus genomes. For example, a unique relationship exists between fowlpox virus and reticuloendotheliosis virus (22–25). It was shown that the integration of reticuloendotheliosis virus into the genome of fowlpox virus may have some benefit to fowlpox virus for infection of poultry previously vaccinated against fowlpox (24). Poxviruses are distributed universally, and zoonoses of many of the orthopoxvirus (OPV) isolates have been reported. For example, VARV was responsible for an estimated 300–500 million human deaths in the 20th century and is nowadays feared as a bioterrorism pathogen (26, 27). A recent data set of 45 complete genomes of VARV and several non-human mammalian strains of OPVs (28) showed that most closely related to VARV are the camelpox virus (29) and TATV, which was isolated in 1968 from Kemp's gerbil (Tatera kempi)† in Benin, West Africa (30).
Results and Discussion
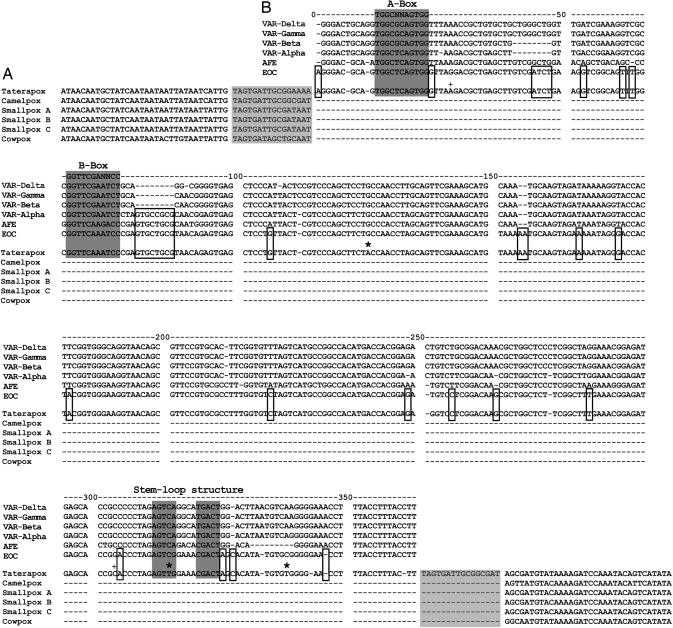
Here, we describe the horizontal transfer of a SINE (Sauria SINE) from a reptilian genome (the snake Echis ocellatus) to the TATV genome. Sauria SINEs are ≈350 nucleotides long and are widely distributed among lizards and snakes, the most diverse group of living reptiles (Squamata). They comprise a typical 5′ tRNA-related region, a tRNA-unrelated region, and a 3′ tail region (containing short tandem repeats) which is derived from ubiquitous squamate Bov-B LINEs (12). Using the complete TATV genome sequence (accession no. DQ437594), we discovered an insertion of a Sauria SINE at position 33368–33720 with clearly recognizable direct repeats, typical 3′ end Sauria SINE characteristic tandem repeats and RNA polymerase III-specific internal promoter sequences. This Sauria SINE is not present in other OPV genomes, yet the orthologous genomic location in all published OPVs is the same as the virus sequence surrounding the Sauria SINE (Fig. 1A). To our knowledge, this is the first discovery of a retroposon insertion in a virus genome. Extensive RepeatMasker database searches did not reveal SINEs in viruses other than taterapox.
Fig. 1.
TATV-SINE locus aligned with consensus sequences of six Sauria SINE subfamilies. (A) Alignment of genomic regions of six OPVs showing the TATV-SINE insertion and 50 bp of flanking regions, including the single insertion site (boxed in light gray), of closely related OPVs (28). (B) SINE sequences isolated from genomes of varanid lizards comprise four different subfamilies (VARα-type, VARβ-type, VARγ-type, and VARδ-type sequences). VARα-type members share an 8-bp insertion (unshaded box, positions 81–88) with both subfamilies isolated from genomes of snakes (AFE and EOC). The EOC Sauria SINE subfamily and the TATV-SINE are closely related (diagnostic nucleotides are boxed). Each CpG site is marked with an asterisk. Nucleotide positions 28 and 304 are marked with a plus (see Fig. 2 for further explanation). The two deletions in the TATV-SINE sequence (positions 344 and 361) could have been created upon retrotransposition of the SINE. Functional regions are boxed in dark gray: the 5′ end RNA polymerase III-specific internal promoter sequences (A-Box and B-Box) and the 3′ end stem-loop structure for RT recognition (12).
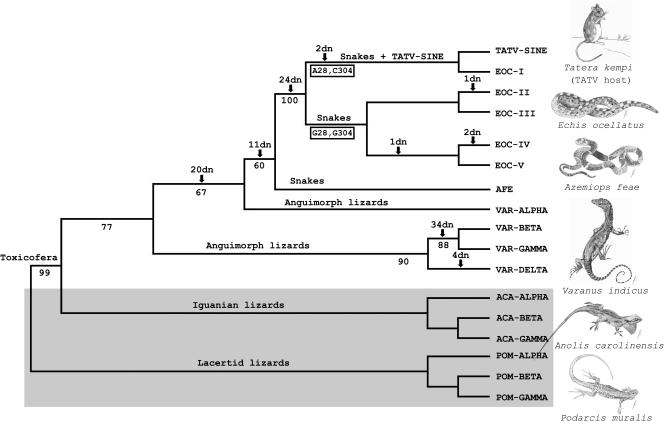
Our initial sequence analyses revealed that sequences of Sauria SINE subfamilies described for varanid lizards [Varanus indicus (mangrove monitor)] and snakes [Azemiops feae (Fea's viper)] are closely related to the TATV-SINE (12). These squamates belong to a clade recently defined as Toxicofera, referring to the presence of venom in this phylogenetic lineage (32, 33). To track the evolution of the TATV-SINE sequence, we investigated new Sauria SINEs in Toxicofera species with different geographical distribution and experimentally isolated 38, 13, and 17 SINE loci from the genomes of E. ocellatus (carpet viper, West African distribution), Varanus griseus (desert monitor, North African and Indo-Asian distribution), and Varanus timorensis (spotted tree monitor, Indo-Australian distribution), respectively. Analyses of these sequences revealed diagnostic nucleotides that made it possible to divide repetitive sequences of the analyzed lizards and snakes into four and two different subfamilies, respectively (Fig. 1B). The VARα-type subfamily closes the gap of Sauria SINE sequence evolution between lizards and snakes, as shown in Fig. 1B (positions 81–88 are boxed), whereas the Sauria SINE subfamily isolated from E. ocellatus, named EOC, is most closely related to the TATV-SINE, as shown by 24 boxed diagnostic nucleotides (Figs. 1B and 2). The carpet viper belongs to the group of advanced snakes (Caenophidia, 2,500+ species) whose radiation dates back at least 100 million years. This evolutionary time frame seems to correlate not only with the evolution of the snake venom system (33) but also with the origin of Sauria SINE-specific EOC members in advanced snakes (Fig. 3 A and B), which represent ancestors of the TATV-SINE (Fig. 3C). Although we cannot rule out the possibility that an Echis species different from E. ocellatus is responsible for the generation of the TATV-SINE, we have good reason to believe that the TATV-SINE was derived from E. ocellatus. The geographical distributions of four Echis species (E. ocellatus, Echis leucogaster, Echis jogeri, and Echis pyramidum) partially overlap with the geographical distribution of T. kempi. However, the overlap with E. jogeri and E. pyramidum is very small and the distribution of E. leucogaster is mostly northwest of E. ocellatus (34).
Fig. 2.
Phylogenetic tree of Sauria SINEs and TATV-SINE obtained by the maximum-likelihood method. The relationships of investigated Sauria SINE subfamily consensus sequences of lizards and snakes are illustrated. Sauria SINEs of lacertid lizards (POM, Podarcis muralis, common wall lizard) and iguanian lizards (ACA, Anolis carolinensis, green anole) served as outgroups and are boxed in gray (12). We categorized EOC SINE members into five subsubfamilies to show the precise origin of the TATV-SINE (EOC-I to EOC-V). Subsubfamily EOC-I differs from the EOC consensus sequence by two diagnostic nucleotides that are shared with the TATV-SINE (positions 28 and 304 in Fig. 1B; see also positions 51 and 330 in SI Fig. 6). Numbers below the branches represent puzzle support values for Sauria SINE subfamilies. Numbers above the branches correspond to diagnostic nucleotides (dn) in subfamilies/subsubfamilies.
Fig. 3.
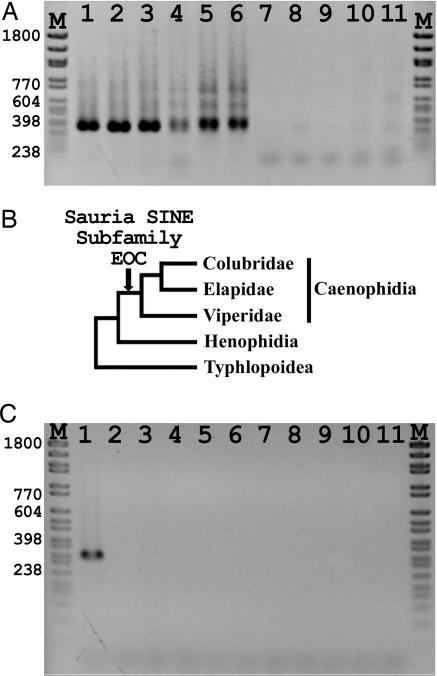
Distribution of Sauria SINE subfamily EOC and subsubfamily EOC-I. (A) Genomic DNA from snakes, lizards and rodents‡ was amplified by PCR, using primers EOCconsF and EOCconsR (see Material and Methods). A discrete PCR product of ≈340 bp was generated in all three major lineages of advanced snakes (lanes 1–4 Viperidae; lane 5 Elapidae; lane 6, Colubridae) but not in other snakes (lane 7), lizards (lanes 8–10), or rodents (lane 11). (B) Phylogenetic relationships of snakes. (C) Genomic DNA from snakes, lizards, and rodents‡ was amplified by PCR, using Sauria SINE-specific subsubfamily primers EOC-I-F and EOC-I-R (see Material and Methods). A discrete PCR product of ≈315 bp was generated only in E. ocellatus (lane 1). Lane 1 E. ocellatus; lane 2, E. coloratus; lane 3, Macrovipera lebetina; lane 4, Crotalus horridus; lane 5, Notechis scutatus; lane 6, Natrix tesselata; lane 7, Boa constrictor; lane 8, V. griseus; lane 9, V. timorensis; lane 10, P. muralis; lane 11, T. kempi. M, size marker.
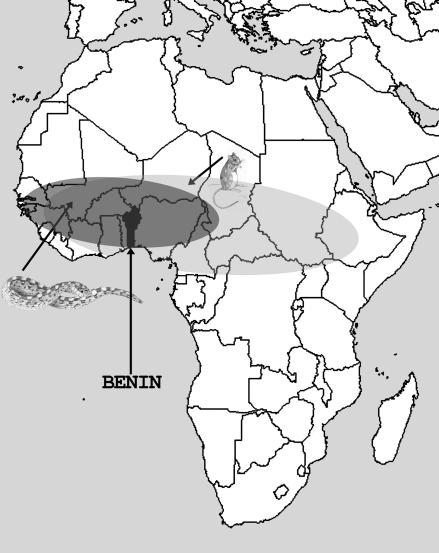
The largest overlap is with E. ocellatus, which also lives in the geographical region where TATV was isolated from Kemp's gerbil (refs. 30 and 34 and Fig. 4). Hence, this snake might even represent a potential source species of VARV-related viruses. Furthermore, the low level of variation among EOC members suggests that the source gene of subfamily members within the genome of the carpet viper is very young and highly active in its retrotransposition of offspring copies [supporting information (SI) Fig. 6]. A low average genetic distance within Echis species and its proposed historically younger radiation in comparison with some other viperid clades (35) supports this observation. We could divide the 38 EOC members into five distinct subsubfamilies based on the distribution of few diagnostic nucleotides (Fig. 2 and SI Fig. 6). In this way, we uncovered the particular source sequence of the TATV-SINE in the genome of E. ocellatus. Four Sauria SINE subsubfamily loci (EOC-I, SI Fig. 6) are identical to the TATV-SINE and cluster together with the TATV snake retroposon in phylogenetic analyses (Fig. 2). Thus, our results demonstrate the genealogy of vertically transferred Sauria SINE members in lizards and snakes and uncover the horizontal transfer of one particular Sauria SINE member to the TATV genome.
Fig. 4.
Overlapping geographical distributions of the rodent T. kempi and the snake E. ocellatus in Africa. The distributions of T. kempi and E. ocellatus are shown in light gray and dark gray ellipses, respectively. TATV was isolated from Kemp's gerbil, caught in Benin (black) at the time of an epidemic of human smallpox (30). Kemp's gerbil and the carpet viper are distributed in the entire Benin region.
The alignment of OPVs revealed that the direct repeats flanking the TATV-SINE were derived from the viral genome (Fig. 1A), indicating that the TPRT was accomplished by using the viral DNA as a target and a Sauria SINE RNA as a template. The fact that Sauria SINEs are only present in the genomes of lizards and snakes constitutes strong evidence that the retrotransposition actually occurred in the reptilian genome. An EN-independent retrotransposition event could have been expected if the TPRT resulted in a retroposon insertion without direct repeats and a 3′ tail truncation of the repetitive element. However, as the complete structure of the TATV-SINE and its flanking regions in the TATV genome are well preserved, we can exclude an EN-independent retrotransposition pathway that was described for human LINE1 insertions in Chinese hamster ovary cells (36, 37). Furthermore, members of the Sauria SINE subsubfamily EOC-I are identical to the TATV-SINE and were identified only in the genome of E. ocellatus (Fig. 3C). The above evidence also indicates that the TATV-SINE was not horizontally transferred at the DNA level in the body of Kemp's gerbil. Instead, the term “horizontal retrotransposition” might be more appropriate (Fig. 5). Thus, it seems most likely that the SINE entered the TATV genome while TATV replicated in snake cells. Although this conclusion is somewhat speculative because it is based on a single retrotransposition event, additional events possibly could be identified upon analysis of SINE insertions in virus genomes after experimental infection of snake cells with TATV, a virological approach certainly of interest in future studies. In this context, it should be noted that the TATV genome that harbored the TATV-SINE was sequenced (28) from the replication-competent gerbil virus that was characterized >30 years ago (30). At that time it was shown that TATV grew well in Vero, LLC-MK2, GMK-AH, and RK-13 cell lines, in primary monkey kidney cells, and in human diploid fibroblasts (FS-9). In these cells TATV produced a cytopathic effect similar to that of VARV but distinct from that of ectromelia, monkeypox, rabbitpox, cowpox, or vaccinia virus. As with VARV, the cytopathic effect produced by TATV started with hypertrophic foci and progressed to the formation of syncytial cells and plaques (30). Overall, most OPVs, especially those closely related to VARV, are presumed capable of establishing infections in humans. Our data provide strong evidence that the carpet viper was infected by a VARV-related virus. Although we cannot rule out the possibility that the snake became temporarily infected by a pathogen, given our data it is reasonable to propose that snakes are possible hosts of TATV and to suggest that snakes should be investigated with regard to whether they may carry variola-like viruses. This may help us to better understand the two closest relatives of VARV in camels and gerbils (28).
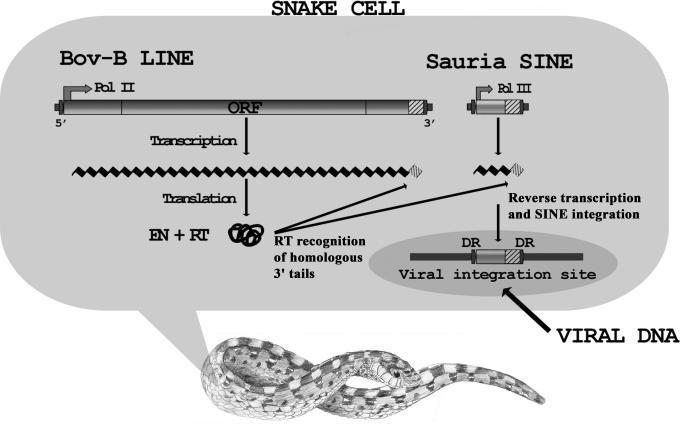
Fig. 5.
Horizontal retrotransposition of a Sauria SINE to a poxvirus in the cell of a highly poisonous snake. SINEs and LINEs are transcribed through internal promoter sequences by RNA polymerases. Each SINE family recruits the enzymatic machinery for retrotransposition (RT and EN) from the corresponding LINE family through an identical 3′ tail sequence (6). Reverse transcription and integration of the Sauria SINE into the viral genome occurred within the body of E. ocellatus. Direct repeats (DR) derived from the virus flanking the Sauria SINE show that the horizontal transfer was actually a horizontal retrotransposition.
If the snake and the rodent is a host of TATV, this virus or related viruses might function as a vector for horizontal transfer of retroposons from reptile genomes to mammalian genomes. Previously, the mite Proctolaelaps regalis was proposed to serve as a vector for the horizontal transfer of mobile P elements between different Drosophila species (38). Here, we propose that poxviruses might similarly be a vector for the horizontal transfer of retroposons from reptiles (specifically advanced snakes) to mammals. Bov-B LINEs were initially discovered in ruminants but later shown to be ubiquitous in squamate genomes. Given the patchy distribution of Bov-B LINEs among mammalian genomes, these LINEs were proposed to have been transferred horizontally from the ancestor of advanced snakes to the ancestor of ruminants (39). Although the issue of horizontal transfer versus vertical inheritance of retroposons has been discussed in detail (40), our data suggest that retroposons might be horizontally transferred from reptiles to mammals, using poxviruses as a vector. In this context, it should be mentioned that many different retroposons, such as Bov-tA and Bov2A (41, 42), are structurally related to Bov-B LINEs that reside in ruminant genomes, whose diversification might have been less dramatic if Bov-B LINEs were not horizontally transferred from reptiles to ruminants. Besides Bov-B LINEs, mammalian genomes contain a large number of mammalian-wide interspersed repeats (MIRs), some of which require the activity of Bov-B LINEs for amplification through the recognition of common tail sequences (43). Therefore, considering that retroposon amplification may underlie accelerated evolution of species (10), the horizontal transfer and insertion of retroposons from reptiles to mammals could have contributed to accelerated evolution of many mammalian lineages.
Materials and Methods
DNA Extraction.
Genomic DNA from lizards, snakes, and rodents was isolated by standard phenol-chloroform extraction (44).
Construction and Screening of Genomic Libraries.
Genomic libraries of two lizards (V. timorensis and V. griseus) and one snake (E. ocellatus) were constructed by complete digestion of genomic DNA with HindIII followed by sedimentation through a sucrose gradient and selection of DNA fragments of up to 2 kbp. The size-fractionated genomic DNA was ligated into HindIII-digested pUC18 plasmids at 37°C overnight. Aliquots of the ligation reactions were transformed into Escherichia coli DH5-α cells. Colonies were transferred to membranes for screening. Sauria SINE loci were screened by using internal SINE primers (TATV-S1F, TWAAGACRCTGAGCTTGTC; TATV-S1R, TGTTACCTTCCCACCGWAGT) labeled by primer extension in the presence of [α-32P]dCTP. Alternatively, [γ-32P]dATP-labeled internal primer sequences were used to screen for novel Sauria SINE sequences. Hybridization was performed at 42°C overnight in a solution of 6× SSC containing 1% SDS, 2× Denhardt's solution, and 100 μg/ml herring sperm DNA and washed 2 times at 45–55°C for 10 min in a solution of 2× SSC containing 1% SDS.
Sequencing of Cloned DNA.
Positive plasmid clones that appeared to contain SINE loci were recolonized, and the inserts cloned and sequenced by using universal primers M4 and RV (TaKaRa Bio, Shiga, Japan). Sequencing was performed with an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
PCR.
Genomic DNA of snakes, lizards and rodents was amplified by PCR, using internal primers specific for the Sauria SINE subfamily EOC (EOCconsF, AGGGACGCAGTGGCTCAGTG; EOCconsR, AAGGTTCCCCCGCACATATG) and for the Sauria SINE subsubfamily EOC-I (EOC-I-F, GCAGTGGCTCAGTGGGTTAA; EOC-IR, TAGTCGTTTCCGACTCTAGGGTG). PCR conditions were as follows: After initial denaturation for 3 min at 94°C, 33 cycles were performed consisting of 30 s of denaturation at 94°C; 50 s of annealing at 60 and 70°C, respectively; and 40 s of elongation at 72°C.
Sequence Analyses.
Multiple sequence alignments were constructed by using CLUSTAL W software (45) followed by visual inspection for errors. Database searches were performed with BLASTN software (46). OPV sequences with the following accession numbers were taken from GenBank: taterapox virus, DQ437594; camelpox virus, AY009089; smallpox strain A virus, DQ441416; smallpox strain B virus, DQ441433; smallpox strain C virus, DQ441419; and cowpox virus, DQ437593. We screened SINE/LINE elements in virus sequences against an enhanced RepeatMasker library (www.repeatmasker.org). We extracted SINE/LINE sequences from the RepeatMasker library and complemented the original version of this library with Sauria SINEs and other novel repetitive elements to construct a new RepeatMasker library that comprised all known SINE/LINE data.
Using TREE-PUZZLE 5.0, a maximum likelihood analysis based on the HKY85 model was performed by using the discrete gamma distribution (eight categories) for site heterogeneity (47). Puzzling supports were based on 25,000 replicates. Frequently encountered CpG sites were not included in the analyses of diagnostic nucleotides and the maximum likelihood analysis.
Supplementary Material
Acknowledgments
We thank Stephan Gimmel (Bern, Switzerland) and Valeria Strebkova (Terrarium, Moscow Zoo, Moscow, Russia) for supplying tissues of snakes and lizards; Paolo Colangelo (University of Rome, Rome, Italy) and Ingo Stürmer (Georg-August-University, Göttingen, Germany) for tissues of rodents; our colleague Hidenori Nishihara for sophisticated RepeatMasker searches and, along with Masaki Kajikawa, for helpful discussions; Melanie Piskurek for art work; and two anonymous reviewers for helpful comments. This work was supported by research grants from the Japan Society for the Promotion of Science (O.P.) and the Ministry of Education, Culture, Sports, Science and Technology of Japan (N.O.).
Abbreviations
- EN
endonuclease
- EOC
Sauria SINE subfamily in advanced snakes isolated from Echis ocellatus
- LINE
long interspersed element
- OPV
Orthopoxvirus
- RT
reverse transcriptase
- SINE
short interspersed element
- TATV
Taterapox virus
- TPRT
target-primed reverse transcription
- VARV
Variola virus.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF033459–EF033488 and EF101172–EF101210).
All Tatera species from Africa have recently been proposed to be distinct from the Asian Tatera indica and were therefore elevated to genus rank (31). The new genus is called Gerbilliscus.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700531104/DC1.
Although only T. kempi is shown in the electrophoresis, several other rodents were tested under these PCR conditions. However, the results were the same (data not shown).
References
- 1.Weiner AM, Deininger PL, Efstratiadis A. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 2.Okada N. Trends Ecol Evol. 1991;6:358–361. doi: 10.1016/0169-5347(91)90226-N. [DOI] [PubMed] [Google Scholar]
- 3.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 4.Ohshima K, Hamada M, Terai Y, Okada N. Mol Cell Biol. 1996;16:3756–3764. doi: 10.1128/mcb.16.7.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kajikawa M, Okada N. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- 6.Ohshima K, Okada N. Cytogenet Genome Res. 2005;110:474–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- 7.Shimamura M, Yasue H, Ohshima K, Abe H, Kato H, Kishiro T, Goto M, Munechika I, Okada N. Nature. 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- 8.Nikaido M, Rooney AP, Okada N. Proc Natl Acad Sci USA. 1999;96:10261–10266. doi: 10.1073/pnas.96.18.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shedlock A, Okada N. BioEssays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 10.Batzer MA, Deininger PL. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz J, Roos C, Zischler H. Cytogenet Genome Res. 2005;108:26–37. doi: 10.1159/000080799. [DOI] [PubMed] [Google Scholar]
- 12.Piskurek O, Austin CC, Okada N. J Mol Evol. 2006;62:630–644. doi: 10.1007/s00239-005-0201-5. [DOI] [PubMed] [Google Scholar]
- 13.Kriegs JO, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishihara H, Hasegawa M, Okada N. Proc Natl Acad Sci USA. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada M, Kido Y, Himberg M, Reist JD, Ying C, Hasegawa M, Okada N. Genetics. 1997;146:355–367. doi: 10.1093/genetics/146.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melamed P, Chong KL, Johansen MV. Nat Genet. 2004;36:786–787. doi: 10.1038/ng0804-786. [DOI] [PubMed] [Google Scholar]
- 17.Matveev V, Nishihara H, Okada N. Mol Biol Evol. 2007 doi: 10.1093/molbev/msm083. in press. [DOI] [PubMed] [Google Scholar]
- 18.Doolittle WF. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 19.Hughes AL, Friedmann R. Mol Phyl Evol. 2005;35:186–195. doi: 10.1016/j.ympev.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 20.McLysaght A, Baldi PF, Gaut BS. Proc Natl Acad Sci USA. 2003;100:15655–15660. doi: 10.1073/pnas.2136653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldo AM, McClure M. J Virol. 1999;73:7710–7721. doi: 10.1128/jvi.73.9.7710-7721.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore KM, Davis JR, Sato T, Yasuda A. Avian Dis. 2000;44:827–841. [PubMed] [Google Scholar]
- 23.Kim TJ, Tripathy DN. Avian Dis. 2001;45:663–669. [PubMed] [Google Scholar]
- 24.Singh P, Schnitzlein WM, Tripathy DN. J Virol. 2003;77:5855–5862. doi: 10.1128/JVI.77.10.5855-5862.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tadese T, Reed WM. J Virol Meth. 2003;110:99–104. doi: 10.1016/s0166-0934(03)00106-x. [DOI] [PubMed] [Google Scholar]
- 26.Henderson DA, Inglesby TV, Bartlett JG, Ascher MS, Eitzen E, Jahrling PB, Hauer J, Layton M, McDade J, Osterholm MT, et al. J Am Med Assoc. 1999;281:2127–2137. doi: 10.1001/jama.281.22.2127. [DOI] [PubMed] [Google Scholar]
- 27.Henderson DA, Fenner F. Clin Inf Dis. 2001;33:1057–1095. doi: 10.1086/323808. [DOI] [PubMed] [Google Scholar]
- 28.Esposito JJ, Sammons SA, Frace AM, Osborne JD, Olsen-Rasmussen M, Zhang M, Govil D, Damon IK, Kline R, Laker M, et al. Science. 2006;313:807–812. doi: 10.1126/science.1125134. [DOI] [PubMed] [Google Scholar]
- 29.Gubser C, Smith GL. J Gen Virol. 2001;83:855–872. doi: 10.1099/0022-1317-83-4-855. [DOI] [PubMed] [Google Scholar]
- 30.Lourie B, Nakano JH, Kemp GE, Setzer HW. J Infect Dis. 1975;132:677–681. doi: 10.1093/infdis/132.6.677. [DOI] [PubMed] [Google Scholar]
- 31.Colangelo P, Granjon L, Taylor PJ, Corti M. Mol Phyl Evol. 2007;42:797–806. doi: 10.1016/j.ympev.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Vidal N, Hedges SB. C R Biol. 2005;328:1000–1008. doi: 10.1016/j.crvi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Fry BG, Vidal N, Norman JA, Vonk FJ, Scheib H, Ramjan SF, Kuruppu S, Fung K, Hedges SB, Richardson MK, et al. Nature. 2006;439:584–588. doi: 10.1038/nature04328. [DOI] [PubMed] [Google Scholar]
- 34.Okuda D, Nozaki C, Sekiya F, Morita T. J Biochem. 2001;129:615–620. doi: 10.1093/oxfordjournals.jbchem.a002898. [DOI] [PubMed] [Google Scholar]
- 35.Lenk P, Kalyabina S, Wink M, Joger U. Mol Phyl Evol. 2001;19:94–104. doi: 10.1006/mpev.2001.0912. [DOI] [PubMed] [Google Scholar]
- 36.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 37.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 38.Houck MA, Clark JB, Peterson KR, Kidwell MG. Science. 1991;253:1125–1129. doi: 10.1126/science.1653453. [DOI] [PubMed] [Google Scholar]
- 39.Kordis D, Gubensek F. Proc Natl Acad Sci USA. 1998;95:10704–10709. doi: 10.1073/pnas.95.18.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malik HS, Burke WD, Eickbush TH. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 41.Okada N, Hamada M. J Mol Evol. 1997;44:52–56. doi: 10.1007/pl00000058. [DOI] [PubMed] [Google Scholar]
- 42.Shimamura M, Abe H, Nikaido M, Ohshima K, Okada N. Mol Biol Evol. 1999;16:1046–1060. doi: 10.1093/oxfordjournals.molbev.a026194. [DOI] [PubMed] [Google Scholar]
- 43.Gilbert N, Labuda D. Proc Natl Acad Sci USA. 1999;96:2869–2874. doi: 10.1073/pnas.96.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blin N, Stafford DW. Nucleic Acid Res. 1976;3:2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson JD, Higgens DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.