Abstract
Prion proteins are key molecules in transmissible spongiform encephalopathies (TSEs), but the precise mechanism of the conversion from the cellular form (PrPC) to the scrapie form (PrPSc) is still unknown. Here we discovered a chemical chaperone to stabilize the PrPC conformation and identified the hot spots to stop the pathogenic conversion. We conducted in silico screening to find compounds that fitted into a “pocket” created by residues undergoing the conformational rearrangements between the native and the sparsely populated high-energy states (PrP*) and that directly bind to those residues. Forty-four selected compounds were tested in a TSE-infected cell culture model, among which one, 2-pyrrolidin-1-yl-N-[4-[4-(2-pyrrolidin-1-yl-acetylamino)-benzyl]-phenyl]-acetamide, termed GN8, efficiently reduced PrPSc. Subsequently, administration of GN8 was found to prolong the survival of TSE-infected mice. Heteronuclear NMR and computer simulation showed that the specific binding sites are the A-S2 loop (N159) and the region from helix B (V189, T192, and K194) to B-C loop (E196), indicating that the intercalation of these distant regions (hot spots) hampers the pathogenic conversion process. Dynamics-based drug discovery strategy, demonstrated here focusing on the hot spots of PrPC, will open the way to the development of novel anti-prion drugs.
Keywords: anti-prion compound, binding sites, chemical chaperone, dynamics-based drug discovery, transmissible spongiform encephalopathy
The accumulation of abnormal protease-resistant prion protein (PrPSc), a conformational isoform of cellular prion protein (PrPC), is a key event in the pathogenesis of transmissible spongiform encephalopathies (TSEs) (1–3), and this host-encoded PrPC has a crucial role in the development of the diseases (4, 5). Because details of the mechanism of conversion from PrPC to PrPSc still remain obscure at this stage, PrPC could be an appropriate molecular target for the drug treatment of TSEs (6) for avoiding the problems associated with the strain differences in PrPSc (7). PrPC is a membrane-anchored glycosylated protein and is well conserved in mammals, and its physiological function is currently argued (8). The three-dimensional structure of recombinant PrPC has been elucidated by NMR (9–13). Briefly, it contains a globular fold with three α-helices (A, B, and C) and a small, imperfectly formed β-sheet (S1 and S2).
The pathogenic conversion process could be related to the thermal stability or the global conformational fluctuation of PrPC. Recently, a metastable state of the PrPC was characterized by using a high-pressure NMR (14), where hydrostatic pressure was elevated up to 2,500 bar in an on-line high-pressure NMR cell. The thermodynamical stability profile shows that diverse residues in helices B and C are less stable, indicating the formation of the intermediate conformation (PrP*) (14). Subsequently, a Carr–Purcell–Meiboom–Gill relaxation–dispersion study revealed that slow fluctuation on a time scale of microseconds to milliseconds occurs at the corresponding regions [supporting information (SI) Fig. 4a], indicating the conformational rearrangements occurring between the native and the sparsely populated high-energy states (15, 16). Interestingly, mutations related to familial forms of the prion diseases are rather concentrated in helices B and C (SI Fig. 4b), and their distribution is somewhat similar to that of slowly fluctuating regions. Moreover, those residues form a major cavity (Fig. 1a, green). Thus, a small substance capable of specifically binding to those residues could stabilize the PrPC conformation because of the decrease in the Gibbs free energy of PrPC upon binding (6), as well as the suppression of the conformational rearrangements by cross-linking of distant regions. We termed this strategy dynamics-based drug discovery. Because PrPSc is gradually degraded in ex vivo experiments (17, 18), such a population shift toward PrPC will result in a decrease in PrPSc population.
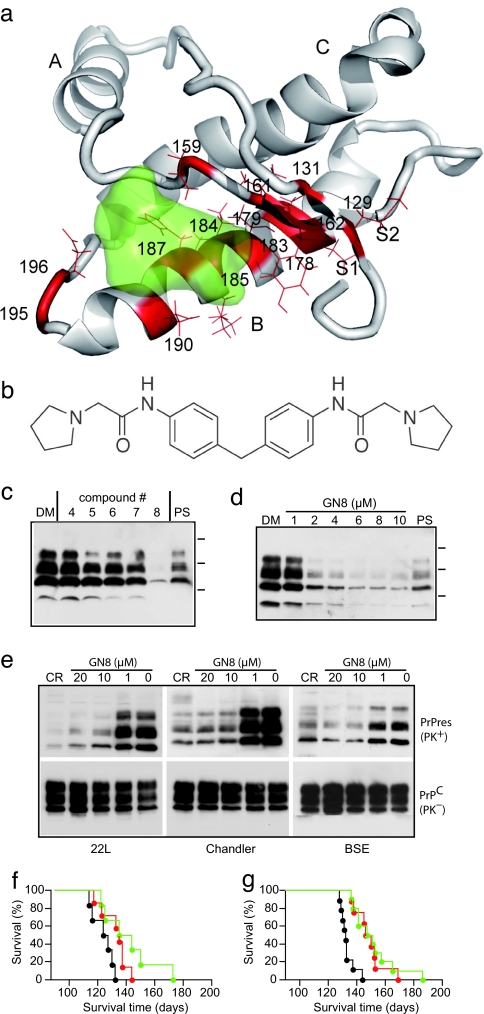
Fig. 1.
In silico and ex vivo screening. (a) Residues undergoing global fluctuation displayed as a wire frame in red mapped on the mouse PrPC structure (residues 124–226) (12), and a binding pocket defined by those residues, colored green. S1, A, S2, B, and C indicate S1 strand, helix A, S2 strand, helix B, and helix C, respectively. The image was created by using PyMol (www.pymol.org). (b) 2-Pyrrolidin-1-yl-N-[4-[4-(2-pyrrolidin-1-yl-acetylamino)-benzyl]-phenyl]-acetamide, termed GN8. (c) Western blotting of PrPSc in GT+FK cells after treatment with different compounds picked up by in silico screening. The cells treated with no. 8 compound showed significant reduction of PrPSc, which was better than that with 10 μg/ml pentosan polysulfate. DM, DMSO at 0.1%; PS, 10 μg/ml pentosan polysulfate. Molecular masses (37, 25, and 15 kDa) are indicated by bars on the right side of the panels. (d) PrPSc signals in serially diluted mock-treated samples and tested samples were scanned and quantified. The IC50 of no. 8 (72-h treatment), as determined by four repeated experiments, was 1.35 μM. (e) GN8 reduced PrPSc also in 22L-, Ch-, and BSE-infected cells. CR, Congo red at 10 μg/ml; PK, proteinase K digestion. (f) Kaplan–Meier's survival curves of FK-infected mice administered GN8 by intraventricular infusion. The control group (n = 6) was killed at 123.8 ± 7.4 days (black line). Average survival time of GN8-treated mice was 132.3 ± 9.2 days (42–70 d.p.i. group, n = 7, red line) and 141.5 ± 18.8 days (70–98 d.p.i. group, n = 6, green line). (g) Kaplan–Meier's survival curves of FK-infected mice administered GN8 subcutaneously. The control group (n = 9) was killed at 133.0 ± 4.9 days (black line). Average survival time of GN8-treated mice was 148.6 ± 10.3 days (67–95 d.p.i. group, n = 8, red line) and 151.4 ± 15.3 days (67–123 d.p.i. group, n = 10, green line).
Based on dynamics-based drug discovery, we conducted a search for chemical compounds that could specifically bind to the unstable residues. We focused on 14 amino acid residues (M129, G131, N159, V161, Y162, D178, C179, T183, I184, L185, H187, T190, G195, and E196, shown in red with side chain in Fig. 1a), located in the loop between helix A and S2 (A-S2 loop) and the loop between helices B and C (B-C loop). A virtual ligand screening program initially picked up 624 chemicals potentially capable of binding to the pocket (Fig. 1a, green) with a binding score better (i.e., less) than −32 (SI Table 1), of 320,000 candidates in a database. We further selected the compounds that formed hydrogen bonds with at least one of the 14 amino acids. With careful examination of binding modes, taking into account Lipinski's rules (19), we then selected the 59 compounds showing the lowest predicted binding free energy.
Results
To evaluate the effect of the selected compounds on the conversion of PrP, we next conducted ex vivo screening. We used a mouse neuronal cell culture uninfected (GT1-7) and persistently infected with human TSE agent (Fukuoka-1 strain), designated GT+FK (20). Of the 59 compounds, we tested the 44 that were commercially available (see SI Table 1). Among these, 2-pyrrolidin-1-yl-N-[4-[4-(2-pyrrolidin-1-yl-acetylamino)-benzyl]-phenyl]-acetamide (compound number 8, molecular weight 420) (Fig. 1b) was found to significantly inhibit the PrPSc production in the GT+FK at 10 μM (Fig. 1c). The other compounds either had little effect even at a higher dose (IC50 > 100 μM) or were highly toxic to the cells at 1 μM. The effect of the compound (now designated GN8) was dose-dependent (Fig. 1d), and by repeating the experiment we established that the effective concentration for 50% reduction of PrPSc (IC50) over 72 h was ≈1.35 μM. The normal PrPC expression in uninfected cells was unaffected. A similar effect was confirmed by using other scrapie-infected GT cells (GT+22L and GT+Ch) (20), and also by using GT+BSE cells stably infected with mouse-adapted bovine spongiform encephalopathy (BSE) (Fig. 1e). This confirms that the action of GN8 is not strain-specific. To see the effect of GN8 in vivo, mice inoculated with 20 μl of 10% FK-1 mouse brain homogenate were given the compound at a dose of 250 μg/kg per day by intraventricular infusion using osmotic pumps (Alzet Durect, Cupertino, CA) during 42–70 days postinoculation (d.p.i.) or 70–98 d.p.i. Although the vehicle-only control (5% glucose/saline) showed that the average survival time was 123.8 ± 7.4 (n = 6), as expected, the GN8-treated mice showed slightly but significantly prolonged survival even after the appearance of clinical signs [132.3 ± 9.2 days (n = 7) in the 42–70 d.p.i. group and 141.5 ± 18.8 days (n = 6) in the 70–98 d.p.i. group; P < 0.05], as shown in Fig. 1f. The effect on the survival of infected mice is limited here because of the transient administration of GN8.
The mice administrated subcutaneously with GN8 at a dose of 8.9 mg/kg per day using infusion pumps survived longer than control mice. Whereas the control [5% glucose/saline (63–120 d.p.i. group)] showed that the average survival time was 133.0 ± 4.9 days (n = 9), the GN8-treated mice showed slightly but significantly prolonged survival [148.6 ± 10.3 days (n = 8) in the 67–95 d.p.i. group and 151.4 ± 15.3 days (n = 10) in the 67–123 d.p.i. group; P < 0.01], as shown in Fig. 1g. Pharmacological analysis using labeled GN8 is currently going on. On the other hand, pentosan polysulfate was not effective at all on the survival time when given peripherally (data not shown). Thus, GN8 could be a potential lead compound for prion diseases.
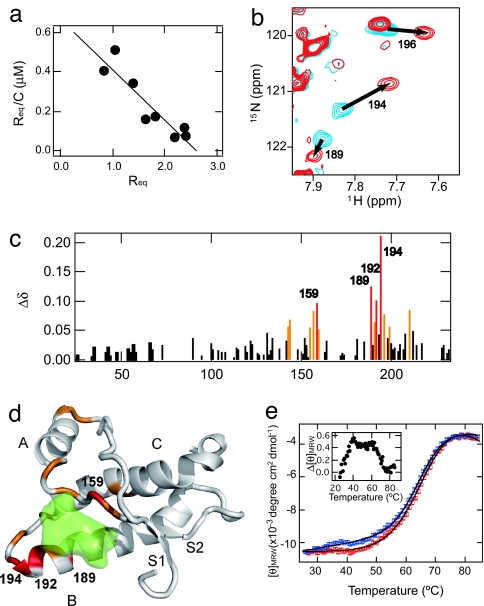
Binding of GN8 to PrPC was confirmed by the surface plasmon resonance (21), and its dissociation constant was estimated to be 3.9 ± 0.2 × 10−6 M from the Scatchard plot (Fig. 2a; see also SI Fig. 5 and SI Methods). To identify the putative sites for interaction of GN8 with PrP, we analyzed the chemical shift perturbation of 1H-15N heteronuclear single quantum coherence NMR spectra (22) of a uniformly 15N-labeled PrP. A comparison of the spectra revealed that three cross peaks (corresponding to V189, K194, and E196) shifted significantly upon the addition of GN8 (Fig. 2b), apparently in a fast-exchange mode. The GN8 concentration did not appear to significantly affect line broadening. Most of the perturbed residues were located in the S2-A loop, the B-helix, or the B-C loop regions, indicating the specific binding between GN8 and PrPC (Fig. 2c). Fig. 2d shows the markedly perturbed residues mapped onto a three-dimensional PrP model.
Fig. 2.
Interaction of an anti-prion compound, GN8, and a recombinant mouse PrPC. (a) Scatchard plot (Req vs. Req/C, where Req and C are the equilibrium response of SPR and the concentration of GN8, respectively) of the specific binding of GN8 with the PrP obtained by a surface plasmon resonance sensorgram. From the slope of the line, Kd was estimated to be 3.9 μM. (Details are shown in SI Fig. 5 and SI Methods.) (b) An overlay of the 1H-15N heteronuclear single quantum coherence NMR spectra of PrP in the absence and presence of GN8. Blue contours show the spectrum of PrPC without GN8, and red contours show the spectrum in the presence of 1.0 mM GN8 at pH 4.5. (c) Plot of the weighted averages of the 1H and 15N chemical shift changes, calculated by using the function Δδ = [(Δδ1H)2 + 0.17(Δδ15N)2]1/2 against the residue number. The absence of bars in the plot indicates unassigned residues, proline residues, or unmeasured shifts due to resonance overlaps. Perturbed residues with Δδ values of >0.9 ppm are shown in red, and those with 0.9 > Δδ > 0.5 ppm are in orange. (d) Mapping of the perturbed residues on the structure of mPrP(121–231) (PDB entry 1AG2). The perturbed residues with Δδ values of >0.9 ppm are shown in red, and those with 0.9 > Δδ > 0.5 ppm are in orange. Binding pocket is overlaid in green. S1, A, S2, B, and C indicate S1 strand, helix A, S2 strand, helix B, and helix C, respectively. The image was created by using PyMol. (e) Thermal unfolding profiles of recombinant mouse PrP (amino acids 23–231, 5 μM) without (blue) or with (red) GN8 (10 μM). CD intensities of PrP in the presence of GN8 were normalized to those of PrP without GN8, and fitted curves (see SI Methods) are also shown. Binding with GN8 stabilizes the conformation of PrPC. (Inset) The difference in extinction coefficients at 222 nm of PrP without GN8 and those in the presence of GN8, as a function of temperature.
To investigate whether GN8 indeed stabilizes the PrPC conformation, we measured the thermal stability using CD. The thermal unfolding curves of recombinant mouse PrP, monitored by the molar ellipticity at 222 nm, representing the helical content of PrP and the overall unfolding behavior with (red) or without (blue) GN8, were compared quantitatively (Fig. 2e and SI Methods). Parameter sets obtained by a nonlinear fit, i.e., melting temperature (Tm) and enthalpy change (ΔH), without GN8 were 65.3 ± 0.4°C and 35.0 ± 1.9 kcal/mol, respectively, whereas those with GN8 were 67.7 ± 0.6°C and 41.8 ± 2.2 kcal/mol, respectively. This indicates that the binding of GN8 stabilizes the PrPC conformation significantly. Intriguingly, the accumulation of the intermediate was demonstrated by an early increase in ellipticity at ≈40°C before the global unfolding as shown in Fig. 2e Inset. However, this is strongly suppressed in the presence of GN8. Thus, GN8 suppressed the production of both PrPSc (Fig. 1 c–e) and PrPU (thermally unfolded state) (Fig. 2e) by reducing the intermediate population (14).
Discussion
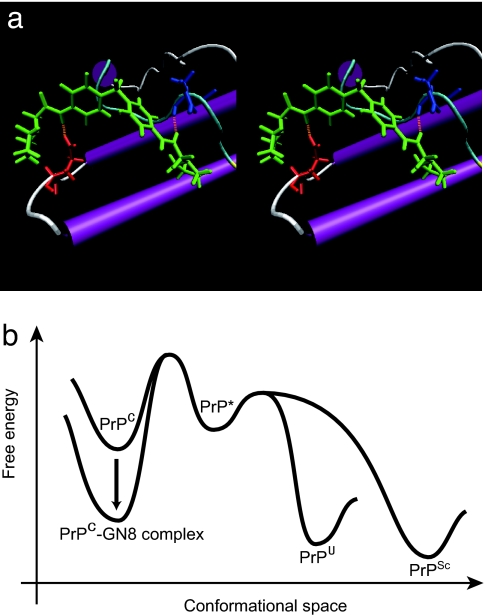
The structure of the PrPC–GN8 complex was further analyzed by computer simulation using the refined energy minimization procedure with flexible receptor side chain. GN8 (Fig. 3a) connected distant residues, N159 (A-S2 loop) and E196 (B-C loop), by hydrogen bonds. The regions with the significant chemical shift changes described above (Fig. 2d) were in good agreement with the binding regions of GN8 in the simulated structure of the prion–GN8 complex (Fig. 3a). Intriguingly, K194 at the C terminus of the helix B and E196 in the B-C loop undergo slow exchange dynamics (16) (SI Fig. 4b). Indeed, the mutation E196K causes a rapidly progressive dementia and ataxia (23) and could be expected to greatly reduce the protein stability because of the elimination of salt bridges between E196, R156, and K194 (23).
Fig. 3.
Inhibitory mechanism of GN8 for pathogenic conversion. (a) Stereoview of the optimized complex structure of the GN8 and mouse PrPC, with putative hydrogen bonds, calculated by using ICM version 3.0. Presumed hydrogen bonds between GN8 (green) and E196 (red) and between GN8 (green) and N159 (blue) are shown in orange (dotted lines). The images were created with VMD (52). (b) Illustration of the Gibbs free energy as a function of the conformational space to explain the inhibitory mechanism of a chemical chaperone, GN8. GN8 stabilizes the PrPC conformation and reduces the population of PrP*, PrPSc, and PrPU. PrP* may not interact with GN8, because the specific conformation around the binding sites would be lost (14). Thus, the free energy level of PrP*, PrPU, and PrPSc would not change much in the presence of GN8.
Intercalation of these two binding regions (A-S2 loop and B-C loop) may be essential to stabilize the PrPC conformation. For example, two representative binding sites, N159 and E196, are close together (distance between Cα atoms ≈ 15.4 Å) in PrPC structure (12), but in the hypothetical PrPSc structure (24) they are considerably more distant (distance between Cα atoms ≈ 45.2 Å). Because GN8 connects distant regions (N159 and E196) in the PrP sequence (36 aa) by hydrogen bonds, large conformational shift may be significantly prohibited, and thus intermediate (PrP*) and further PrPSc or PrPU formation may be also blocked.
Matsuda et al. (25) reported about the chemical chaperone therapy for GM1-gangliosidosis, but there has been no direct evidence for such a mechanism working on the anti-prion compounds. Experimental evidences presented here fully support the concept that GN8 acts as a chemical chaperone to stabilize the normal prion protein (PrP) conformation. As illustrated in Fig. 3b, free energy of PrPC–GN8 complex is significantly less than that of PrPC, so the populations of the transition state, PrP*, PrPU, and PrPSc may be reduced accordingly.
Over the past 10 years there have been various efforts to find out small compounds to reduce PrPSc population. These include porphyrins (26, 27), Congo red and its derivatives (28–30), acridine and phenothiazine derivatives (17, 31, 32), heparan sulfate (33), aminoglycan, and polyamines (34, 35). Simultaneously, various technological developments have been reported including structure-based drug design (36) followed by the structure–activity relationship study (37), small interfering RNA (38), library screening (18), high-throughput screening (39), chimeric ligand approach (40), and so on. Although strategies for drug discovery used in these studies have a broad spectrum from empirical to rational preponderance, there has been no report on the residue-specific evidences for the binding regions of anti-prion compounds. For instance, the effect of BF-168 depends on the strains (41), suggesting that BF-168 may not interact with PrPC but with PrPSc in a strain-dependent manner, but this is still indirect evidence.
The structure of GN8 somewhat resembles a number of other small PrPSc inhibitors, such as the Congo red, which is able to interact with the N-terminal domain of PrPC (42). However, we could not find out any evidence for the interaction between GN8 and the N-terminal domain. The inhibitory mechanism of GN8 and that of Congo red seem to be quite different because of the following reasons: (i) Congo red has two sulfonates on the edge of the molecule, but GN8 does not have negative charge. Thus, GN8 may not strongly interact with His+ at the octapeptide repeats of the N-terminal domain. (ii) Although Congo red can cause aggregation of recombinant PrPC (42), GN8 never causes aggregation even at a relatively high concentration (≈0.03 mM) in NMR tube. (iii) Chemical shift changes caused by binding with GN8 are not significant at the N-terminal half region as shown in Fig. 2c. (iv) SPR affinity profile between GN8 and the C-terminal half of PrPC(120–230) and that between GN8 and the full-length PrPC(23–231) were quite similar, and the calculated dissociation constants were also close (≈5 μM), supporting our conclusion that the major binding regions of GN8 locate at the C-terminal domain. On the other hand, the interaction sites of GN8 with the GPI-anchored PrPC on the cell surface could be different from those with the free PrPC. However, a carboxymethyl moiety on the SPR sensor chip has a negative charge like a phosphate moiety on the cell membrane. Because electrostatic environments surrounding GN8 and prion on these surfaces are similar, GN8 may also interact with the C-terminal domain of the GPI-anchored PrPC.
We found here the effective anti-prion compound GN8, which specifically binds with the hot spots undergoing the slow fluctuation on the time scale of microseconds to milliseconds and exclusively interferes with the pathogenic conversion. According to the dynamics-based drug discovery strategy, we found >20 compounds with any anti-prion activity, and the hit rate is ≈10% at present (data not shown). However, no compound has been more effective than GN8. For instance, we found a compound, GN4 (see SI Table 1), whose structure is quite similar to GN8, but the computer simulation suggested that the binding sites are R156 and N159, which are quite close (2 aa). Thus, GN4 is expected to have less inhibitory effect on the global fluctuation of PrPC, and indeed IC50 of GN4 was >100 μM (data not shown).
To potentially become of any use clinically, GN8 will need to clear many pharmacological hurdles; however, our basic principle presented here constitutes a promising strategy with which to approach the discovery of therapeutic compounds for TSE. Additionally, application of the dynamics-based drug discovery approach, based on the experimentally identified hot spot (43), will make the mass screening of chemical compounds more efficient, especially for diseases related to protein misfolding (44).
Materials and Methods
Virtual Ligand Screening.
We performed in silico screening of ligands on 320,000 compounds in the Available Chemicals Directory (MDL Information Systems, San Leandro, CA) for specific binding to mouse PrPC (12). Residues with the exchange time constant, τex, between two sites of >10 ms are displayed in SI Fig. 4a (16). The software used was ICM version 3.0 (Molsoft, La Jolla, CA). The program, by global optimization of the entire flexible ligand in the receptor (mouse PrPC) field (45), came up with 624 candidates potentially capable of binding to the pocket with a binding score better (i.e., less) than −32, which roughly corresponds to binding energy (kcal/mol). Then, a more refined energy-minimization procedure using a flexible receptor side chain was conducted and further selected the compounds that formed hydrogen bonds with at least one of the 14 amino acids.
Chemical Compounds.
Chemical compounds selected from our virtual ligand screening simulation were purchased from Aldrich Chemical Company (Milwaukee, WI), G & J Research Chemicals (Devon, U.K.), Wako Pure Chemical (Osaka, Japan), Interchim (Montlucon, France), Labotest (Niederschoena, Germany), Florida Center for Heterocyclic Compounds (Gainesville, FL), ChemBridge (San Diego, CA), Maybridge Chemical Company (Cornwall, U.K.), TimTec (Newark, NJ), Ambinter (Paris, France), Oak Samples (Kier, U.K.), Scientific Exchange (Center Ossipee, NH), ChemStar (Moscow, Russia), ChemDiv (San Diego, CA), and AsInEx (Moscow, Russia), or kindly provided by the Drug Synthesis and Chemistry Branch, Developmental Therapeutic Program, Division of Cancer Treatments and Diagnosis, National Cancer Institute. Detailed information on the sources for all of the compounds is shown in SI Table 1. The compounds were dissolved with DMSO for the in vitro screening and with distilled water for the spectral measurements. GN8 hydrochloride salt was prepared for in vivo test as follows: GN8 was first dissolved with dioxane and added to 3 N HCl in dioxane. The solvent was evaporated in vacuo, and the residue was recrystallized from acetone to give the hydrochloride salt of GN8. GN8 hydrochloride salt (2-pyrrolidin-1-yl-N-[4-[4-(2-pyrrolidin-1-yl-acetylamino)-benzyl]-phenyl]-acetamide dihydrochloride); white solid; anal. calcd. for C25H34Cl2N4O2: C, 60.85; H, 6.94; Cl, 14.37; N, 11.35; O, 6.48. %Found: C, 60.83; H, 6.95; N, 11.35.
Recombinant Mouse PrP.
The DNA of mouse PrP(23–231) was amplified by PCR and cloned into the expression vector pET101/D-TOPO (Invitrogen, Carlsbad, CA). As shown in SI Methods, the 15N-labeled recombinant PrP for NMR measurements was expressed in Escherichia coli strain BL21 Star (DE3) (Invitrogen), grown in 15N-labeled minimum medium Spectra9 (Spectra Gases, Branchburg, NJ), and purified by a Ni-chelating affinity chromatography method (46). Oxidization and refolding of the purified protein (47) were performed in buffer containing 4 M urea at pH 8. A recombinant PrP sample for the surface plasmon resonance sensorgram experiment was obtained by a similar procedure using a non-isotope-labeled LB medium instead of the Spectra9.
Cell Culture and Antibodies.
The immortalized mouse neuronal cell line GT1-7 was cultured as described (20). GT1-7 cells stably infected with Fukuoka-1, 22L, or Chandler/RML (designated GT+FK, GT+22L, or GT+Ch, respectively) were maintained for more than a year in our laboratory. GT+BSE cells were infected ex vivo in our laboratory with mouse-adapted BSE agent (a kind gift from T. Yokoyama, National Institute of Animal Health, Tsukuba, Japan). Stock solutions of compounds were prepared fresh in 100% DMSO at 100 mM and stored at 4°C. Before use, compounds were diluted with medium as indicated. Control cells were treated with medium containing solvent alone (0.1%). Approximately 2 × 105 cells were plated in each well of a six-well plate, and drug treatment was started 15 h later. After 72 h of incubation, cells were lysed in 150 μl of 1× Triton X-100/DOC lysis buffer (48), and samples normalized to 2 mg of protein per milliliter. Western blotting for PrPSc was done as described previously (48). Anti-mouse PrP antisera (SS28) (49) and SAF32 antibody (SPI-BIO, Montigny le Bretonneux, France) were used for PrPSc and PrPC, respectively, as the primary antibody. The signals were visualized by ECL-plus (Amersham, Buckinghamshire, U.K.) and scanned by using Fluor Chem (Alpha Innotech, San Leandro, CA).
NMR Measurements and Data Analysis.
For NMR measurements, 0.6 mg/ml 15N-uniformly labeled mouse PrP(23–231) was prepared in 30 mM acetate-d3 buffer (pH 4.5) containing 1 mM NaN3, 4.5 μM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride, 20 μM EDTA, 0.4 μM Bestatin, 0.06 μM pepstatin, 0.06 μM E-64, and 1 nM sodium 2,2-dimethyl-2-silapentane-5-sulfonate dissolved in 90% H2O/10% D2O. NMR spectra were recorded at 20.0°C on an Avance600 spectrometer (Bruker, Rheinstetten, Germany) at Gifu University. The spectrometer operates at 1H frequency of 600.13 MHz and 15N frequency of 60.81 MHz. A 5-mm 1H inverse detection probe with triple-axis gradient coils was used for all measurements. 1H-15N heteronuclear single quantum coherence spectra were acquired with 2,048 complex points covering 9,600 Hz for 1H and 256 complex points covering 1,200 Hz for 15N. NMR data were processed by using the XWIN-NMR software package (Bruker) and Sparky (50). Resonance frequencies in these spectra were identified by using the chemical shift lists on mouse PrP(23–231) and PrP (121–231) (51). For the chemical shift perturbation experiments, aliquots of 2.5 μl of 0, 10, or 100 mM GN8 solutions in DMSO-d6 were added to 0.6 mg/ml 15N PrP(23–231) in a 5-mm-diameter Shigemi microtube; the final concentration of GN8 is 0, 1, and 10 mM, respectively. The backbone 1H and 15N chemical shifts for the GN8-bound protein were assigned by tracing the corresponding peaks in 1H-15N heteronuclear single quantum coherence spectra measured at various concentrations of GN8.
Preparation of GN8 for Injection and in Vivo Test.
The GN8 hydrochloride salt was dissolved in saline with 5% glucose and sterilized by passing through a 0.2-μm filter. The concentration of the stock solution was adjusted to 10 mg/ml and kept at 4°C until use. Ten percent FK-infected brain homogenate was inoculated into right temporal lobes of 4-week-old male mice (ddY), followed by intraventricular infusion of GN8 using an osmotic pump. Six or 10 weeks after inoculation, an osmotic pump was inserted subcutaneously and the infusion cannula was implanted into the right ventricle. The pump was filled with GN8 (1.4 mg/ml) or saline with 5% glucose. In a parallel experiment, pentosan polysulfate (Bene, Munich, Germany) was administered at 200 μg/kg per day by intraventricular or i.p. infusion. All mice were carefully examined daily for neurological signs, and the incubation period was monitored. Survival data were statistically evaluated according to Kaplan–Meier's method using StatMateIII (ATMS, Tokyo, Japan).
In case of subcutaneous infusion of GN8, the concentration of the stock solution was adjusted to 50 mg/ml. One percent FK-infected brain homogenate was inoculated in the same way, and an osmotic pump including GN8 was inserted subcutaneously at 9 weeks after inoculation.
Supplementary Material
Acknowledgments
Some compounds were kindly provided by the Drug Synthesis and Chemistry Branch, Developmental Therapeutic Program, Division of Cancer Treatments and Diagnosis, National Cancer Institute. We thank Ms. Sakiko Morishita and Mr. Yuki Matsui for technical help. K. Kuwata was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and grants from the Ministry of Health, Labour, and Welfare. This study was supported by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation.
Abbreviations
- TSE
transmissible spongiform encephalopathy
- PrP
prion protein
- PrPC
cellular isoform of PrP
- PrPSc
scrapie isoform of PrP
- PrP*
sparsely populated high-energy state of PrP
- BSE
bovine spongiform encephalopathy
- d.p.i.
days postinoculation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702671104/DC1.
References
- 1.Prusiner SB. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. Crit Rev Biochem Mol Biol. 1991;26:397–438. doi: 10.3109/10409239109086789. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 5.Sailer A, Bueler H, Fischer M, Aguzzi A, Weissmann C. Cell. 1994;77:967–968. doi: 10.1016/0092-8674(94)90436-7. [DOI] [PubMed] [Google Scholar]
- 6.Cohen FE, Pan KM, Huang Z, Baldwin M, Fletterick RJ, Prusiner SB. Science. 1994;264:530–531. doi: 10.1126/science.7909169. [DOI] [PubMed] [Google Scholar]
- 7.Peretz D, Williamson RA, Legname G, Matsunaga Y, Vergara J, Burton DR, DeArmond SJ, Prusiner SB, Scott MR. Neuron. 2002;34:921–932. doi: 10.1016/s0896-6273(02)00726-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhang CC, Steele AD, Lindquist S, Lodish HF. Proc Natl Acad Sci USA. 2006;103:2184–2189. doi: 10.1073/pnas.0510577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James TL, Liu H, Ulyanov NB, Farr-Jones S, Zhang H, Donne DG, Kaneko K, Groth D, Mehlhorn I, Prusiner SB, Cohen FE. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wuthrich K. Proc Natl Acad Sci USA. 2005;102:646–650. doi: 10.1073/pnas.0409008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 13.Lysek DA, Schorn C, Nivon LG, Esteve-Moya V, Christen B, Calzolai L, von Schroetter C, Fiorito F, Herrmann T, Guntert P, Wuthrich K. Proc Natl Acad Sci USA. 2005;102:640–645. doi: 10.1073/pnas.0408937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuwata K, Li H, Yamada H, Legname G, Prusiner SB, Akasaka K, James TL. Biochemistry. 2002;41:12277–12283. doi: 10.1021/bi026129y. [DOI] [PubMed] [Google Scholar]
- 15.Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 16.Kuwata K, Kamatari YO, Akasaka K, James TL. Biochemistry. 2004;43:4439–4446. doi: 10.1021/bi036123o. [DOI] [PubMed] [Google Scholar]
- 17.Doh-Ura K, Iwaki T, Caughey B. J Virol. 2000;74:4894–4897. doi: 10.1128/jvi.74.10.4894-4897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kocisko DA, Baron GS, Rubenstein R, Chen J, Kuizon S, Caughey B. J Virol. 2003;77:10288–10294. doi: 10.1128/JVI.77.19.10288-10294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 20.Milhavet O, McMahon HE, Rachidi W, Nishida N, Katamine S, Mange A, Arlotto M, Casanova D, Riondel J, Favier A, Lehmann S. Proc Natl Acad Sci USA. 2000;97:13937–13942. doi: 10.1073/pnas.250289197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Touil F, Pratt S, Mutter R, Chen B. J Pharm Biomed Anal. 2006;40:822–832. doi: 10.1016/j.jpba.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Zuiderweg ER. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 23.Peoc'h K, Manivet P, Beaudry P, Attane F, Besson G, Hannequin D, Delasnerie-Laupretre N, Laplanche JL. Hum Mutat. 2000;15:482. doi: 10.1002/(SICI)1098-1004(200005)15:5<482::AID-HUMU16>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 24.Govaerts C, Wille H, Prusiner SB, Cohen FE. Proc Natl Acad Sci USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuda J, Suzuki O, Oshima A, Yamamoto Y, Noguchi A, Takimoto K, Itoh M, Matsuzaki Y, Yasuda Y, Ogawa S, et al. Proc Natl Acad Sci USA. 2003;100:15912–15917. doi: 10.1073/pnas.2536657100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caughey WS, Raymond LD, Horiuchi M, Caughey B. Proc Natl Acad Sci USA. 1998;95:12117–12122. doi: 10.1073/pnas.95.21.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Priola SA, Raines A, Caughey WS. Science. 2000;287:1503–1506. doi: 10.1126/science.287.5457.1503. [DOI] [PubMed] [Google Scholar]
- 28.Caspi S, Halimi M, Yanai A, Sasson SB, Taraboulos A, Gabizon R. J Biol Chem. 1998;273:3484–3489. doi: 10.1074/jbc.273.6.3484. [DOI] [PubMed] [Google Scholar]
- 29.Milhavet O, Mange A, Casanova D, Lehmann S. J Neurochem. 2000;74:222–230. doi: 10.1046/j.1471-4159.2000.0740222.x. [DOI] [PubMed] [Google Scholar]
- 30.Sellarajah S, Lekishvili T, Bowring C, Thompsett AR, Rudyk H, Birkett CR, Brown DR, Gilbert IH. J Med Chem. 2004;47:5515–5534. doi: 10.1021/jm049922t. [DOI] [PubMed] [Google Scholar]
- 31.Korth C, May BC, Cohen FE, Prusiner SB. Proc Natl Acad Sci USA. 2001;98:9836–9841. doi: 10.1073/pnas.161274798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.May BC, Fafarman AT, Hong SB, Rogers M, Deady LW, Prusiner SB, Cohen FE. Proc Natl Acad Sci USA. 2003;100:3416–3421. doi: 10.1073/pnas.2627988100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adjou KT, Simoneau S, Sales N, Lamoury F, Dormont D, Papy-Garcia D, Barritault D, Deslys JP, Lasmezas CI. J Gen Virol. 2003;84:2595–2603. doi: 10.1099/vir.0.19073-0. [DOI] [PubMed] [Google Scholar]
- 34.Perez M, Wandosell F, Colaco C, Avila J. Biochem J. 1998;335:369–374. doi: 10.1042/bj3350369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Supattapone S, Nguyen HO, Cohen FE, Prusiner SB, Scott MR. Proc Natl Acad Sci USA. 1999;96:14529–14534. doi: 10.1073/pnas.96.25.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrier V, Wallace AC, Kaneko K, Safar J, Prusiner SB, Cohen FE. Proc Natl Acad Sci USA. 2000;97:6073–6078. doi: 10.1073/pnas.97.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.May BC, Zorn JA, Witkop J, Sherrill J, Wallace AC, Legname G, Prusiner SB, Cohen FE. J Med Chem. 2007;50:65–73. doi: 10.1021/jm061045z. [DOI] [PubMed] [Google Scholar]
- 38.Daude N, Marella M, Chabry J. J Cell Sci. 2003;116:2775–2779. doi: 10.1242/jcs.00494. [DOI] [PubMed] [Google Scholar]
- 39.Bertsch U, Winklhofer KF, Hirschberger T, Bieschke J, Weber P, Hartl FU, Tavan P, Tatzelt J, Kretzschmar HA, Giese A. J Virol. 2005;79:7785–7791. doi: 10.1128/JVI.79.12.7785-7791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dollinger S, Lober S, Klingenstein R, Korth C, Gmeiner P. J Med Chem. 2006;49:6591–6595. doi: 10.1021/jm060773j. [DOI] [PubMed] [Google Scholar]
- 41.Ishikawa K, Kudo Y, Nishida N, Suemoto T, Sawada T, Iwaki T, Doh-ura K. J Neurochem. 2006;99:198–205. doi: 10.1111/j.1471-4159.2006.04035.x. [DOI] [PubMed] [Google Scholar]
- 42.Caughey B, Caughey WS, Kocisko DA, Lee KS, Silveira JR, Morrey JD. Acc Chem Res. 2006;39:646–653. doi: 10.1021/ar050068p. [DOI] [PubMed] [Google Scholar]
- 43.Clackson T, Wells JA. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 44.Soto C, Estrada L, Castilla J. Trends Biochem Sci. 2006;31:150–155. doi: 10.1016/j.tibs.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Schapira M, Raaka BM, Samuels HH, Abagyan R. Proc Natl Acad Sci USA. 2000;97:1008–1013. doi: 10.1073/pnas.97.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bocharova OV, Breydo L, Parfenov AS, Salnikov VV, Baskakov IV. J Mol Biol. 2005;346:645–659. doi: 10.1016/j.jmb.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 47.Lu BY, Beck PJ, Chang JY. Eur J Biochem. 2001;268:3767–3773. doi: 10.1046/j.1432-1327.2001.02283.x. [DOI] [PubMed] [Google Scholar]
- 48.Nishida N, Harris DA, Vilette D, Laude H, Frobert Y, Grassi J, Casanova D, Milhavet O, Lehmann S. J Virol. 2000;74:320–325. doi: 10.1128/jvi.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishida N, Tremblay P, Sugimoto T, Shigematsu K, Shirabe S, Petromilli C, Erpel SP, Nakaoke R, Atarashi R, Houtani T, et al. Lab Invest. 1999;79:689–697. [PubMed] [Google Scholar]
- 50.Goddard TD, Kneller DG. SPARKY. San Francisco: Univ of California; 2001. Version 3. [Google Scholar]
- 51.Riek R. Zürich, Switzerland: Swiss Federal Institute of Technology; 1988. PhD thesis. [Google Scholar]
- 52.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:27–28. 33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.