Abstract
Site-directed spin labeling (SDSL) was used to examine and compare transmembrane signaling events in the bacterial outer-membrane transport proteins BtuB, FecA, and FhuA. These proteins extract energy for transport by coupling to the transperiplasmic protein TonB, an interaction that is thought to be mediated by the Ton box, a highly conserved energy-coupling motif in these transporters. In the ferric citrate transporter, FecA, SDSL indicates that the Ton box undergoes a substrate-induced disorder transition similar to that seen for BtuB, the vitamin B12 transporter. This conformational change produces an aqueous exposed, highly disordered protein fragment, which likely regulates transporter–TonB interactions. However, in the ferrichrome transporter, FhuA, SDSL does not reveal a substrate-induced unfolding transition. In this protein, with or without substrate, the Ton box conformation is found to be highly dynamic and constitutively unfolded. In addition, SDSL indicates that structural features seen in high-resolution models are not found in membrane-associated FhuA. Taken together, these data indicate that the Ton box of FhuA may always be available for interactions with TonB, implying that transporter–TonB interactions in FhuA are either constitutive or not regulated by the Ton box configuration.
Keywords: EPR spectroscopy, membrane protein, site-directed spin labeling
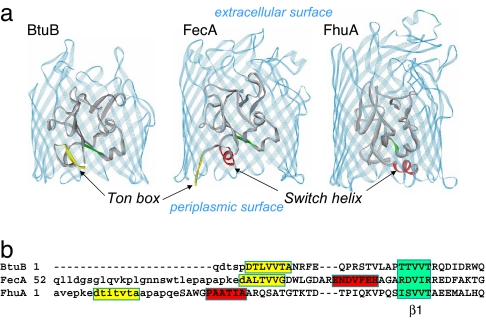
The bacterial outer membrane contains a series of high-affinity transporters that function in the uptake of rare nutrients, such as iron and vitamin B12. These transporters extract energy for transport from the inner membrane by coupling to the transperiplasmic protein TonB and are therefore termed TonB-dependent (1–5). High-resolution crystal structures have been obtained for a number of TonB-dependent transport proteins, and structures for three members of this family, the vitamin B12 transporter, BtuB (6), the ferric citrate transporter, FecA (7, 8), and the ferrichrome transporter, FhuA (9, 10), are shown in Fig. 1a. These proteins have homologous structures based on a 22-stranded β-barrel, extracellular binding loops, and an N-terminal region that is folded into the barrel. Substrate binding occurs in the extracellular binding loops and triggers conformational changes in the N-terminal region near the periplasmic surface, thus transducing a signal across the outer membrane (11).
Fig. 1.
Crystal structures of TonB-dependent transporters. (a) The structures of BtuB [Protein Data Bank (PDB) ID code 1NQE], FecA (PDB ID code 1PNZ), and FhuA (PDB ID code 1BY3) in the absence of substrate. The N-terminal region of these transporters is folded into the protein β-barrel (solid ribbon in gray). The Ton box for BtuB and FecA is shown in yellow. The Ton box is not resolved in FhuA. For FecA and FhuA, the H1 or switch helix is also shown in red. Both the Ton box and switch helix lie on the periplasmic surface of the transporter. (b) Structure-based sequence alignment of the N-terminal regions of BtuB, FecA, and FhuA, given previously (8). The Ton box is highlighted in yellow, and the switch helix in FecA and FhuA is in red. Segments showing the position of the first β-strand (β1) in the N-terminal fold are shown in green. Lowercase sequences indicate residues that were not resolved in the structural models.
An important step in this transport cycle involves the reversible association between the transporter and TonB, which is mediated by a highly conserved seven-residue segment in these transporters termed the Ton box (Fig. 1b). Site-directed spin labeling (SDSL) and chemical derivatization studies of BtuB (11–14) demonstrate that the Ton box undergoes a substrate-dependent conformational change and an order-to-disorder transition. In the absence of substrate, the Ton box adopts a folded conformation within the interior of the BtuB barrel. Upon binding of vitamin B12, the Ton box unfolds and extends 20–30 Å from the periplasmic face of the barrel (15). The exposure of the Ton box into the periplasm is a likely mechanism to regulate transporter–TonB interactions because conserved, dynamic regions of proteins have been shown to mediate protein–protein interactions and play critical roles in cellular control (16, 17).
Crystallographic models of BtuB present a different picture of the Ton box conformational change. In these models, the Ton box is folded into the barrel in the absence and presence of substrate (6). The different conformations observed with SDSL and crystallography in the presence of substrate are a result of solutes, which osmotically trap the Ton box in a folded conformation (18, 19). A recent crystal structure of BtuB complexed with a C-terminal fragment of TonB places the Ton box in an extended conformation so that it interacts with TonB (20), consistent with the extension of the N terminus detected by SDSL and chemical derivatization.
At the present time it is not known whether a common mechanism for substrate-induced structural transitions occurs in the Ton box of all TonB-dependent transporters. Structural transitions have been reported in the crystal structures of various transporters, but no clear common mechanism has emerged. For example, in the crystal structure of the ferrichrome transporter FhuA, with or without substrate, the Ton box is not resolved (10). In addition to the Ton box, there is an N-terminal helix in the crystal structures of both FhuA and FecA termed the switch helix (Fig. 1a), which undergoes a conformational change in the presence of substrate. Like the Ton box unfolding transition, this switch helix transition has been proposed to act as a mechanism by which TonB senses substrate binding (10).
The work presented here investigates whether there is a common structural mechanism of substrate-induced unfolding of the Ton box in FhuA and FecA, as has been seen by SDSL in BtuB. More than two dozen spin-labeled cysteine mutants were purified and reconstituted into lipid bilayers to examine the conformation and structure of the Ton box and switch helices in the presence and absence of their respective substrates. In FecA a transition similar to that observed for BtuB was detected. However, the results for FhuA are quite surprising and indicate that the Ton box of FhuA is one of the most dynamic protein segments studied to date by SDSL. This segment is constitutively unfolded for all conditions and all spin-labeled mutants investigated here. These results are discussed in terms of the regulation of transporter–TonB interactions in this family of outer-membrane transporters.
Results
The Ton Box in FecA Undergoes a Substrate-Dependent Order-to-Disorder Transition.
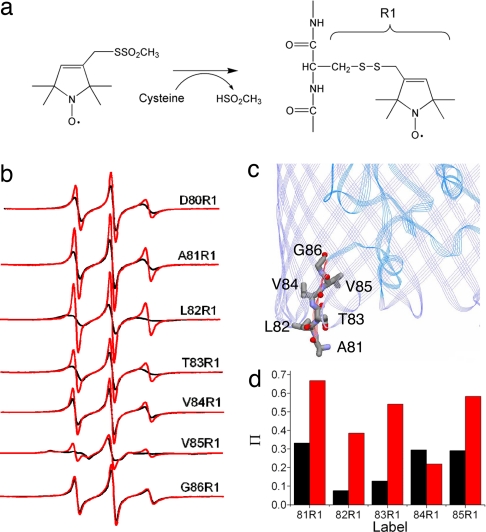
EPR line shapes encode information regarding the rate and amplitude of motion (ordering) of the spin-labeled side chain on the nanosecond timescale. The label motion results from the rotation of bonds within the nitroxide linkage as well as local backbone fluctuations, and the line shapes that result are uniquely defined by the local structure and protein dynamics at the labeled site (21, 22). We used EPR to examine the state of the Ton box in FecA by introducing unique cysteines and incorporating the spin-labeled side chain R1 (Fig. 2a) into the Ton box (amino acid positions 80–86) one at a time. Shown in Fig. 2b are X-band EPR spectra in the presence and absence of the FecA substrate ferric citrate.
Fig. 2.
Order-to-disorder transition in the FecA Ton box. (a) Structure of the spin-labeled side chain R1 incorporated into FecA. (b) X-band EPR spectra of single sites labeled with R1 along the Ton box in FecA (residues 80–86) both without (black line) and with (red line) the substrate ferric citrate. (c) Crystal structure (PDB ID code 1PNZ) showing the placement of the side chains in the Ton box in FecA. Residue 80 is not resolved in the crystal structure. (d) Ni(II)EDDA collision parameters, Π, for the FecA Ton box mutants in the absence (black) and presence (red) of ferric citrate. Values of Π are proportional to the frequency of collisions between the aqueous soluble Ni(II)EDDA and the R1 side chain (25).
In the absence of substrate, several EPR spectra from the FecA Ton box result from nitroxide labels that are undergoing fast isotropic motion with little ordering. For example, the spectrum resulting from the label at position 80 is similar to those from positions 81, 84, and 86 and corresponds to a nitroxide having a correlation time (τc) of 1.2 ns. Such highly mobile nitroxides are typical of those found at relatively flexible protein segments such as loops (23). The label at position L82R1 exhibits slightly slower motion, and the EPR line shape from V85R1 reflects immobilization of the nitroxide due to tertiary contact within the protein structure.
These spectra indicate that the Ton box in FecA is interacting within the protein but retains a degree of flexibility. Several of the contacts seen by EPR can be rationalized in terms of the crystal structure (Fig. 2c) (7, 8). For example, the slightly broader line shape from L82R1 might be the result of an interaction with the FecA barrel, and the more immobile spectrum obtained for V85R1 might result from an interaction with Asp-96 on the switch helix; however, the spectra for 84 and 86 indicate that this face of the Ton box is not in strong contact with the barrel as suggested by the crystal structure. The data indicate that the Ton box is more dynamic or has a different configuration in bilayers than indicated by the crystal structure.
Upon the addition of substrate, labels along the Ton box undergo an increase in motion, and the EPR spectra indicate that the Ton box is now unstructured. For example, the spectrum obtained from D80R1 reflects a 20% decrease in rotational correlation time upon the addition of substrate. The spectrum from V85R1, which was highly ordered, now yields two components, one of which resembles the original spectrum and a second that results from a nitroxide undergoing fast isotropic motion. The two components in this spectrum could be the result of two different rotameric states of the label, or two different conformations of the Ton box, one folded and another that is dynamic or unfolded. To test this possibility, we added 25% wt/vol PEG 3350 to the membrane suspension. This polymer is expected to be excluded from hydrated protein surfaces (24), and it should alter protein conformational equilibria that involve significant changes in hydration; however, it should not alter the rotameric states of the label. Addition of PEG 3350 eliminates the more mobile component [see supporting information (SI) Fig. 7], indicating that this spectrum reports an equilibrium, probably between folded and unfolded forms.
The results for FecA resemble those found previously for BtuB, where the Ton box was also found to be in equilibrium between folded and unfolded states. For BtuB, substrate addition shifted the equilibrium from one where a folded state was highly favored to one where a disordered or unfolded state was favored (11). Like BtuB, power saturation measurements (25) in the presence of Ni(II)EDDA indicate that labels in the FecA Ton box, with the exception of V84R1, increase their aqueous exposure when substrate is bound (Fig. 2d).
In addition to binding ferric citrate, FecA is also known to bind citrate. EPR spectra were obtained in the presence of citrate (see SI Fig. 8). The binding of citrate produces very minor changes in the EPR spectra of the FecA Ton box and slightly increases the dynamics of labels along the Ton box. These relatively minor changes in Ton box dynamics are consistent with the conclusion that the binding of iron-free citrate results in a nonproductive transport complex (8).
The Ton Box in FhuA Is Highly Dynamic and Constitutively Unfolded.
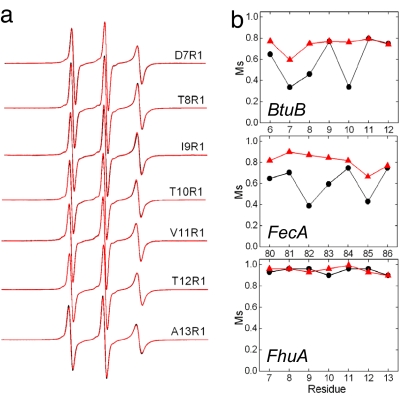
The configuration of the FhuA Ton box was also examined in palmitoyloleoylphosphatidylcholine bilayers, and X-band spectra from the Ton box of FhuA, sites 7–13, with and without substrate are shown in Fig. 3a. In each case the spectra arise from labels that are highly mobile and attached to unfolded, unstructured, and highly dynamic protein segments. These spectra result from labels that are executing fast isotropic motion, and the spectra can be simulated by fast motion having a correlation time that is ≈0.5 ns or less. When substrate is added the spectra undergo little change and exhibit the same high rate of motion characteristic of an unfolded protein segment.
Fig. 3.
Ton box dynamics in FhuA, BtuB, and FecA. (a) X-band EPR spectra of single sites labeled with R1 (Fig. 2a) along the FhuA Ton box (residues 7–13) without (black line) and with (red line) the substrate ferrichrome. In every case the spectra from apo and ligand-bound states overlap. (b) A plot of the Ms obtained from the EPR spectra of R1 when placed along the Ton box in the absence (●, black) and presence (▴, red) of substrate for BtuB, FecA, and FhuA. The Ms is defined as Ms = (δ−1 − δi−1)/(δm−1 − δi−1), where δ is the central resonance linewidth and δi and δm are the least and most mobile R1 side chains found in proteins (42). Values for δi and δm were taken as 8.5 and 1.8 G, respectively.
Several experiments were performed to ensure that there was no adventitious substrate already bound to FhuA. First, when the protein was grown in minimal media or in the presence of an iron chelator, 2,2′-dipyridyl, results identical to those shown in Fig. 3a were obtained. Second, atomic absorption on the isolated and reconstituted FhuA indicated that the protein was iron-free. Escherichia coli does not produce this siderophore (26), which is consistent with the finding that our isolated FhuA is ferrichrome-free. To determine that added ferrichrome would bind to our FhuA preparation, a fluorescence-based assay was used and yielded a substrate affinity comparable to that previously found (27). Finally, to ensure that the effects seen in Fig. 3 were not an artifact of the protein preparation, several sites in the Ton box of FhuA were labeled in the intact outer membrane as described previously (13). In these samples no evidence for a substrate-induced unfolding of the Ton box was found, indicating that detergent extraction, purification, and reconstitution did not alter the result seen in Fig. 3a (data not shown).
The scaled mobilities (Ms) of the Ton box on FhuA and FecA are plotted in Fig. 3b with and without substrate, along with Ms previously found for BtuB (11). The Ms provides a semiquantitative comparison of motion of protein spin-labeled sites, where Ms = 1 represents the most mobile and Ms = 0 represents the least mobile sites found in proteins (28). This parameter is obtained from the central EPR linewidth, and it is dominated by the correlation time of the nitroxide. From Fig. 3b it can be seen that the Ton boxes in BtuB and FecA undergo similar changes in Ms upon substrate addition and that the segment becomes highly mobile in the presence of substrate. However, the values of Ms for the Ton box of FhuA are unchanged by substrate and represent the highest vales of Ms that are seen in proteins labeled with R1. The data indicate that the Ton box of FhuA is a highly mobile, unstructured protein segment with or without substrate.
The lack of any substrate-induced change in the Ton box spectra from FhuA is surprising. FhuA is structurally homologous to BtuB and FecA, both of which exhibit a substrate-dependent folding-to-unfolding transition. As discussed below, this suggests that the interactions between TonB-dependent transporters and TonB are not regulated in the same manner.
The FhuA “Switch” Helix Does Not Switch upon the Addition of Substrate.
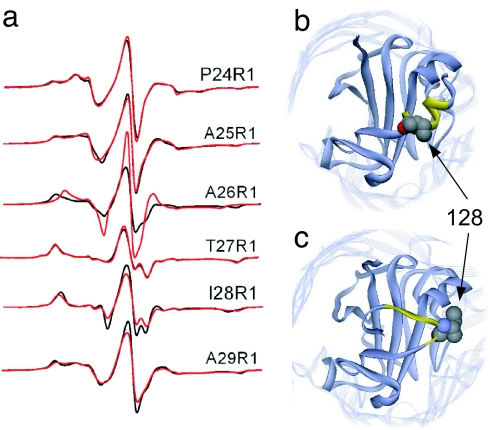
In the crystal structure of FhuA, a helix that is C-terminal to the Ton box is seen to undergo a substrate-dependent conformational change to a more extended structure (Fig. 1) (9, 10). It is somewhat surprising that this structural transition, just 10 residues from the Ton box, does not produce changes in the EPR spectra along the Ton box (Fig. 3a). To investigate the switch helix in FhuA in bilayers, single spin labels were incorporated along the H1 or switch helix (positions 24–29) of FhuA, and the EPR spectra were recorded in the absence and presence of ferrichrome (Fig. 4a). The spectra indicate that significant structural changes are not taking place in this segment of membrane-reconstituted FhuA upon the addition of substrate. In almost every case the rate and modes of motion of the labels are unchanged. The most significant change is seen for A26R1. In the absence of substrate, the spectrum for A26R1 is due to a label that is immobile and in strong tertiary contact. Upon substrate addition, the EPR spectrum indicates additional motional averaging of A26R1, perhaps because of an increase in protein dynamics, but the label at this site remains in tertiary contact.
Fig. 4.
Structure of the switch helix in FhuA. (a) X-band EPR spectra of the R1 label incorporated along the H1 or switch helix in FhuA without (black line) and with (red line) substrate. (b and c) Crystal structure of FhuA showing the switch helix without (b) and with (c) substrate. In the crystal structure, the I28 side chain (highlighted in Corey–Pauling–Koltun rendering) is a surface-exposed helical site in the absence of substrate but is buried in the presence of substrate.
Further examination of the switch helix spectra indicates that they are not consistent with the helical conformation observed in the crystal structure of the apo form of FhuA (Fig. 4b). T27R1 and I28R1 have spectra that are the result of nitroxide motion near or at the EPR rigid limit (correlation times over ≈50 ns). As indicated in Fig. 5, these spectra have line widths and second moments (29) that are found only for labels buried within the protein interior. Both oxygen and Ni(II)EDDA collision parameters (Π) are very low (Π values ≲0.1), consistent with buried sites. The remainder of the spectra P24R1, A25R1, and A29R1 contain at least one component that is strongly immobilized. In the crystal structure (Fig. 4b) one face of the switch helix is exposed to the periplasm, but spectra characteristic of labels on exposed helical surfaces are not seen. For example, I28R1 is on a helix surface in the apo structure. An estimate of the percentage of solvent accessibility for this side chain from the crystal structure is 43%, and this value is similar to that for other exposed helical sites in proteins. In the presence of substrate, this drops to 1% in the crystal structure (Fig. 4c). However, in both cases, the spectrum from this position is rigidly immobilized (Figs. 4a and 5). The spectrum from I28R1 is more consistent with the ligand-bound structure in Fig. 4c, as are spectra from labels at a number of other positions in this segment. Thus, the switch helix does not appear to be in the conformation shown in the crystal structure of the apo form, and, of the two crystal structures, the structure found in the presence of substrate (Fig. 4c) is in closer agreement with the spectroscopic result for FhuA in palmitoyloleoylphosphatidylcholine bilayers.
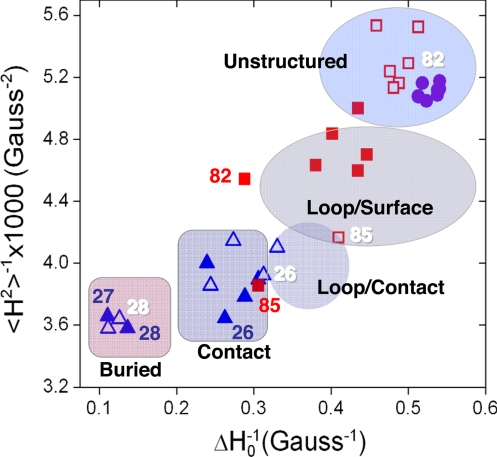
Fig. 5.
Nitroxide line shape and local side chain environment. A plot of the reciprocal of the central linewidth (ΔH0−1) versus the reciprocal of the second moment (〈H2〉−1) of the EPR spectrum for sites in the FhuA Ton box (●), the FecA Ton box without (■) and with (□) ferric citrate, and in the switch helix of FhuA without (▴) and with (▵) ferrichrome. Highly mobile regions having narrow line widths, and low second moments are unstructured. Other regions are defined based on earlier work on spin-labeled annexins and T4 lysozyme (23, 29) and are defined as follows. Loop/surface sites are on flexible regions of proteins. Loop/contact sites are on flexible loops that are in tertiary contact. Contact sites are in regions of defined secondary structure with some surface exposure but in tertiary contact. Buried sites are within the protein interior having no surface exposure. Selected residues are indicated. Embossed numbers indicate sites in the presence of substrate.
Like FhuA, FecA also contains a switch helix (7). We placed labels into two positions of the switch helix of FecA, at positions 96 and 100, and recorded the X-band EPR spectra for these labeled mutants in the presence and absence of substrate (see SI Fig. 9). In neither case are substrate-dependent changes seen in the EPR spectra. Thus, under the conditions used here, where the protein is reconstituted into lipid bilayers, the switch or H1 helices in FhuA and FecA do not appear to switch, and in FhuA it may be in a different configuration than that indicated by the crystal structure. The basis for these differences is not presently known but may be a result of the different conditions used for the crystallographic and spectroscopic experiments (see Discussion).
Several additional positions in the N-terminal region of FhuA were examined by using SDSL. FhuA contains a turn near its N terminus (the H1/β1 connecting loop) that is structurally homologous to the turn in BtuB that unfolds and contains the Ton box (residues 32–39 in FhuA). SDSL on this segment indicates that it is folded and does not change structure upon the addition of ferrichrome (see SI Fig. 10). Thus, upon the binding of substrate, there is no evidence for a significant conformational change in either the switch helix or the H1/β1 connecting loop in FhuA, both of which are C-terminal to the Ton box.
Discussion
Substrate-dependent transmembrane signaling events in TonB-dependent transporters are reported to include changes in the Ton box and other regions at the periplasmic surface. The data obtained here allow for a comparison of the signaling events that take place in FecA, FhuA, and BtuB in lipid bilayers. In FecA, the EPR spectra are consistent with a disorder transition in the Ton box upon the addition of substrate. These spectra are similar to those obtained previously for the Ton box of BtuB in both native and reconstituted membranes, which also result from a disorder transition upon the binding of substrate (11, 13). In BtuB, long-range distance measurements made by using pulse EPR demonstrate that this substrate-induced disordering is associated with an extension of the Ton box, by 20–30 Å, into the periplasmic space (see Fig. 6) (15). The EPR spectra obtained here for FecA suggest that a similar projection of the Ton box into the periplasmic space accompanies substrate addition in this transporter.
Fig. 6.
Structures of BtuB in three different configurations. (a) The apo form of BtuB (PDB ID code 1NQE), where the Ton box is shown in red and the N-terminal fold is shown in yellow. (b) A structure based on the substrate-bound crystal structure (PDB ID code 1NQH), with the Ton box configuration adjusted to fit EPR-derived distance constraints (15). The substrate, vitamin B12, is shown in a Corey–Pauling–Koltun representation. (c) Structure of BtuB bound to a C-terminal fragment of TonB (PDB ID code 2GSK), where the TonB fragment is shown in light gray (20).
In FhuA, the Ton box acts differently than it does in BtuB or FecA. In FhuA, the EPR spectra are consistent with a Ton box that is highly dynamic and unfolded in either the absence or presence of substrate. This is consistent with the absence of electron density for these residues in the crystal structures of FhuA (10). This result is not entirely surprising, because the Ton box in FhuA is not located in a homologous region on the N terminus when compared with BtuB or FecA (see Fig. 1b). However, the finding that the Ton box of FhuA is constitutively unfolded implies that TonB–transporter interactions may not be regulated in the same way in this family of transporters.
At the present time the role of these signaling events in TonB-dependent transport is not known. However, the Ton box is believed to be an energy-coupling segment, and conformation changes in the Ton box have been proposed to regulate interactions with TonB. Chemical cross-linking (12, 30) and recent protein crystal structures (20, 31) indicate that the Ton box interacts with TonB. There also is evidence that this Ton box–TonB interaction occurs in a substrate-dependent manner (12, 32); thus, some signal transduction event must initiate this interaction. Genetic studies suggest that iron siderophore and vitamin B12 transport systems compete for TonB (33), and regulation of the transporter–TonB interaction by a substrate-dependent signaling event would account for this observation and optimize the use of TonB, which is stoichiometrically limiting.
It has been observed that highly conserved, dynamic, or unfolded regions of proteins facilitate protein–protein interactions (17), and the substrate-induced unfolding and extension of the Ton box in BtuB and FecA nicely fit this paradigm. However, in recent crystal structures TonB interacts not only with the Ton box but with several regions at the periplasmic surface. The data obtained here suggest that the Ton box is not regulating transporter–TonB interactions in FhuA because it is always unfolded. However, other regions at the periplasmic surface of FhuA may change structure or dynamics and regulate this interaction. Alternatively, FhuA–TonB interactions may not be regulated by substrate. Indeed it has been reported that 1:1 and 2:1 complexes are formed between TonB and FhuA without substrate; however, it is claimed that the formation of the 2:1 complex is enhanced by substrate (34). Interestingly, the recent crystal structure of FhuA in complex with TonB had a 1:1 stoichiometry and was obtained in the presence of substrate (31).
Spectroscopic, chemical derivatization, and crystallographic methods do not provide a consistent view of substrate-induced structural changes in the periplasmic face of these transporters. In the case of the Ton box of BtuB, this has been shown to be the result of the precipitants used for protein crystallization (18, 19). One of the more notable substrate-induced changes seen by crystallography in FhuA is the unwinding of the switch helix on the periplasmic surface. This unwinding causes the N terminus to translocate by ≈17 Å, presumably moving the Ton box further into the periplasm. It has been suggested this conformational change is a signal indicating substrate binding and that it facilitates an interaction between the Ton box and TonB (10). However, the results of SDSL indicate that there are at most minor changes in this segment, probably involving an increase in dynamics on the nanosecond timescale, with no rearrangement of secondary structure. Furthermore, EPR spectra provide a strong signature for local structure (21), and in FhuA the spectra are not consistent with those expected from the switch helix as seen in the apo form of FhuA. We have not fully investigated the basis for the differences between crystallography and SDSL, but work on BtuB indicates that protein conformations can be shifted depending on the detergents and precipitants used in different experimental approaches (18, 19, 35). Presently it is not clear whether the switch helix is necessary for TonB-dependent transport. Deletion of the switch helix impairs both binding and transport of ferrichrome, but by only 20% compared with wild-type FhuA (36).
There is clearly a variability seen in the N-terminal region of TonB-dependent transporters when different transporters and methods are compared. This variability may reflect a functionally important instability or dynamics in this region of these proteins. As indicated above, dynamics or instability in the protein fold would be expected for a region involved in protein–protein contact. Furthermore, it has been proposed that transport may be facilitated by an unfolding of the N-terminal region of BtuB (37), a property that would be consistent with a dynamic or inherently unstable region.
In summary, in membrane bilayers SDSL indicates that the Ton box of both FecA and BtuB undergoes a disorder transition. This structural change may function to regulate transporter–TonB interactions and the utilization of TonB. The Ton box of FhuA is different. It appears to be disordered and highly dynamic in either the presence or absence of substrate. Furthermore, there do not appear to be any significant substrate-dependent structural changes taking place near the N terminus of the transporter. These data demonstrate that the Ton box in FhuA is always available for interaction with TonB and that structural changes in the Ton box of FhuA are not involved in regulating FhuA–TonB interactions.
Materials and Methods
Mutagenesis, Protein Expression, and Outer Membrane Isolation.
The plasmids harboring wild-type FhuA and FecA, which are pHK763 (38) and pIS711 (39), respectively, were generously provided by Volkmar Braun (University of Tübingen, Tübingen, Germany). FhuA and FecA were individually mutated to cysteine by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) to attach a thiol-reactive spin label. Proteins containing a single cysteine mutation were overexpressed in an E. coli BL21(DE3) strain lacking all of the outer membrane porins such as OmpA, OmpC, and LamB (40). Cells were grown in LB media containing 0.1 mg/ml ampicillin, and the transcription of FhuA and FecA was initiated by the addition of isopropyl-β-d-thiogalactoside. Outer membranes were prepared as described previously (37).
Spin Labeling, Purification, and Reconstitution.
The prepared outer membrane was solubilized, spin-labeled, and purified as described previously except that different gradient profiles of LiCl were generated for FhuA and FecA (37, 41). The purified proteins were reconstituted into palmitoyloleoylphosphatidylcholine vesicles by dialysis (41), and the reconstituted vesicles were sedimented by using a Beckman (Fullerton, CA) Airfuge. For spin labeling, 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanethiosulfonate (Toronto Research Chemicals, North York, ON, Canada) was used.
EPR Measurements.
EPR measurements were performed on 4–5 μl of sample loaded into glass capillaries with 0.60 mm i.d. × 0.84 mm o.d. (VitroCom, Mountain Lakes, NJ). EPR spectra were obtained on a modified Varian E-line 102 series X-band spectrometer with a loop gap resonator (Medical Advances, Milwaukee, WI). All spectra were taken at 2-mW incident microwave power and 1-G modulation amplitude. Scan range was 100 G. For comparison, all spectra were normalized within a plot.
In FhuA, the native protein has four cysteines in the extracellular loops that are involved in disulfide bonds. To determine whether any labeling of the native cysteines by the 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanethiosulfonate reagent took place, wild-type FhuA was treated and measured by EPR as described above. The results demonstrate that background labeling is negligible indicating that the cysteines in FhuA are not susceptible to spin labeling. Similar measurements on wild-type FecA also indicated that there was no significant contribution to the EPR signal due to nonspecific labeling.
For EPR measurements in the presence of substrate, 5 mM stock solutions of ferric citrate (Sigma, St. Louis, MO) and ferrichrome (Biophore Research Products, Tübingen, Germany) were prepared in the EPR reconstitution buffer. Final concentration of 2 mM substrate was directly added to liposome samples. For measurements with PEG 3350 (Fluka, Buchs, Switzerland), appropriate amounts of 50% wt/vol PEG 3350 stock solution, reconstitution buffer, and liposome sample were mixed. Because the protein is likely to be symmetrically disposed across the reconstituted liposomes, the samples were taken through five freeze–thaw cycles in liquid N2 in all measurements where substrate or PEG was added to saturate all possible binding sites.
Supplementary Material
Acknowledgments
We thank Dr. Volkmar Braun for the FecA and FhuA strains and plasmids, Drs. Wayne Hubbell and Christian Altenbach for LabView software used in the EPR data analysis, and Dr. Robert Nakamoto for helpful discussions and reading of the manuscript. This work was supported by the National Institutes of Health, National Institute of General Medical Sciences (Grant GM 035215).
Abbreviations
- SDSL
site-directed spin labeling
- Ms
scaled mobility
- PDB
Protein Data Bank.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702172104/DC1.
References
- 1.Wiener MC. Curr Opin Struct Biol. 2005;15:394–400. doi: 10.1016/j.sbi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Faraldo-Gómez JD, Sansom MSP. Nat Rev Mol Cell Biol. 2003;4:105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 3.Postle K, Kadner R. Mol Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 4.Klebba PE. Front Biosci. 2003;8:s1422–s1436. doi: 10.2741/1233. [DOI] [PubMed] [Google Scholar]
- 5.Braun V. Front Biosci. 2003;8:s1409–s1421. doi: 10.2741/1232. [DOI] [PubMed] [Google Scholar]
- 6.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Nat Struct Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, Deisenhofer J. Science. 2002;295:1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]
- 8.Yue WW, Grizot S, Buchanan SK. J Mol Biol. 2003;332:353–368. doi: 10.1016/s0022-2836(03)00855-6. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 10.Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, Moras D. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 11.Fanucci GE, Coggshall KA, Cadieux N, Kim M, Kadner RJ, Cafiso DS. Biochemistry. 2003;42:1391–1400. doi: 10.1021/bi027120z. [DOI] [PubMed] [Google Scholar]
- 12.Cadieux N, Kadner RJ. Proc Natl Acad Sci USA. 1999;96:10673–10678. doi: 10.1073/pnas.96.19.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merianos HJ, Cadieux N, Lin CH, Kadner R, Cafiso DS. Nat Struct Biol. 2000;7:205–209. doi: 10.1038/73309. [DOI] [PubMed] [Google Scholar]
- 14.Cadieux N, Phan PG, Cafiso DS, Kadner RJ. Proc Natl Acad Sci USA. 2003;100:10688–10693. doi: 10.1073/pnas.1932538100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q, Ellena JF, Kim M, Cafiso DS. Biochemistry. 2006;45:10847–10854. doi: 10.1021/bi061051x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Febs J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 17.Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 18.Fanucci GE, Lee JY, Cafiso DS. Biochemistry. 2003;42:13106–13112. doi: 10.1021/bi035439t. [DOI] [PubMed] [Google Scholar]
- 19.Kim M, Xu Q, Fanucci GE, Cafiso DS. Biophys J. 2006;90:2922–2929. doi: 10.1529/biophysj.105.078246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 21.Fanucci GE, Cafiso DS. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Hubbell WL, Cafiso DS, Altenbach CA. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 23.Isas JM, Langen R, Haigler HT, Hubbell WL. Biochemistry. 2002;41:1464–1473. doi: 10.1021/bi011856z. [DOI] [PubMed] [Google Scholar]
- 24.Timasheff SN. Biochemistry. 2002;41:13473–13482. doi: 10.1021/bi020316e. [DOI] [PubMed] [Google Scholar]
- 25.Farahbakhsh ZT, Altenbach C, Hubbell WL. Photochem Photobiol. 1992;56:1019–1033. doi: 10.1111/j.1751-1097.1992.tb09725.x. [DOI] [PubMed] [Google Scholar]
- 26.van der Helm D, Baker JR, Engwilmot DL, Hossain MB, Loghry RA. J Am Chem Soc. 1980;102:4224–4231. [Google Scholar]
- 27.Locher KP, Rosenbusch JP. Eur J Biochem. 1997;247:770–775. doi: 10.1111/j.1432-1033.1997.t01-1-00770.x. [DOI] [PubMed] [Google Scholar]
- 28.Columbus L, Hubbell WL. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 29.Mchaourab H, Lietzow M, Hideg K, Hubbell W. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 30.Ogierman M, Braun V. J Bacteriol. 2003;185:1870–1885. doi: 10.1128/JB.185.6.1870-1885.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 32.Moeck GS, Postle K, Coulton JW. J Biol Chem. 1997;272:28391–28397. doi: 10.1074/jbc.272.45.28391. [DOI] [PubMed] [Google Scholar]
- 33.Kadner RJ, Heller KJ. J Bacteriol. 1995;177:4829–4835. doi: 10.1128/jb.177.17.4829-4835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khursigara CM, De Crescenzo G, Pawelek PD, Coulton JW. Biochemistry. 2005;44:3441–3453. doi: 10.1021/bi047882p. [DOI] [PubMed] [Google Scholar]
- 35.Fanucci GE, Lee JY, Cafiso DS. J Am Chem Soc. 2003;125:13932–13933. doi: 10.1021/ja0376442. [DOI] [PubMed] [Google Scholar]
- 36.Endriss F, Braun M, Killmann H, Braun V. J Bacteriol. 2003;185:4683–4692. doi: 10.1128/JB.185.16.4683-4692.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coggshall KA, Cadieux N, Piedmont C, Kadner R, Cafiso DS. Biochemistry. 2001;40:13946–13971. doi: 10.1021/bi015602p. [DOI] [PubMed] [Google Scholar]
- 38.Killmann H, Herrmann C, Torun A, Jung G, Braun V. Microbiology. 2002;148:3497–3509. doi: 10.1099/00221287-148-11-3497. [DOI] [PubMed] [Google Scholar]
- 39.Kim I, Stiefel A, Plantor S, Angerer A, Braun V. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 40.Prilipov A, Phale PS, Van Gelder P, Rosenbusch JP, Koebnik R. FEMS Microbiol Lett. 1998;163:65–72. doi: 10.1111/j.1574-6968.1998.tb13027.x. [DOI] [PubMed] [Google Scholar]
- 41.Fanucci GE, Cadieux N, Piedmont CA, Kadner RJ, Cafiso DS. Biochemistry. 2002;41:11543–11551. doi: 10.1021/bi0259397. [DOI] [PubMed] [Google Scholar]
- 42.Columbus L, Hubbell WL. Trends Biochem Sci. 2002;6:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.