Abstract
The process of transcript elongation by RNA polymerase II (Pol II) involves transcription-dependent exchange and displacement of all core histones and is tightly controlled by numerous protein complexes modifying chromatin structure. These processes can contribute to regulation of transcription initiation and elongation, as well as the chromatin state. Recent data suggest that the histone octamer is displaced from DNA at a high rate of transcription, but can survive less frequent transcription that is accompanied only by partial loss of H2A/H2B histones. Here we propose that critical density of Pol II molecules could be required for displacement of the histone octamer and discuss mechanisms that are most likely involved in the processes of histone exchange.
Keywords: chromatin, nucleosome, histones, exchange, transcription, elongation
1. Introduction
The vast majority of eukaryotic genome is organized into chromatin structure that highly compacts DNA in the nuclei, allows regulated access of various protein complexes to DNA and efficient progression of processive enzymes (such as DNA and RNA polymerases) along the template. Chromatin consists of repeating subunits called nucleosomes. Each nucleosome core includes 147 bp of DNA wrapped 1 2/3 times around a histone octamer containing two each of histones H2A, H2B, H3, and H4 [1]. The core histones are arranged in a tripartite manner: a central (H3/H4)2 tetramer is flanked on each side by an H2A/H2B dimer ([1,2], Fig. 1, insert). One molecule of linker histone, H1, binds to the DNA linking adjacent nucleosomes. Nucleosomes are further compacted into a 30 nm chromatin fiber that most likely is formed by coiled dinucleosomes [3]; these fibers are, in turn, further compacted into structures not fully understood [4].
Figure 1. Possible histone octamer fates during intense and moderate transcription by PolII in vivo.

During intense transcription a considerable fraction of the octamers can be displaced from DNA or exchanged (1). The octamer most likely dissociates into chaperone-bound H3/H4 tetramer and two H2A/H2B dimers; the octamers are re-assembled on DNA very soon after the efficiency of transcription is decreased. During moderate transcription the octamer could be transiently displaced from DNA (2); the octamer immediately re-binds to DNA behind the transcribing PolII. Alternatively, during moderate transcription one H2A/H2B dimer could be transiently displaced by PolII from the nucleosome or exchanged (2’). Insert: Side view of the structure of nucleosome core [1];. DNA is shown in white, the H3/H4 tetramer in purple, and two H2A/H2B dimers – in green and yellow. The histone displacement and exchange pathways are indicated by dashed and solid arrows, respectively.
Compact nucleoprotein organization causes severe problems for processes such as DNA replication, recombination, repair, and transcription in vitro. Therefore it is not surprising that many of these processes are accompanied by changes in chromatin structure (chromatin remodeling). Chromatin remodeling is conducted by numerous protein complexes that include multiple ATP-dependent chromatin remodelers, as well as DNA and RNA polymerases [5]. It has become increasingly evident that modulation of chromatin structure plays a central role in numerous intranuclear processes and in their regulation [6]. In particular, in addition to PolII itself, numerous enzymes involved in modifying chromatin structure (such as ATP-dependent chromatin remodelers, histone-modifying enzymes and histone chaperones) are associated with active genes in transcription-dependent manner (see below) suggesting that at very least chromatin is an ultimate player during transcript elongation. Furthermore, nucleosomes remain associated with transcribed genes unless the level of transcription is extremely high [7–9] and can participate in regulation of the rate of transcript elongation both in vivo and in vitro [10,11].
This review focuses on the recent progress towards elucidating the mechanistic aspects of transcript elongation by RNA polymerase II (PolII) through nucleosomal templates. Earlier findings and other aspects of transcription of chromatin by PolII are covered in several excellent recent reviews [5,12–16].
2. Chromatin transcription by Pol II in vivo
2.1. Histone displacement from DNA and nucleosome recovery during transcription in vivo
The structures of the 30 nm chromatin fiber and nucleosome are clearly incompatible with ongoing transcription and have to be disrupted to allow Pol II movement along DNA (reviewed in [12]). Disruption of the higher order chromatin structure is transient and reversible: when Pol II molecules are spaced by more than 200–400 bp, nucleosomal 10 nm and 30 nm filaments are observed between them [17].
Nucleosome structure can also be disrupted during very intense transcription; this can be accompanied by partial loss of all core histones (Fig. 1, model 1). Using chromatin immunoprecipitation technique (ChIP) it has been demonstrated that up to 80 % of all core histones can be removed from the transcribed regions of yeast genes [7–9]; the extent of the removal is directly proportional to the efficiency of transcription [8,9]. In earlier studies using micrococcal nuclease (MNase) it has also been shown that some nucleosomes can be lost [18] during very active transcription in yeast. Maximal removal of core histones from the transcribed regions occurs only at high density of Pol II molecules (approximately one molecule per 150 bp [7]). All core histones can be depleted from actively transcribed yeast genes, although in one study it was found that the extent of histone loss is higher for H2A/H2B histones [9]. While the results of studies of yeast genes by different groups are consistent, contradictory data on transcriptional-dependent histone displacement were obtained in Drosophila. In some cases nucleosome displacement at the high density of Pol II molecules was observed using electron microscopy and ChIP in Drosophila [17,19]. However other groups did not detect core histone depletion even from very intensely transcribed Drosophila hsp70 gene (Pol II molecules are spaced by ~100 bp) using an experimental procedure that is very similar to ChIP [20,21]. In this case, only partial (~50 %) and selective depletion of H2A/H2B histones was observed [21]. In summary, in most cases partial transcription-dependent displacement of all core histones occurs when the density of Pol II approaches one molecule per 100–150 bp; histones H2A/H2B are displaced preferentially to H3/H4 histones.
Electron microscopy data suggest that nucleosomes can be recovered immediately behind transcribing Pol II [17,22]. Once very transcriptionally active yeast genes that are partially depleted of core histones are turned off, the histones are re-bound to DNA at a very high rate [9]. Based on these observations it has been proposed that all core histones are reversibly displaced during each passage of PolII molecule both at low and at high efficiency of transcription (Fig. 1, models 1 and 2 [17]). According to this proposal, displacement of the complete octamer is an obligatory feature of the mechanism of PolII transcription through the nucleosome that does not depend on the activity of a gene. Then the observation that a complete set of core histones remain associated with genes transcribed with lower efficiency [7–9,21] is explained by the fast recovery of nucleosomes behind transcribing PolII [17]. However this simple model does not explain numerous experimental observations suggesting that histones H2A/H2B and H3/H4 are functionally and structurally distinct components of nucleosomes that behave differently during moderate transcription. In particular, it has been observed using electron microscopy that PolII elongation complexes and nucleosomes can co-exist on the same nucleosome-covered DNA fragment [22]. Furthermore, the model does not explain the higher exchange rates of H2A/H2B histones as compared with H3/H4 histones.
2.2. Transcription-dependent histone exchange in vivo
The H2A/H2B histone dimers and the H3/H4 histone tetramer occupy distinct positions in the nucleosome ([1], Fig. 1, inset). The H3/H4 histone tetramer organizes central 80 bp of nucleosomal DNA and is tightly bound to DNA [23]. Two H2A/H2B dimers flank the tetramer and organize the ends of nucleosomal DNA; the dimers are bound to DNA less tightly than the tetramer. The weaker binding of the dimers to DNA and their location close to the ends of nucleosomal DNA make them primary candidates for displacement from DNA on an encounter of processive enzymes traveling along DNA (such as DNA and RNA polymerases) with a nucleosome. Indeed, there is fast and extensive transcription-dependent, replication-independent exchange of H2A/H2B histones [24,25] that is localized to transcribed regions of the genes [25]. About 3% of H2B histone exchanged within several minutes in living human cells in transcription-dependent manner [24]. The high extent of the exchange suggests that it occurs not only on very active genes (they constitute a very small fraction of the genome), but also on moderately transcribed genes. Consistent with this view, extensive and fast H2A/H2B exchange occurs during moderate-level transcription of “housekeeping” genes in Physarum [25]. The high rate and the efficiency of the exchange suggest that it could occur very frequently during transcription, perhaps during every round of transcription; all originally DNA-bound H2A/H2B dimers can be exchanged [25]. While the functional meaning of the exchange is currently unknown, one study has suggested that transcription-dependent exchange of the dimers containing a histone variant (H2A.Z/2B) to H2A/H2B dimers could stabilize the active state of the genes [26]. The rate of transcription-dependent exchange of H3/H4 histones was considerably (at least 20-fold) lower than the rate of H2A/H2B exchange. In agreement with these results, the cross-linking studies suggest that even during very active transcription interactions of H2A/H2B histones with DNA are disrupted preferentially relative to H3/H4-DNA interactions [17,21]. Taken together, these studies suggest that H3/H4 tetramers are much less mobile than the H2A/H2B dimers during transcription at moderate and perhaps even at high level when all core histones are extensively exchanged. However this view has been apparently challenged by recent discovery of replication-independent (RI) incorporation of H3.3 histone.
H3.3 is a variant of histone H3 that is constitutively synthesized in low amounts in all analyzed eukaryotes [27]. There are four differences in the amino acid sequences between major H3s and H3.3 in Drosophila; three of them are located within the histone fold and hidden within the nucleosome and all four changes are important for the efficient RI incorporation [28]. It has been shown that in Drosophila H3.3, in contrast to major versions of H3, can be assembled into chromatin in RI process; both major H3s and H3.3 can be incorporated into chromatin during replication-coupled (RC) deposition [28]. RC and RI histone deposition is directed by different human histone chaperones – CAF-1 [29] and HirA, respectively [30]. CAF-1 targets acetylated forms of major H3/H4 histones [29] while HirA binds to H3.3/4 histone complexes [30]. H3.3 is associated with transcriptionally active chromatin regions [28] and enriched in modifications associated with active loci [31–33]. Based on these data, it has been suggested that H3/H4 tetramers that contain primarily the major forms of H3 histone (H3.1 and H3.2) after replication-coupled (RC) deposition can be exchanged in transcription-dependent way to H3.3/4-containing tetramers [28]. In agreement with this suggestion, later studies have shown that RI incorporation of H3.3 into chromatin is transcription-dependent, localized to transcribed regions of genes and is accompanied by displacement of the major H3 forms [33–36]. These features make transcription-dependent incorporation of H3.3/H4 tetramers and H2A/H2B dimers into chromatin similar. However there are also very important differences between these processes.
The rate and/or efficiency of transcription-dependent exchange of bulk human H3/H4 histones (measured separately for H3 and H4 histones and including exchange of both H3.3 and the major H3s) is considerably lower than the rate of H2A/H2B dimer exchange [24]. Similarly, the overall exchange rate of H3 localized on moderately transcribed “housekeeping” genes of Physarum are at least 20-fold lower than the rate of H2A/H2B exchange on the same genes [25]. This view is consistent with the results of earlier pulse-chase experiments where RI incorporation of nascent H2A/H2B histones into chromatin in the S phase in vivo occurs preferentially as compared with incorporation of nascent H3/H4 histones [37–40]. It has been proposed that nascent H2A/H2B dimers were incorporated into chromatin in a transcription-dependent process [38] although no direct evidence for this mechanism was provided. These experiments suggest that overall extents and rates of transcription-dependent incorporation/exchange of H3/H4 (including H3.3) and H2A/H2B histones are considerably different. The mechanisms of the exchange are also likely to be different.
One attractive possible mechanism that explains the observed difference in the rates of transcription-dependent exchange of H3/H4 tetramers and H2A/H2B dimers is based on high similarity of the processes of H3/H4 histone exchange and transient transcription-dependent displacement of all core histones (part 2.1). Unlike H2A/H2B exchange that occurs efficiently even on moderately transcribed genes [25], the extents of both H3/H4 histone exchange and transient displacement of the octamer are proportional to the rate of transcription by PolII [8,9,33–35]. Similarly, active ribosomal genes that are transcribed very efficiently can lose complete histone octamer [41,42]; this correlates with extensive H3.3 histone incorporation into rDNA locus [25,28,35]. In fact, the majority of accumulation of the H3.3 occurs within the rDNA locus in Drosophila [27].
The studies described above suggest that all core histones can be exchanged in transcription-dependent way. However the H3/H4 tetramer is much less mobile than the H2A/H2B dimers during moderate transcription that occurs on the majority of genes, but could be efficiently exchanged on very active genes; the mechanisms of histone exchange on moderately and highly transcribed genes are likely to be considerably different. On highly transcribed genes that constitute a small fraction of eukaryotic genome the complete histone octamer can be displaced from DNA and all histones can be exchanged very frequently (Fig. 1, model 1). During this process the H3/H4 tetramer containing major forms of H3 histone is exchanged with H3.3/4 tetramer. In contrast, during moderate-level transcription by PolII (that occurs on the majority of genes) the complete histone octamer is displaced/exchanged very rarely, but H2A/H2B exchange occurs at a high rate (Fig. 1, model 2’).
2.3. Nucleosome unfolding during PolII transcription
It has been proposed that nucleosomes could also be unfolded (with the histone-histone interaction within the nucleosomes being transiently disrupted) to accommodate the passage of PolII. Nucleosomes enriched in actively transcribed sequences have increased accessibility of the sulfhydryl groups of the cysteine residues of histone H3 [43,44]. Non-transcribed, intact nucleosomes do not have reactive SH groups because the cysteines of H3 are buried in the histone octamer structure. Moreover, accessibility of these SH groups to external probes is higher in nucleosomes containing acetylated histones [45]. Electron spectroscopic imaging revealed that the reactive nucleosomes are predominantly U-shaped, and the DNA wrapped around the core histones is S-shaped like a stretched out spring [46]. The presence of reactive nucleosomes, like the partial depletion of H2A/H2B histones [21], closely correlates with ongoing transcription suggesting that they are transient intermediates generated by transcription through chromatin [43,44]. One possibility that is consistent with these data and with the structural studies [46] is that transcribed nucleosomes are reactive because they are missing one or both H2A/H2B dimer(s). This remains to be directly analyzed.
3. Mechanism of transcription through chromatin by PolII in vitro
In vitro studies have demonstrated existence of at least two considerably different mechanisms of transcription through chromatin. The mechanism used by bacteriophage RNA polymerases (RNAPses) and eukaryotic PolIII is characterized by a low nucleosomal barrier and is accompanied by direct transfer of the complete histone octamer from in front to behind the polymerase accompanied by formation of an intranucleosomal DNA loop ([47–49], reviewed in [5]). More recently it was discovered that PolII handles chromatin in a considerably different way [50,51]. The PolIII-related mechanism is similar to the mechanism of action of ATP-dependent chromatin remodelers [5]; the PolII-related mechanism is utilized by PolII and by E. coli RNAPse [52].
3.1. The mechanism of chromatin remodeling by transcribing PolII
Studies from many laboratories have shown that nucleosomes can survive the passage by various RNA polymerases without being liberated into solution in vitro [see [53,54] for review]. In the case of PolII, the progress of the field was severely limited by the lack of an appropriate experimental system in vitro. A novel technique for assembly of authentic elongation complexes from synthetic oligonucleotides and purified RNAPses was developed recently [55,56]. The assembled and promoter-initiated elongation complexes (ECs) transcribe chromatin using the same mechanism [52]. Using this new approach, it was determined that during transcription of mononucleosomal templates one of the two H2A/H2B dimers is quantitatively displaced from the histone octamer [50]. As a result, nucleosomes are converted to DNA-bound histone hexamers (“hexasomes”) during transcription. The nucleosomal barrier for PolII is much higher than for other RNAPses [50,51,57,58] and nucleosomes are not translocated along DNA during transcription [50].
Perhaps the most intriguing and interesting feature of the PolII-related mechanism is the lack of nucleosome translocation along DNA during transcription, in particular because this part of the mechanism could be essential for proper transcription of nucleosome arrays (part 6). PolIII-related mechanism of transcription through chromatin provides almost immediate solution for nucleosome survival and bypass by the enzymes: during transcription the histone octamer is partially displaced from DNA and the open octamer surface can re-bind to DNA behind the enzyme forming an intranucleosomal DNA loop [59]. Thus the octamer is transferred out of the way of the transcription machinery to behind the enzyme. However it is much more difficult to explain how the hexasomes can survive transcription by PolII and remain at the original positions on DNA after transcription. One possible model is based on well-established mechanism of transcription through a nucleosome by bacteriophage SP6 RNAP [48]; it could explain survival of the hexasomes during transcription ([5], Fig. 2). According to the model, after PolII enters a nucleosome and partially displaces nucleosomal DNA from the surface of histone octamer (intermediate 1), nucleosomal DNA containing the elongation complex could re-bind to the octamer that remains at the same position on DNA (intermediate 2). Thus DNA-histone contacts are formed both in front and behind transcribing PolII at the same time and PolII is bound within a small DNA loop on the nucleosome [48]. Formation of the intermediate 2 could be facilitated by the 90 degrees DNA bend introduced by PolII into DNA during elongation [60,61]; it has been shown that at least SP6 RNAP can form such complexes with nucleosomes [48]. However PolII is about six times bigger than SP6 RNAP; moreover, the structure of PolII EC could prevent formation of a tight PolII-nucleosome complex, unless conformation of PolII or histone octamer is considerably changed (Fig. 2). If the intermediate 2 can be formed, eventually transcribing PolII may escape into productive transcription after disruption of DNA-histone contacts in front of the EC and re-formation of the hexasome at the original position on DNA behind the enzyme (intermediate 3). This mechanism explains critical features of PolII transcription through chromatin: hexasome survival during transcription and preservation of the original locations of histones on DNA. An alternative mechanism including transient complete displacement of the octamer from DNA to a chaperone was proposed [12]. However it has become less likely in the light of the observation indicating that nucleosome-specific pausing extends along the whole length of nucleosome-covered DNA [51]. These latter observation strongly suggest that histones remain DNA-bound and interfere with PolII transcription along whole nucleosomal DNA, before the enzyme leaves the boundaries of the transcribed nucleosome.
Figure 2. Hypothetical mechanism of PolII transcription through nucleosome.

Insert: Top - Yeast PolII elongation complex structure [60] (the two largest subunits are in blue, other subunits – in grey. DNA and RNA paths are partially modeled after [60]; DNA and RNA regions hidden in the structure are shown by dashed lines; RNA is in red). The direction of PolII movement is indicated by arrows. Bottom - structure of nucleosome core [1]; view from the top (only one DNA turn is shown). As PolII enters the nucleosome (1), DNA from the surface of the octamer is partially uncoiled. After PolII enters the nucleosome, DNA could re-coil on the surface of the octamer with PolII that remains associated with DNA (intermediate 2, the histone octamer is not shown). It is proposed that after this re-coiling the octamer is re-bound to DNA behind the PolII at the same position as before transcription (dashed arrow). The structure of the octamer and/or PolII may need to be changed to form the complex. Eventually DNA-histone interactions could be disrupted in front of PolII (dotted arrow), and (3) the enzyme can continue elongation leaving behind the hexasome (nucleosome missing one H2A/H2B dimer).
In the studies of transcription through the nucleosome by PolII in vitro [50] a single-round assay was used because the experimental system does not support multiple rounds of transcription [62]. Therefore this in vitro system most likely recapitulates conditions characteristic for moderately transcribed genes where molecules of elongating PolII are widely spaced along DNA, do not communicate with each other and move as independent entities. Therefore it is not surprising that the data on the displacement of H2A/H2B dimer by PolII in vitro are consistent with multiple evidence of transcription-dependent H2A/H2B dimer dissociation/exchange that occurs during moderate transcription in vivo (part 2). According to this view, H2A/H2B dimer displacement is transient in vivo and irreversible in vitro, perhaps because some factors facilitating re-binding of the dimer to the hexasome are missing in the in vitro system. No displacement of H3/H4 histone tetramer was observed in vitro [50]; if displacement of complete octamer did occur in vitro, it would be irreversible because DNA-free histone octamer immediately and irreversibly disrupted in the transcription buffer [63]. Since the displacement/exchange of H3/H4 tetramer was observed in vivo only at the high rate of transcription, the data in vivo (part 2) and in vitro are also consistent. The studies in vitro further suggest that the mechanisms of moderate (less than one molecule of PolII per 500 bp) and highly efficient transcription (one molecule of PolII per 100–200 bp) are likely to be mechanistically distinct. How could the high density of PolII molecules change the fate of nucleosomes on transcription?
The mechanism proposed for PolII transcription through the nucleosomes in vitro ([50], Fig. 2) suggests two possible explanations for the observed displacement of the histone octamer from highly transcribed genes (Fig. 3). The model 1 suggests that when PolII molecules are closely spaced, the second molecule could sterically prevent DNA-octamer re-association behind the first transcribing PolII molecule and thus prevent formation of the key “survival” intermediate 2 (Fig. 2). This would substantially decrease the number of DNA-histone interactions and could cause dissociationof complete histone octamer from DNA instead of recovery of the hexasome after transcription. Alternatively, the model 2 proposes that the second molecule of PolII could approach the hexasome that was just transcribed and therefore the complete nucleosome was not recovered yet. Since the hexasome is stabilized by considerably smaller number of DNA-histone interactions, the intermediate 2 (Fig. 2) cannot be formed and the histone hexamer could be displaced from DNA by the second molecule of transcribing PolII. The second model does not require very close spacing of PolII molecules and therefore can better explain the gradual inverse correlation between the rate of transcription and the extent of octamer displacement [8,9].
Figure 3. Octamer dissociation from DNA could be caused by closely spaced PolII molecules.
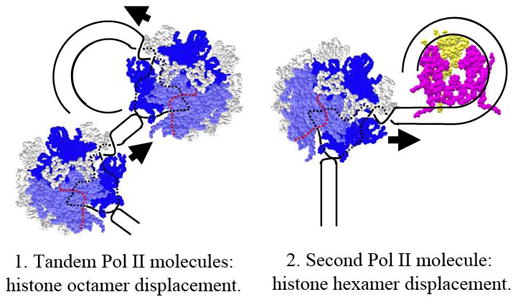
Dissociation of the octamer could occur by the following mechanisms: (1) When PolII molecules are closely spaced, the second molecule could prevent DNA-octamer re-association behind the first transcribing PolII molecule and thus prevent formation of the intermediate 2 (Fig. 2). This would substantially decrease the number of DNA-histone interactions and could cause octamer dissociation from DNA. (2) Alternatively, second molecule of PolII could approach the hexasome that was just transcribed and therefore the complete nucleosome was not recovered yet. Since the hexasome is stabilized by considerably smaller number of DNA-histone interactions, the histone hexamer could be displaced from DNA by the second molecule of PolII. The second model does not require very close spacing of PolII molecules.
3.2. Nature of the strong nucleosomal barrier to PolII
It has been shown that a nucleosome presents much higher barrier for PolII than for PolIII or SP6 RNAPses in vitro [50]; the height of the barrier can be regulated in vitro and in vivo [10,64–66]. Even a single nucleosome can be an absolute barrier for PolII at physiological salt concentration [50,57]. Numerous studies have suggested that nucleosomal barrier to PolII is characterized by its exceptional height and close similarity of DNA- and nucleosome-specific pausing patterns [50,51,57,58]; transient DNA sequence-specific pauses can be strongly amplified along the entire length of nucleosomal DNA. The height and DNA sequence-specificity of the barrier are explained in part by the observation that presence of nucleosomes results in more severe arrests that occur mostly at the positions of intrinsic pause sites observed on histone-free DNA [51]. It has been shown that this sequence-specific transient pausing in a nucleosome makes PolII highly vulnerable to arrest that involves backtracking of the elongation complex for a considerable distance (more than 10 bp) along DNA. After backtracking of PolII histone-DNA contacts are reestablished in front of PolII [51]; this could prevent escape of the polymerase into productive elongation. It is also likely that the intermediates that must be formed during transcription through a nucleosome to allow survival of the hexasome at the original position on DNA (intermediate 2, Fig. 2) strongly contribute to the height of the nucleosomal barrier. Thus for bacteriophage SP6 RNAP it has been shown that all complexes that are paused/arrested in the nucleosome have structures similar to the intermediate 2 [48]. DNA sequence-specificity of the barrier could also be explained in part by heterogeneity of nucleosome positioning on the templates used in some previous experiments [50,57,58]; there is no doubt that future studies using more accurately positioned nucleosomes will reveal nucleosome-specific pausing/arrest sites.
4. Factors involved in histone displacement/exchange during transcription through chromatin
Nucleosomes form a very high barrier for transcription by PolII in vitro (part 3.2). At the same time, in spite of the presence of nucleosomes on transcribed genes (part 2), the average rate of transcription in vivo is similar to the rate of transcription of histone-free DNA in vitro [57,67,68]. The potency of the nucleosomal barrier necessitates the involvement of various factors facilitating progression of PolII through the nucleosome. Indeed, numerous protein factors capable of modifying chromatin properties are associated with genes in transcription-dependent manner. These include histone chaperones, chromatin assembly factors, elongation factors, ATP-dependent chromatin remodelers, and covalent modifications of the histones. Many of the factors that can facilitate transcription through chromatin are also involved in transcription-dependent histone exchange/displacement. In this review we will discuss only the factors that directly participate in histone displacement/exchange or increase the rate of transcription through chromatin. Histone modifications and corresponding factors associated with transcribed genes were extensively reviewed recently [15,16] and will not be covered here.
4.1. FACT
The first discovered factor having an ability to stimulate transcription through chromatin in a highly purified system is the heterodimeric protein complex FACT (facilitates chromatin transcription [69]). FACT displays kinetics of recruitment and chromosome tracking that are similar to PolII in vivo [70,71]. There is also considerable genetic evidence connecting FACT with transcript elongation in vivo [72]. FACT specifically interacts with all core histones in vitro and possesses intrinsic histone chaperone activity [72,73]. Recent data suggest that FACT facilitates PolII-induced displacement of H2A/H2B dimer from the nucleosome in vitro, probably via direct interaction with the H2A/H2B histones [73]. This activity may explain the in vivo observations suggesting that FACT not only participates in PolII-induced nucleosome disruption, but also is required for nucleosome re-assembly behind the transcribing enzyme in vivo [9,71,74,75]. Furthermore, yeast FACT genetically interacts with Hir/Hpc proteins involved into transcription-dependent chromatin assembly [74,76].
4.2. TFIIS
TFIIS is a PolII-specific elongation factor facilitating transcription of DNA sequences or through DNA-bound proteins that cause transcription arrest (reviewed in [77]). Arrested ECs cannot resume transcription spontaneously because before the arrest PolII molecule backtracks along DNA and its active site is set out of the register with the 3′-end of growing RNA [78,79]. TFIIS rescues arrested complexes by stimulating endonucleolytic RNA cleavage close to the PolII active center that generates a new 3′-end and restores the catalytic activity of the enzyme [77]. TFIIS is associated with early transcribed regions [80,81], facilitates transcription through chromatin in vivo [82], and functionally interacts with Set2 histone methyltransferase that is involved in transcription through chromatin [83,84].
A stimulatory effect of TFIIS on overcoming the nucleosome barrier has been reported in several studies from Luse laboratory [57,58]. In a more recent study it has been shown that TFIIS can reactivate the backtracked PolII complexes arrested during transcription through the nucleosome and thus promotes transcription through chromatin [51,85]. In yeast TFIIS genetically interacts with FACT [72] suggesting that it could also participate in transcription-dependent histone exchange/displacement.
4.3. Spt proteins
Spt6 (Suppressor of Ty) was identified as a gene which product facilitates recovery of chromatin structure at promoters; the protein can act as a histone chaperone that promotes nucleosome assembly in vitro [86]. In the absence of Spt6-mediated nucleosome reassembly, nucleosomes cannot be recovered on the PHO5 yeast promoter and the promoter becomes activator-independent – it remains active even under repressing conditions when transcriptional activator proteins (Pho2 and Pho4) dissociate from the promoter [87]. Yeast genetics and biochemical analysis of human Spt 2, 4–6 proteins have suggested that these proteins are also involved in transcript elongation in chromatin [88,89]. In agreement with these studies, Drosophila homologs of Spt5 and Spt6 proteins co-localize with transcriptionally active chromosomal sites and with phosphorylated, actively elongating form of PolII on polytene chromosomes [90,91]. Genetic studies have identified interactions between Spt6 and elongation factor TFIIS [92]. Spt6 physically interacts with PolII, FACT and the Spt5 subunit of elongation factor DSIF in vitro and in vivo [93,94], and can enhance the rate of PolII elongation in vitro [94]. Futhermore, Spt6 and 2 proteins, like FACT, participate in recovery of chromatin structure during transcript elongation in vivo [75,95]. Finally, Spt 2 and 6 genetically interact with other factors participating in recovery of chromatin structure during elongation: FACT and Hir complexes [72,75,95]. Taken together, the available data suggest that Spt2 and 4–6 proteins are closely associated with PolII elongation complex and participate in recovery of chromatin structure that is transiently disrupted during transcription.
4.4. ATP-dependent chromatin remodelers
Some ATP-dependent chromatin remodelers have the ability to perturb nucleosomes in ways that could assist polymerases both in initiation and elongation (see [5,96] for review). Thus human SWI/SNF complex is required for both initiation and elongation on the human hsp 70 gene in vitro [11]. Analysis of the human heat shock factor (hHSF1) provided further evidence for the role of SWI/SNF during elongation, suggesting that chromatin remodeling occurs in this system as part of the process of transcript elongation in vivo [10,66]. Additional evidence for a role of SWI/SNF in elongation in vivo comes from studies of mutant SWI2/SNF2 subunits in yeast, which are synthetic lethal in combination with disruption of the PPR2 gene encoding elongation factor TFIIS [97]. Yeast and human SWI/SNF complexes are associated with PolII [98,99] suggesting that they could be recruited to transcribed genes together.
The ATPase CHD1 remodels nucleosomes in vitro and functions in elongation [100]. Studies performed on Drosophila polytene chromosomes revealed that Chd1 associates with highly active sites of transcription [101]. Chd1 mutant alleles in yeast genetically interact with the Set2 histone methyltransferase and the ISWI family, both implicated in elongation [102,103]. Chd1 physically associates with the elongation factors Paf, DSIF, and FACT [76,103] and with histone H3 that is methylated in transcription-dependent way [104]. In summary, some ATP-dependent remodelers can facilitate transcript elongation in vivo. The questions remain of how SWI/SNF-mediated chromatin remodeling facilitates elongation, and how SWI/SNF activities are coordinated with other known chromatin-remodeling elongation factors, such as FACT and Spt6.
4.5. Factor-dependent PolII transcription and histone displacement/exchange
The data discussed above suggest that numerous protein factors can facilitate transcription through chromatin and participate in transcription-dependent histone displacement/exchange. Most likely candidates that can facilitate transcription through chromatin both in vivo and in vitro include FACT, TFIIS and SWI/SNF factors. FACT most likely works by facilitating PolII-induced displacement of the H2A/H2B histones [73]. TFIIS reactivates PolII complexes arrested during transcription through the nucleosome [51]. The molecular mechanism of SWI/SNF action remains to be established.
The best candidates for factors involved in histone displacement/exchange include Spt2, Spt6, FACT and Hir proteins. Histone displacement/exchange most likely is a two-step process including transient H2A/H2B displacement during moderate transcription and displacement of the complete histone octamer during efficient transcription (part 2). It could be difficult to identify the players participating in each of the processes because H2A/H2B displacement most likely would facilitate the loss of the octamer (part 3.1). This view is supported by the studies from Winston laboratory where considerable transcription-dependent nucleosome disruption was observed in yeast containing mutations in genes encoding Spt2, Spt6, FACT and Hir [75,95]. Chromatin disruption was observed even on moderately transcribed genes where only H2A/H2B dimer is likely to be lost during transcription; at the same time, the extent of disruption and histone loss in the mutant strains was dependent on the rate of transcription [95] suggesting that Spt2, Spt6, FACT and Hir proteins could facilitate re-binding of all core histones to DNA.
5. Transcription of eukaryotic genes
The current view of transcription through chromatin that is accompanied by histone loss and exchange is shown in Fig. 4. Most likely, during moderate transcription (low density of PolII) single molecules of PolII encounter 30 nm chromatin fiber. Disruption of the higher order chromatin structure and partial uncoiling of DNA from the surface of the histone octamer is required for transcription through a nucleosome. It is achieved by PolII in cooperation with several elongation factors (FACT, TFIIS and possibly SWI/SNF). During transcription the H2A/H2B dimer is transiently displaced from DNA by PolII and FACT, and rebinds to the hexasomes immediately after PolII passes the nucleosome; the rebinding of the dimers is assisted by Spt2, Spt6, FACT and Hir proteins. During moderate transcription only H2A/H2B histone dimers are displaced/exchanged. When density of PolII approaches one molecule per 200 bp, the complete histone octamer is transiently and reversibly displaced from DNA, most likely with the help of Spt2, Spt6, FACT and Hir proteins. The nucleosomal structure recovers immediately after the efficiency of transcription is decreased; this results in transcription-dependent exchange of all core histones, with major forms of H3 histone being replaced by the H3.3 variant.
Figure 4. Gene transcription at low and high PolII densities.
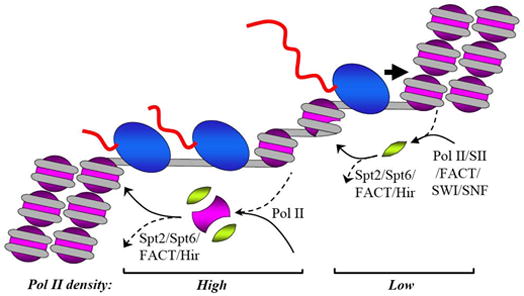
During transcription at a moderate or low level the histone octamer is not likely to be displaced from DNA even transiently. However transcription by PolII is accompanied by frequent transient displacement of H2A/H2B dimer(s); this results in extensive and fast transcription-dependent exchange of H2A/H2B histones and possibly in rare exchange of H3/H4 histones. Transcription through nucleosomes and displacement of the dimers could be accomplished by PolII itself; however the rate of elongation is strongly increased by TFIIS and FACT. Re-binding of displaced H2A/H2B dimer to DNA occurs almost immediately after PolII passage and most likely is facilitated by FACT. At a higher density of PolII molecules complete histone octamer can be displaced (by one of the mechanisms described in Fig. 3). Since efficiency of this process depends only on the density of PolII molecules, PolII is the primary player in the reaction. If the rate of transcription is decreased, the octamer rebinds to DNA almost immediately; the recovery of chromatin structure depends upon activity of Spt2, Spt6 FACT and Hir proteins. Other designations are as in Fig. 1.
6. Conclusions and perspectives
Recently it has become apparent that transcript elongation in chromatin environment is an extremely complex and finely tuned process that involves a complex orchestrated action of many players including chromatin remodeling enzymes, the enzymes involved in covalent histone modifications (such as acetylation and methylation), elongation factors and a special transcription-dependent chromatin assembly system. One important reason for development of this complicated machinery is the need to keep the DNA in the compact state provided by the chromatin structure. Transcription-dependent disruption of chromatin structure can result in severe decompaction of DNA. The need to keep DNA in a compact state may explain the acrobatic abilities of both PolII and the nucleosome core that allow transcription through nucleosomes without histone displacement into solution, as well as rapid reformation of higher order chromatin structure after transcription. All transcription-dependent changes in chromatin structure seem to be minimal, reversible and fast. These considerations could be especially important for higher eukaryotic organisms where the DNA size is larger and genes on average are transcribed less frequently than in prokaryotes.
The ability of nucleosomes to remain at the original positions on DNA [50] during transcription by PolII could be quite important for preservation of certain histone modifications within the nucleosomal arrays. Indeed, the alternative mechanism (characteristic for PolIII and SP6 RNAPses) includes obligatory nucleosome translocation during transcription that would result in movement of the whole array towards the promoter and disruption of the promoter-proximal nucleosome during every round of transcription [59,105]. This would result in very extensive and obligatory exchange/loss of all core histones. At the same time the PolII-related mechanism allows relatively slow exchange of H3/H4 histones on moderately transcribed genes.
At the same time, the discovery that PolII elongation complex can remodel higher order chromatin structure, displace H2A/H2B histone dimer or complete histone octamer from DNA and deliver numerous chromatin-modifying activities underscores the ability of transcription to serve as a possible modifier of chromatin structure and perhaps participate in establishing of various chromatin states that could be important for proper regulation of eukaryotic genes. Thus it has been suggested that intergenic transcription by “pioneering” PolII across a chromatin domain could make the domain accessible for subsequent activation of transcription of specific genes [106,107]. Transcription-dependent methylation and acetylation of histone H3 can participate in maintenance of a “memory” of the active state of the gene ([108], reviewed in [15,16]) or epigenetic state of X-chromosomes [109]. It is quite possible that survival of H3/H4 histones during moderate transcription contributes to preservation of the “memory”.
It has also been proposed that the high nucleosomal barrier could be used for regulation of the rate of transcript elongation [11]. Indeed, transcript elongation blocks located 20–200 bp downstream of the promoters that are relieved during gene activation have been identified in a growing number of eukaryotic genes, including proto-oncogenes c-myc and c-fos, and HIV-1 polyprotein gene (see [110] for review). One interesting recent example of such regulation is gene activation in yeast resting in the G0 phase of the cell cycle; hundreds of genes that are induced immediately upon exit to the lag phase contain PolII bound immediately upstream or at the beginning of their transcribed regions [111]. These DNA-pre-bound, transcriptionally active PolII molecules most likely mediate fast response of the genes to the cell cycle transition. At least in the case of human hsp 70 gene it was established that the first nucleosome positioned early in the transcribed region of the gene presents a strong barrier for elongating PolII that is regulated by the activator both in vivo and in vitro [11].
Further analysis of the mechanisms involved in transcription through chromatin clearly requires development of an in vitro systems recapitulating chromatin behavior observed in vivo. To be most meaningful, much of this work will have to be carried out with nucleosomal arrays capable of folding into higher order structures and under physiologically relevant conditions.
Acknowledgments
This work was supported by the NIH GM58650 grant to V.M.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 3.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 4.Widom J. Chromatin structure: linking structure to function with histone H1. Curr Biol. 1998;8:R788–791. doi: 10.1016/s0960-9822(07)00500-3. [DOI] [PubMed] [Google Scholar]
- 5.Studitsky VM, Walter W, Kireeva M, Kashlev M, Felsenfeld G. Chromatin remodeling by RNA polymerases. Trends Biochem Sci. 2004;29:127–135. doi: 10.1016/j.tibs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 7.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 9.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corey LL, Weirich CS, Benjamin IJ, Kingston RE. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 2003;17:1392–1401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 12.Studitsky VM. Chromatin remodeling by RNA polymerase II. Mol Biol. 2005;39:639–654. [PubMed] [Google Scholar]
- 13.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 14.Svejstrup JQ. The RNA polymerase II transcription cycle: cycling through chromatin. Biochim Biophys Acta. 2004;1677:64–73. doi: 10.1016/j.bbaexp.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Shilatifard A. Chromatin Modifications by Methylation and Ubiquitination: Implications in the Regulation of Gene Expression. Annu Rev Biochem. 2006 doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 16.Eissenberg JC, Shilatifard A. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr Opin Genet Dev. 2006;16:184–190. doi: 10.1016/j.gde.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Daneholt B. The transcribed template and the transcription loop in Balbiani rings. Cell Biol Int Rep. 1992;16:709–715. doi: 10.1016/s0309-1651(05)80015-3. [DOI] [PubMed] [Google Scholar]
- 18.Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose- regulated genes in yeast. EMBO J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daury L, Chailleux C, Bonvallet J, Trouche D. Histone H3.3 deposition at E2F-regulated genes is linked to transcription. EMBO Rep. 2006;7:66–71. doi: 10.1038/sj.embor.7400561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 21.Nacheva GA, Guschin DY, Preobrazhenskaya OV, Karpov VL, Ebralidse KK, Mirzabekov AD. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989;58:27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- 22.De Bernardin W, Koller T, Sogo JM. Structure of in-vivo transcribing chromatin as studied in simian virus 40 minichromosomes. J Mol Biol. 1986;191:469–482. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- 23.Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- 24.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farris SD, Rubio ED, Moon JJ, Gombert WM, Nelson BH, Krumm A. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J Biol Chem. 2005;280:25298–25303. doi: 10.1074/jbc.M501784200. [DOI] [PubMed] [Google Scholar]
- 27.Thatcher TH, MacGaffey J, Bowen J, Horowitz S, Shapiro DL, Gorovsky MA. Independent evolutionary origin of histone H3.3-like variants of animals and Tetrahymena. Nucleic Acids Res. 1994;22:180–186. doi: 10.1093/nar/22.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 29.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 30.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 31.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci U S A. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 33.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 34.Wirbelauer C, Bell O, Schubeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson V, Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981;23:121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- 38.Jackson V, Chalkley R. Histone synthesis and deposition in the G1 and S phases of hepatoma tissue culture cells. Biochemistry. 1985;24:6921–6930. doi: 10.1021/bi00345a026. [DOI] [PubMed] [Google Scholar]
- 39.Louters L, Chalkley R. Exchange of histones H1, H2A, and H2B in vivo. Biochemistry. 1985;24:3080–3085. doi: 10.1021/bi00334a002. [DOI] [PubMed] [Google Scholar]
- 40.Perry CA, Dadd CA, Allis CD, Annunziato AT. Analysis of nucleosome assembly and histone exchange using antibodies specific for acetylated H4. Biochemistry. 1993;32:13605–13614. doi: 10.1021/bi00212a028. [DOI] [PubMed] [Google Scholar]
- 41.Banditt M, Koller T, Sogo JM. Transcriptional activity and chromatin structure of enhancer-deleted rRNA genes in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4953–4960. doi: 10.1128/mcb.19.7.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dammann R, Lucchini R, Koller T, Sogo JM. Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol Cell Biol. 1995;15:5294–5303. doi: 10.1128/mcb.15.10.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen TA, Allfrey VG. Rapid and reversible changes in nucleosome structure accompany the activation, repression, and superinduction of murine fibroblast protooncogenes c-fos and c-myc. Proc Natl Acad Sci U S A. 1987;84:5252–5256. doi: 10.1073/pnas.84.15.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen TA, Smith MM, Le SY, Sternglanz R, Allfrey VG. Nucleosome fractionation by mercury affinity chromatography. Contrasting distribution of transcriptionally active DNA sequences and acetylated histones in nucleosome fractions of wild-type yeast cells and cells expressing a histone H3 gene altered to encode a cysteine 110 residue. J Biol Chem. 1991;266:6489–6498. [PubMed] [Google Scholar]
- 45.Walia H, Chen HY, Sun JM, Holth LT, Davie JR. Histone acetylation is required to maintain the unfolded nucleosome structure associated with transcribing DNA. J Biol Chem. 1998;273:14516–14522. doi: 10.1074/jbc.273.23.14516. [DOI] [PubMed] [Google Scholar]
- 46.Bazett-Jones DP, Mendez E, Czarnota GJ, Ottensmeyer FP, Allfrey VG. Visualization and analysis of unfolded nucleosomes associated with transcribing chromatin. Nucleic Acids Res. 1996;24:321–329. doi: 10.1093/nar/24.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 48.Bednar J, Studitsky VM, Grigoryev SA, Felsenfeld G, Woodcock CL. The nature of the nucleosomal barrier to transcription: direct observation of paused intermediates by electron cryomicroscopy. Mol Cell. 1999;4:377–386. doi: 10.1016/s1097-2765(00)80339-1. [DOI] [PubMed] [Google Scholar]
- 49.Walter W, Studitsky VM. Facilitated transcription through the nucleosome at high ionic strength occurs via a histone octamer transfer mechanism. J Biol Chem. 2001;276:29104–29110. doi: 10.1074/jbc.M103704200. [DOI] [PubMed] [Google Scholar]
- 50.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II. Loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 51.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 52.Walter W, Kireeva ML, Studitsky VM, Kashlev M. Bacterial polymerase and yeast polymerase II use similar mechanisms for transcription through nucleosomes. J Biol Chem. 2003;278:36148–36156. doi: 10.1074/jbc.M305647200. [DOI] [PubMed] [Google Scholar]
- 53.Luse D, Felsenfeld G. Chromatin Structure and Gene Expression. Oxford University Press; New York: 1995. Transcription through chromatin; pp. 104–122. [Google Scholar]
- 54.Studitsky VM. Transcription through chromatin. Mol Biol (Mosk) 2001;35:235–247. [PubMed] [Google Scholar]
- 55.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 56.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 57.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 58.Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 59.Studitsky VM, Clark DJ, Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 60.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 61.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 62.Walter W, Kireeva ML, Tchernajenko V, Kashlev M, Studitsky VM. Assay of the fate of the nucleosome during transcription by RNA polymerase II. Methods Enzymol. 2003;371:564–577. doi: 10.1016/S0076-6879(03)71042-8. [DOI] [PubMed] [Google Scholar]
- 63.Feng HP, Scherl DS, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–7831. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 64.Brown SA, Kingston RE. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 1997;11:3116–3121. doi: 10.1101/gad.11.23.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown SA, Weirich CS, Newton EM, Kingston RE. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 1998;17:3146–3154. doi: 10.1093/emboj/17.11.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol. 2001;21:5826–5837. doi: 10.1128/MCB.21.17.5826-5837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tennyson CN, Klamut HJ, Worton RG. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 68.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 69.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 70.Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, Lis JT. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 71.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 73.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 74.Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics. 2002;162:1557–1571. doi: 10.1093/genetics/162.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 76.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 78.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci U S A. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mote J, Jr, Reines D. Recognition of a human arrest site is conserved between RNA polymerase II and prokaryotic RNA polymerases. J Biol Chem. 1998;273:16843–16852. doi: 10.1074/jbc.273.27.16843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 81.Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 82.Kulish D, Struhl K. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol Cell Biol. 2001;21:4162–4168. doi: 10.1128/MCB.21.13.4162-4168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 84.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 85.Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Synergistic Functions of SII and p300 in Productive Activator-Dependent Transcription of Chromatin Templates. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 86.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 87.Adkins MW, Tyler JK. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell. 2006;21:405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 88.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila spt5 and spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hartzog GA, Speer JL, Lindstrom DL. Transcript elongation on a nucleoprotein template. Biochim Biophys Acta. 2002;1577:276–286. doi: 10.1016/s0167-4781(02)00458-x. [DOI] [PubMed] [Google Scholar]
- 93.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim DK, Aida M, Inukai N, Narita T, Yamada T, Furuya A, Sato H, Yamaguchi Y, Mandal SS, Reinberg D, Wada T, Handa H. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol Cell Biol. 2004;24:3324–3336. doi: 10.1128/MCB.24.8.3324-3336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nourani A, Robert F, Winston F. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:1496–1509. doi: 10.1128/MCB.26.4.1496-1509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Langst G, Becker PB. Nucleosome remodeling: one mechanism, many phenomena? Biochim Biophys Acta. 2004;1677:58–63. doi: 10.1016/j.bbaexp.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 97.Davie JK, Kane CM. Genetic interactions between TFIIS and the SWI-SNF chromatin-remodeling complex. Mol Cell Biol. 2000;20:5960–5973. doi: 10.1128/mcb.20.16.5960-5973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 99.Cho H, Orphanides G, Sun X, Yang XJ, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stokes DG, Tartof KD, Perry RP. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc Natl Acad Sci U S A. 1996;93:7137–7142. doi: 10.1073/pnas.93.14.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Studitsky VM. Preparation and analysis of positioned nucleosomes. Methods Mol Biol. 1999;119:17–26. doi: 10.1385/1-59259-681-9:17. [DOI] [PubMed] [Google Scholar]
- 106.Travers A. Chromatin modification by DNA tracking. Proc Natl Acad Sci U S A. 1999;96:13634–13637. doi: 10.1073/pnas.96.24.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 108.Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Conaway JW, Conaway RC. Transcription elongation and human disease. Annu Rev Biochem. 1999;68:301–319. doi: 10.1146/annurev.biochem.68.1.301. [DOI] [PubMed] [Google Scholar]
- 111.Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]


