Abstract
Protein 4.1B is a 4.1/ezrin/radixin/moesin domain-containing protein whose expression is frequently lost in a variety of human tumors, including meningiomas, non-small-cell lung cancers, and breast carcinomas. However, its potential tumor-suppressive function under in vivo conditions remains to be validated. In a screen for genes involved with prostate cancer metastasis, we found that 4.1B expression is reduced in highly metastatic tumors. Down-regulation of 4.1B increased the metastatic propensity of poorly metastatic cells in an orthotopic model of prostate cancer. Furthermore, 4.1B-deficient mice displayed increased susceptibility for developing aggressive, spontaneous prostate carcinomas. In both cases, enhanced tumor malignancy was associated with reduced apoptosis. Because expression of Protein 4.1B is frequently down-regulated in human clinical prostate cancer, as well as in a spectrum of other tumor types, these results suggest a more general role for Protein 4.1B as a negative regulator of cancer progression to metastatic disease.
Protein 4.1B is a member of the Protein 4.1 superfamily of proteins, which is characterized by the presence of a conserved N-terminal 4.1/ezrin/radixin/moesin domain. Many of these proteins link transmembrane glycoproteins such as CD44 to the actin cytoskeleton and have been shown to affect numerous processes, including cell polarization, migration, and proliferation, among other functions (1). Based on sequence homology, the Protein 4.1 superfamily of proteins can be further divided into five subgroups: Protein 4.1 molecules, ezrin-radixin-moesin (ERM) proteins, talin-related molecules, protein tyrosine phosphatase proteins, and novel band 4.1-like 4 (2). Given their roles in numerous cellular processes, it is not surprising that some members of these subgroups have also been implicated in tumor progression. In particular, the ERM-like protein, merlin (the product of the NF2 gene), is a critical suppressor of meningiomas and schwannomas (3, 4), and NF2-heterozygous mice develop a variety of spontaneous and highly metastatic tumors (5). In addition, another ERM protein, ezrin, has recently been shown to enhance metastasis of bone and soft tissue sarcomas (6, 7).
The Protein 4.1 subgroup, of which 4.1B is a member, includes at least three additional proteins (4.1G, 4.1N, and 4.1R), and each member possesses N-terminal 4.1/ezrin/radixin/moesin, spectrin-actin-binding, and C-terminal domains (2). Interspersed among these highly conserved domains are three unique regions that likely confer functional specificity to these proteins. However, although 4.1R has been found to be a regulator of erythroid cytoskeletal morphology (8), the precise roles of the other Protein 4.1 subgroup proteins have thus far remained unclear.
In a screen for genes involved with prostate cancer metastasis, we found that 4.1B was down-regulated in highly metastatic tumor cells. Previous studies have shown that 4.1B, or a truncated form of this protein (known as Deleted in Adenocarcinoma of the Lung-1), is frequently lost in brain, lung, and breast cancers, and that overexpression of 4.1B can inhibit the in vitro growth of tumor cell lines (9–14). In some cases, growth suppression was associated with increased apoptosis. However, these results were obtained from overexpression experiments conducted in vitro, and the putative role of 4.1B as a tumor suppressor in vivo has yet to be validated. In addition, 4.1B-deficient mice are healthy and do not develop spontaneous tumors above background levels (15).
In this study, we show that loss of 4.1B promotes metastasis in an orthotopic xenotransplant model of prostate cancer. In addition, by using the transgenic adenocarcinoma of the mouse prostate (TRAMP) tumor model, we observed that 4.1B-deficient mice developed aggressive, spontaneous carcinomas at a significantly higher frequency than did 4.1B-heterozygous mice and that these tumors often metastasized to local lymph nodes. In both models, loss of 4.1B was associated with reduced apoptosis. Combined with clinical data showing that 4.1B expression is down-regulated in four independent studies of human prostate cancer, these results provide in vivo evidence that 4.1B acts as a negative regulator of tumor progression.
Results
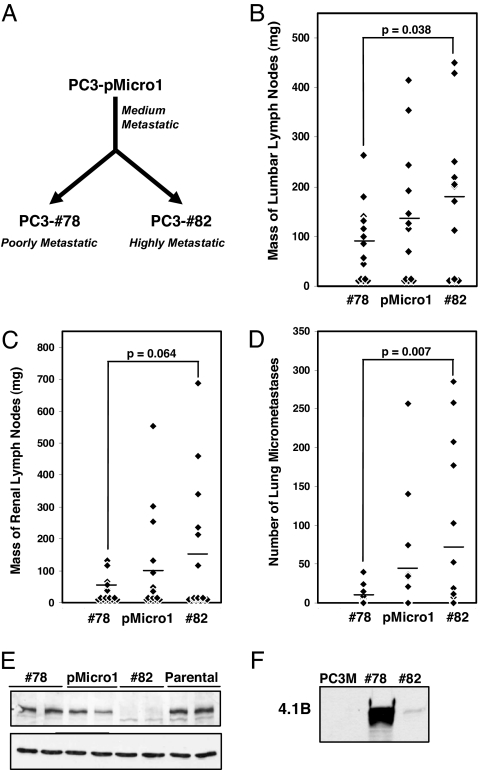
We used a technique known as surgical orthotopic implantation (SOI) to introduce PC-3 cells, a human prostate adenocarcinoma cell line, into immunodeficient mice (16). SOI involves grafting solid, s.c. tumor-derived tissue into the mouse prostate and models many of the initial steps of metastasis, including de-adhesion of malignant cells from the primary tumor, intravasation into lymphatics, and invasion of lymph nodes (17). Repeated in vivo passaging of PC-3 cells using SOI yielded PC3-pMicro-1 cells, from which PC3–#78 cells and PC3–#82 cells were subsequently derived, following an additional passage in vivo (referred to as pMicro-1, #78, and #82 cells, respectively) (Fig. 1 A and Materials and Methods). Further characterization of these three cell lines using SOI revealed that #82 cells formed orthotopic prostate tumors that displayed increased metastasis to the draining paraaortic/lumbar lymph nodes, to the more distal pararenal lymph nodes, and to the lung, relative to tumors derived from #78 cells (Fig. 1 B–D). These differences were seen despite the fact that these cells exhibited similar rates of proliferation in vitro (data not shown) and formed primary tumors of roughly the same size [supporting information (SI) Fig. 6]. Tumors derived from pMicro-1 cells exhibited an intermediate metastatic phenotype.
Fig. 1.
Derivation of metastatic variant prostate cancer cell lines and identification of 4.1B as a protein that is down-regulated in highly metastatic cells. (A) Highly metastatic #82 cells and poorly metastatic #78 cells were isolated after repeated in vivo passaging of PC-3 cells by using surgical orthotopic implantation. (B–D) #82 cells formed orthotopic primary tumors that exhibited an increased propensity to metastasize to paraaortic/lumbar lymph nodes (B), pararenal lymph nodes (C), and lung (D) relative to tumors from #78 cells (horizontal bar, mean). (E) Western blotting for 4.1B (Upper) or GAPDH loading control (Lower) confirmed gene expression analyses indicating that 4.1B was specifically down-regulated in highly metastatic #82 and PC-3M cells (F).
Given the significant differences in metastatic potential between #78 and #82 cells, we decided to perform gene expression analyses on tumors derived from these cells. We found that Protein 4.1B was among the genes most significantly down-regulated in highly metastatic #82 cells (SI Fig. 7). We validated this result at the protein level by performing Western blot analysis for 4.1B on our derived cell lines, the original starting population of PC-3 cells (parental) obtained from American Type Culture Collection (Manassas, VA) (Fig. 1E), and PC-3M cells, a highly metastatic subline independently derived by Kozlowski et al. (18) (Fig. 1F). Consistent with our gene expression results, 4.1B protein levels were specifically down-regulated in highly metastatic #82 and PC-3M cells, relative to poorly metastatic #78 and parental cells. Protein 4.1B was also present at an intermediate level in pMicro-1 cells. However, expression of the related proteins, 4.1G and 4.1N, was unchanged (SI Fig. 8).
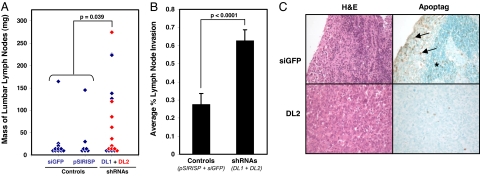
Because Protein 4.1B has been previously implicated in tumor cell migration, adhesion, apoptosis, and growth inhibition in vitro (9–14), we examined this gene candidate further as a potential suppressor of prostate cancer metastasis. We began by down-regulating 4.1B in poorly metastatic #78 cells by stable RNAi (SI Fig. 9). Cells expressing any of four different short-hairpin RNAs (shRNAs) against 4.1B (78-DL1–4) did not exhibit apparent morphological or growth differences in vitro, relative to control cells expressing either the vector only (78-pSIRISP) or an irrelevant shRNA against GFP (78-siGFP) (data not shown). Using SOI, we implanted the shRNA-expressing #78 cells that had exhibited the two most effective knockdowns of 4.1B expression (78-DL1 and 78-DL2), as well as the 78-pSIRISP and 78-siGFP control cells. Although the resulting primary tumor masses in the prostate were roughly equivalent (SI Fig. 10), the draining paraaortic/lumbar lymph nodes from animals implanted with 78-DL1 and 78-DL2 tumors were significantly larger than nodes from mice implanted with control tumors (Fig. 2A; P = 0.039). Subsequent analyses also revealed that, of the two control tumors that exhibited significant lymphatic metastasis in Fig. 2A, one of these tumors (derived from the 78-pSIRISP cell line) had spontaneously down-regulated 4.1B (SI Fig. 11).
Fig. 2.
Down-regulation of 4.1B increases the metastatic potential of poorly metastatic #78 cells. (A and B) The average mass of the draining paraaortic/lumbar lymph nodes (A), and the percentage node area that had been infiltrated by tumor cells (B) were both significantly increased in mice bearing #78 orthotopic tumors expressing DL1 and DL2 shRNAs against 4.1B relative to those bearing #78 control tumors. (C) Paraaortic/lumbar lymph nodes from mice bearing #78 tumors expressing DL1 and DL2 were often completely infiltrated by tumor cells, with few apoptotic cells present (Lower). In contrast, nodes from mice bearing control #78 tumors (Upper) more commonly possessed areas of subcapsular invasion, where apoptotic cells were frequently observed (arrows). In most cases, the interior regions of these nodes were relatively uninvaded (*). Representative serial lymph node sections stained either by H&E (Left) or for apoptosis (Right) are shown at ×20 magnification.
Histological examination revealed that paraaortic/lumbar lymph nodes from mice implanted with 78-DL1 and 78-DL2 tumors were often completely infiltrated with tumor cells (Fig. 2B; n = 46 nodes examined). However, nodes from animals implanted with control tumors frequently exhibited only partial, subcapsular invasion (n = 42 nodes examined; P < 0.0001), and this site was where many tumor cells appeared apoptotic (Fig. 2C Upper, arrows). In contrast, apoptosis was rare in nodes that were completely replaced by tumor cells (Fig. 2C Lower), as was often the case with shRNA-expressing tumors. Mice implanted with tumors expressing shRNAs against Protein 4.1B also developed somewhat more lung metastases than those implanted with control tumors, although these differences did not reach statistical significance because of the limited number of animals that had exhibited systemic spread (SI Fig. 12). Overall, these results indicate that Protein 4.1B suppresses prostate cancer metastasis, and down-regulation of 4.1B is sufficient to increase the metastatic potential of poorly metastatic cells.
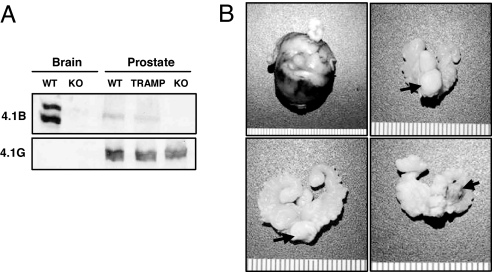
We next determined whether loss of Protein 4.1B could also affect tumor progression in a spontaneous model of prostate cancer. We obtained 4.1B knockout mice (15) and confirmed the absence of this protein by performing Western blotting on brain and prostate lysates from 4.1B+/+ or 4.1B−/− animals (Fig. 3A). We then crossed these animals with TRAMP mice expressing SV40 in the prostate (19). At 26 weeks of age, palpable carcinomas in the prostate were observed in 11 of 19 (58%) of 4.1B−/−;TRAMP+/− mice, compared with 2 of 26 (7.7%) of 4.1B+/−;TRAMP+/− mice (Fig. 3B and Table 1; P = 0.0002).
Fig. 3.
4.1B−/− mice develop aggressive adenocarcinomas in a spontaneous tumor model of prostate cancer. (A) Western blotting for 4.1B (Upper) or 4.1G (Lower) in brain or prostate tissues shows that 4.1B is specifically lost in knockout animals. (B) 4.1B−/−;TRAMP mice more commonly developed a variety of palpable, high-grade carcinomas in the prostate (arrows), including (starting from Upper Left and proceeding clockwise) compound, multilobed carcinomas; ventral-lobed carcinomas; anterior-lobed carcinomas; and dorsal-lobed carcinomas. These results are tabulated in Table 1.
Table 1.
Effects of protein 4.1B on tumor progression in a spontaneous model of prostate cancer
| Type of cancer | Incidence (%) |
P value | |
|---|---|---|---|
| 4.1B+/−/TRAMP+/− | 4.1B−/−/TRAMP+/− | ||
| Grade 6 carcinoma | 4/26 (15) | 11/19 (58) | 0.003 |
| Palpable grade 6 carcinoma | 2/26 (7.7) | 11/19 (58) | 0.0002 |
| Lymph node metastasis | 2/26 (7.7) | 6/19 (32) | 0.04 |
Results are given as the number affected from the total number of mice of each genotype. Percentages are given in parentheses.
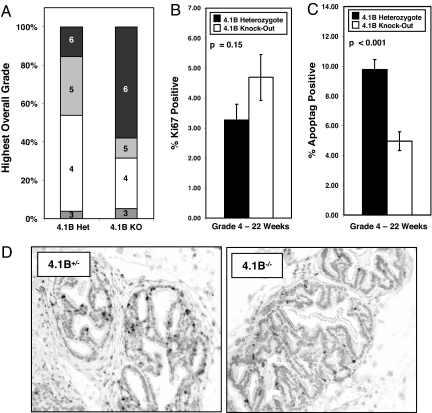
Using the system described by Hurwitz et al. (20), we then performed histopathological grading on both the ventral and dorsolateral prostate lobes for all animals (see Materials and Methods). Sections were scored on a scale of 1 to 6 (1, normal prostate; 6, poorly differentiated adenocarcinoma). Because tumorigenic prostates often exhibited heterogeneity with regard to severity, even within the same section, two grades were assigned for each prostate lobe of each animal: a highest grade and a predominant grade (SI Fig. 13). Although most mice, regardless of genotype, developed histologically detectable prostate cancer with a grade of 4 or higher, we invariably found that 4.1B−/−;TRAMP+/− mice developed the highest, least differentiated, and most aggressive grade of prostate cancer (grade 6) at a significantly increased frequency relative to heterozygotes [Table 1 and Fig. 4A (P = 0.003) and SI Fig. 14]. As was seen in the xenotransplant model, 4.1B−/− TRAMP tumorigenic prostates also displayed an increased propensity to metastasize to draining lymph nodes relative to 4.1B+/− TRAMP prostates (Table 1). The presence of metastatic tumor cells in the nodes was confirmed by cytokeratin 8 staining (SI Fig. 15). These results indicate that loss of Protein 4.1B promotes progression of spontaneous prostate cancer.
Fig. 4.
Higher grade and less differentiated 4.1B−/−;TRAMP prostate carcinomas likely arise from tumorigenic lesions that exhibit less apoptosis than those of heterozygous lesions. (A) Each entire prostate was assigned a single highest grade identical to the highest grade assigned to any one constituent lobe. Please refer to SI Fig. 14 for additional histologic scoring results organized by prostate lobe. (B) Proliferation rates were similar regardless of 4.1B status in 22-week-old, grade 4 TRAMP ventral prostates, but 4.1B−/− TRAMP ventral prostates were significantly less apoptotic relative to grade- and age-matched 4.1B+/− TRAMP prostates (C). (D) Representative sections stained for apoptotic cells.
We next stained prostate sections for proliferation and apoptotic markers. Because tumorigenesis in the TRAMP model begins with normal prostate tissue and increases with severity over time, we reasoned that if proliferative or apoptotic differences were manifested in the lower graded prostatic tumors, the overall rate of progression to grade 6 adenocarcinoma would be affected. Therefore, we began by staining 26-week-old, grade 4-matched prostate sections with Ki67 antibody and TUNEL. We found no differences between 4.1B−/− or 4.1B+/− TRAMP prostates for either proliferation or apoptosis, and we also found no differences in 26-week-old, grade 5-matched sections (SI Fig. 16). However, especially for the 4.1B−/− cohort, these samples were biased in that many tumors had already progressed to grade 6 and could not be used for this analysis. In other words, what had been analyzed were the remaining tumorigenic prostates that had not progressed to the higher grades. To perform a more unbiased study, we stained grade 4-matched prostate sections from a younger cohort of 22-week-old TRAMP mice. Although there was again largely no difference in proliferation (Fig. 4B; P = 0.15), we found that 4.1B−/−;TRAMP prostates displayed significantly lower rates of apoptosis compared with heterozygous TRAMP mice (Fig. 4 C and D; P < 0.001). These results suggest that loss of 4.1B protects cancerous lesions from undergoing cell death at 22 weeks, which may facilitate progression to higher grade, less differentiated, and more invasive tumors by 26 weeks.
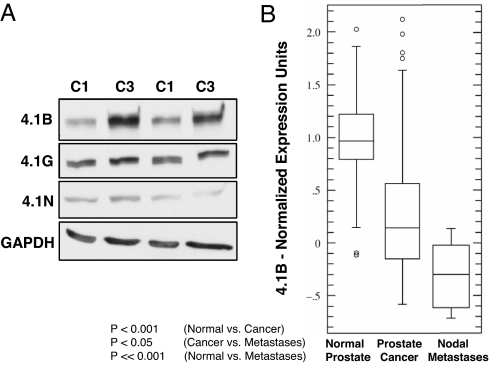
To determine whether down-regulation of 4.1B might contribute to prostate cancer progression in other experimental systems, we examined the TRAMP-C series of cell lines, which were derived by Foster et al. (21). Consistent with our xenotransplant and spontaneous tumor results, we found that 4.1B protein levels were specifically reduced in malignant TRAMP-C1 cells relative to nonmalignant TRAMP-C3 cells (Fig. 5A). In addition, we found that 4.1B gene expression was significantly down-regulated in four studies of human clinical prostate cancer. For instance, Oncomine analysis of a data set originally obtained by Lapointe et al. (22) revealed that 4.1B expression was significantly down-regulated in prostate tumor samples relative to normal prostate tissues (Fig. 5B; P < 0.001) and further down-regulated in lymph node metastases compared with normal prostate (P < 0.001). Oncomine analysis also revealed three additional studies where 4.1B expression was significantly down-regulated (P ≤ 0.001) in prostate cancer. These data sets were originally collected by Welsch et al., Singh et al., and Yu et al. (23–25) and are displayed in SI Fig. 17.
Fig. 5.
4.1B is down-regulated in aggressive TRAMP tumor cell lines and in human clinical prostate cancer. (A) 4.1B protein levels are reduced in malignant TRAMP-C1 cells (C1) relative to nonmalignant TRAMP-C3 cells (C3); independent replicate samples are shown. (B) 4.1B expression is significantly down-regulated during human clinical prostate cancer progression, from normal prostate tissue, to prostate cancer, and finally to prostate cancer lymph node metastases [data obtained by Lapointe et al. (22) and processed by Oncomine 3.0]. Three additional human clinical prostate cancer studies displaying down-regulation of 4.1B can be found in SI Fig. 17.
Discussion
We have shown here that Protein 4.1B acts as a negative modulator of the aggressive tumor phenotype in two different in vivo models of prostate cancer. We found that 4.1B expression was significantly reduced in highly metastatic human prostate adenocarcinoma cells, and that down-regulation of 4.1B in poorly metastatic cells was sufficient to increase their metastatic propensity in a xenotransplant orthotopic model. Furthermore, 4.1B-deficient TRAMP mice displayed increased susceptibility for developing invasive, undifferentiated adenocarcinomas relative to 4.1B-heterozygous TRAMP mice. In both systems, tumorigenic cells lacking 4.1B displayed reduced apoptosis, which may have promoted the invasive phenotype.
Our findings are concordant with in vitro studies showing that overexpression of 4.1B can, in some cases, induce cell death (11, 14). Although the mechanisms by which 4.1B promotes apoptosis remain unclear, one study recently showed that overexpression of 4.1B increases caspase-8 activity in MCF-7 cells (14). Others have reported that overexpression of 4.1B induces Rac1-dependent JNK signaling (13). Recent work from our laboratory has also shown that 4.1B can interact with a potential tumor suppressor, integrin β8 (26), and this interaction has been confirmed in our PC-3 cells (data not shown).
Although 4.1B was originally identified as a protein whose expression was reduced in human non-small-cell lung carcinomas (9), subsequent studies have shown that down-regulation occurs across many different tumor cell types, including meningiomas, and also carcinomas arising from the breast, kidney, and colon/rectum. For instance, 4.1B has been reported to be lost in ≤60% of sporadic meningiomas (27), whereas allelic loss of region 18p11.3, where the 4.1B gene resides, was detected in 55% of ductal carcinomas in situ, as well as in 67% of invasive and metastatic breast cancers (28). In addition, 4.1B protein expression has been observed to be down-regulated during tumorigenesis in spontaneous mouse models of pancreatic and intestinal cancer (29, 30). Consistent with these findings, we have observed that 4.1B expression is significantly reduced in four studies of human clinical prostate cancer. We have also found on Oncomine that 4.1B levels are significantly down-regulated (P < 0.001) in human clinical lung and liver cancers (data not shown; see refs. 31 and 32). Even more interestingly, the Oncomine database revealed that prostate tumors which overexpressed the Ets family transcription factor ERG exhibited significantly lower levels of 4.1B expression relative to tumors that did not overexpress Ets family transcription factors (SI Fig. 18; P < 0.001; see ref. 22). Because recent studies have shown that the genes encoding ERG and another Ets family transcription factor, ETV1, are translocated and, consequently, overexpressed in as many as 80% of human prostate cancers, it is important in future work to determine whether these genes are inversely coregulated (33).
Because Protein 4.1B has been frequently observed to be lost in both low- and high-grade meningiomas and breast carcinomas, these findings have led some to propose that loss of this protein is a critical early step for tumorigenesis (27, 28). However, as one study examining breast cancer has noted, there were also instances where the chromosomal region 18p11.3 was specifically lost in invasive tumor foci, but not in regions of ductal carcinomas in situ in samples derived from the same patients (28). These results suggest that down-regulation of 4.1B during either the early or later stages of tumorigenesis can promote cancer progression. Our parental PC-3 prostate cells expressed high levels of 4.1B and loss of this protein was only observed in aggressive variant cells (PC3–#82 and PC-3M) derived from this original starting population, suggesting that down-regulation of 4.1B enhanced the later stages of tumor progression to metastatic disease. Similarly, because genetic ablation of 4.1B in mice did not increase their predisposition to spontaneous tumor formation (15), but instead promoted the progression of prostatic tumor lesions already initiated by SV40, this observation again suggests that loss of 4.1B is important during the later stages of prostate tumorigenesis. In addition, as we have shown with the gene expression data set obtained from Lapointe et al. (22), prostate tumors might down-regulate 4.1B expression relative to normal tissue early on and then further down-regulate expression during progression to metastatic disease. Whether 4.1B expression is also correlated with other clinical parameters for assessing prostate cancer severity, such as Gleason grade, remains to be seen.
In summary, our results obtained from several independently derived prostate cancer cell lines, two different in vivo tumor models, and four human clinical studies of prostate cancer together suggest that down-regulation of 4.1B is a frequent and important event in prostate cancer that causally contributes to an aggressive tumor phenotype. Our findings offer in vivo evidence that a member of the Protein 4.1 subgroup can act as a suppressor of tumor progression. Because protein 4.1B expression is down-regulated in a variety of human cancers, in addition to those arising from the prostate, these findings suggest that 4.1B may serve a more general role as a negative modulator of malignancy across a spectrum of tumor cell types.
Materials and Methods
Cells.
All derivatives of the human cell line PC-3 (American Type Culture Collection) were cultured as previously described (17, 34). The metastatic variant cell lines were derived by passaging PC-3 cells in vivo: The cells were grafted by SOI into the prostate and allowed to form palpable tumors over a period of 2 to 3 months, after which metastatic cells were isolated from lung tissues physically dissociated in culture. These cells were expanded in vitro and then reimplanted into mice for additional rounds of in vivo passage. Implantation of PC-3 cells originally yielded the pMicro-1 cell line, and subsequent reimplantation of pMicro-1 cells yielded the PC3–#78 and PC3–#82 cell lines, which were isolated from separately grafted mice. TRAMP-C1 and -C3 cells (American Type Culture Collection) were cultured as previously described (21). PC-3M cells were obtained from I. J. Fidler (M. D. Anderson Cancer Center, Houston, TX) (18).
Plasmids and Antibodies.
shRNA sequences were cloned into the retroviral vector, pSIRISP (W. C. Hahn, Dana–Farber Cancer Institute, Boston, MA) (35). The plasmids were transfected into Phoenix cells (American Tissue Culture Collection), and viral supernatant was used to stably infect PC3–#78 cells expressing ecotropic receptor (H. Lodish, Massachusetts Institute of Technology). See SI Materials and Methods for shRNA sequences. Antibodies for Western blot included rabbit anti-4.1B (26), as well as rabbit anti-4.1G and anti-4.1N (Protein Express, Chiba, Japan). Total soluble protein was extracted from frozen tumors or cells/tissues by using lysis buffer containing Nonidet 40 and protease inhibitors (Roche, Mannheim, Germany). For immunohistochemistry, zinc- (Becton Dickinson, San Jose, CA) or formalin-fixed sections were stained with rat anti-cytokeratin 8 (TROMA-1; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) (36).
Orthotopic Xenograft and Spontaneous Mouse Tumor Models.
SOI of PC-3 cell derivatives was performed as described previously (17). Briefly, cells were injected as s.c. tumors and harvested ≈3.5 weeks later to obtain graft material for SOI. The procedure was performed in accordance with the Massachusetts Institute of Technology Division of Comparative Medicine animal care guidelines. Between 2 and 3 months after implantation, mice were analyzed when moribund. Lymph nodes were removed, fixed in formalin, and later weighed and sectioned. Lymph node mass was recorded as the combined fixed mass of all nodes from a particular site (paraaortic or renal lymph node site) for each animal. Combined lymph node masses that were <30 mg could not be reliably weighed and were given a mass of 10 mg. To determine percent metastatic invasion of lymph nodes, two sections at different levels were obtained for each node, percent invasion was scored blindly by a pathologist (R.T.B.), and the higher of the two scores was recorded for every individual node.
For spontaneous tumor studies, 4.1B−/− animals were back-crossed into a C57BL/6 background for two to three generations. These mice were crossed with TRAMP+/− mice in a pure C57BL/6 background. In TRAMP studies, individual prostate lobes were sectioned and blindly graded by a pathologist (M.B.) (20). A highest grade was assigned based on the area of greatest pathological severity. A predominant grade was assigned based on the most common grade seen in each section. See SI Fig. 13 for an example of this scoring approach.
Proliferation and Apoptosis.
Zinc-fixed prostate lobes from TRAMP mice and formalin-fixed lymph nodes from mice implanted by SOI were stained with Ki67 antibody (Sigma–Aldrich, St. Louis, MO) for proliferation and/or by TUNEL for apoptosis (ApopTag Plus; Chemicon International, Temecula, CA). Quantitation was performed on grade- and age-matched sections (22 or 26 weeks old) by counting the number of positively stained cells, as well as the total number of cells, from two different fields per section imaged at ×20 magnification, except for grade 5 sections, which were imaged at ×40 magnification. Grade 4 sections were all derived from the ventral lobes of the prostate, whereas grade 5 sections were all derived from dorsolateral lobes. All staining, image capturing with OpenLab 5.0.0 software (Improvision, Lexington, MA), and quantitation were blindly performed by using four to five independent prostate samples per genotype.
Microarrays and Bioinformatics.
For gene expression analyses, total RNA was extracted by RNeasy Midi kit (Qiagen, Valencia, CA) from flash-frozen s.c. tumors. Biotinylated cRNA was hybridized to human U133A chips (Affymetrix, Santa Clara, CA). Data were analyzed by dChip software (Harvard Medical School, Boston, MA) (37) and Gene Pattern (The Broad Institute, Cambridge, MA) (38). The entire MIAME-compliant data set can be downloaded from National Center for Biotechnology Information GEO (www.ncbi.nlm.nih.gov/geo/index.cgi), accession no. GSE7930. Oncomine analyses were performed August 2006 by using Oncomine 3.0 (www.oncomine.org) (39).
Statistics.
Statistics were performed by using an unpaired Student's t test (www.physics.csbsju.edu/stats/Index.html), except in the case of TRAMP tumor incidence data, which were assessed by chi square (www.psych.ku.edu/preacher/chisq/chisq.htm). All error bars shown are standard error.
Supplementary Material
Acknowledgments
We thank Dr. J.H. McCarty (M. D. Anderson Cancer Center, Houston, TX) for anti-4.1B antibodies; the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA) for anti-cytokeratin 8 antibodies; Dr. A. Bai (Massachusetts Institute of Technology) for TRAMP mice; the Massachusetts Institute of Technology BioMicro Center; and the Massachusetts Institute of Technology Division of Comparative Medicine. This work was supported by National Institutes of Health Grant R01CA17007 (to R.O.H.); the Virginia and D.K. Ludwig Fund for Cancer Research; the Prostate Cancer Foundation; National Cancer Institute's Integrative Cancer Biology Program Grant U54-CA112967 (to R.O.H.); the Howard Hughes Medical Institute, of which R.O.H. is an Investigator; a National Institute of General Medical Sciences Predoctoral Training Grant (to S.Y.W.); and a David H. Koch Research Fellowship from the Center for Cancer Research.
Abbreviations
- ERM
ezrin-radixin-moesin
- shRNA
short-hairpin RNA
- SOI
surgical orthotopic implantation
- TRAMP
transgenic adenocarcinoma of the mouse prostate.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE7930).
This article contains supporting information online at www.pnas.org/cgi/content/full/0705499104/DC1.
References
- 1.Bretscher A, Edwards K, Fehon RG. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 2.Sun CX, Robb VA, Gutmann DH. J Cell Sci. 2002;115:3991–4000. doi: 10.1242/jcs.00094. [DOI] [PubMed] [Google Scholar]
- 3.Gutmann DH, Giordano MJ, Fishback AS, Guha A. Neurology. 1997;49:267–270. doi: 10.1212/wnl.49.1.267. [DOI] [PubMed] [Google Scholar]
- 4.McClatchey AI, Giovannini M. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- 5.McClatchey AI, Saotome I, Mercer K, Crowley D, Gusella JF, Bronson RT, Jacks T. Genes Dev. 1998;12:1121–1133. doi: 10.1101/gad.12.8.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khanna C, Wan X, Bose S, Cassaday R, Olomu O, Mendoza A, Yeung C, Gorlick R, Hewitt SM, Helman LJ. Nat Med. 2004;10:182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 7.Yu Y, Khan J, Khanna C, Helman L, Meltzer PS, Merlino G. Nat Med. 2004;10:175–181. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
- 8.Tchernia G, Mohandas N, Shohet SB. J Clin Invest. 1981;68:454–460. doi: 10.1172/JCI110275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran YK, Bogler O, Gorse KM, Wieland I, Green MR, Newsham IF. Cancer Res. 1999;59:35–43. [PubMed] [Google Scholar]
- 10.Gutmann DH, Hirbe AC, Huang ZY, Haipek CA. Neurobiol Dis. 2001;8:266–278. doi: 10.1006/nbdi.2000.0376. [DOI] [PubMed] [Google Scholar]
- 11.Charboneau AL, Singh V, Yu T, Newsham IF. Int J Cancer. 2002;100:181–188. doi: 10.1002/ijc.10470. [DOI] [PubMed] [Google Scholar]
- 12.Robb VA, Gerber MA, Hart-Mahon EK, Gutmann DH. Oncogene. 2005;24:1946–1957. doi: 10.1038/sj.onc.1208335. [DOI] [PubMed] [Google Scholar]
- 13.Gerber MA, Bahr SM, Gutmann DH. Cancer Res. 2006;66:5295–5303. doi: 10.1158/0008-5472.CAN-05-1628. [DOI] [PubMed] [Google Scholar]
- 14.Jiang W, Newsham IF. Mol Cancer. 2006;5:1–8. doi: 10.1186/1476-4598-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi C, McCarty JH, Troutman SA, Eckman MS, Bronson RT, Kissil JL. Mol Cell Biol. 2005;25:10052–10059. doi: 10.1128/MCB.25.22.10052-10059.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An Z, Wang X, Geller J, Moossa AR, Hoffman RM. Prostate. 1998;34:169–174. doi: 10.1002/(sici)1097-0045(19980215)34:3<169::aid-pros3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 17.Wong SY, Haack H, Crowley D, Barry M, Bronson RT, Hynes RO. Cancer Res. 2005;65:9789–9798. doi: 10.1158/0008-5472.CAN-05-0901. [DOI] [PubMed] [Google Scholar]
- 18.Kozlowski JM, Fidler IJ, Campbell D, Xu ZL, Kaighn ME, Hart IR. Cancer Res. 1984;44:3522–3529. [PubMed] [Google Scholar]
- 19.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, Cunha GR, Donjacour AA, Matusik RJ, Rosen JM. Proc Natl Acad Sci USA. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurwitz AA, Foster BA, Allison JP, Greenberg NM, Kwon ED. Current Protocols in Immunology. New York: Wiley; 2001. pp. 20.5.1–20.5.23. [DOI] [PubMed] [Google Scholar]
- 21.Foster BA, Gingrich JR, Kwon ED, Madias C, Greenberg NM. Cancer Res. 1997;57:3325–3330. [PubMed] [Google Scholar]
- 22.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson J, Hampton GM. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 24.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie, et al. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 25.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 26.McCarty JH, Cook AA, Hynes RO. Proc Natl Acad Sci USA. 2005;102:13479–13483. doi: 10.1073/pnas.0506068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutmann DH, Donahoe J, Perry A, Lemke N, Gorse K, Kittiniyom K, Rempel SA, Gutierrez JA, Newsham IF. Hum Mol Genet. 2000;9:1495–1500. doi: 10.1093/hmg/9.10.1495. [DOI] [PubMed] [Google Scholar]
- 28.Kittiniyom K, Gorse KM, Dalbegue F, Lichy JH, Taubenberger JK, Newsham IF. Breast Cancer Res. 2001;3:192–198. doi: 10.1186/bcr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohno N, Terada N, Murata S, Yamakawa H, Newsham IF, Katoh R, Ohara O, Ohno S. Histochem Cell Biol. 2004;122:579–586. doi: 10.1007/s00418-004-0716-7. [DOI] [PubMed] [Google Scholar]
- 30.Terada N, Ohno N, Yamakawa H, Baba T, Fujii Y, Christofori G, Ohara O, Ohno S. Histochem Cell Biol. 2003;120:277–283. doi: 10.1007/s00418-003-0573-9. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 34.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Invest Urol. 1979;17:16–23. [PubMed] [Google Scholar]
- 35.Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, et al. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 36.Brulet P, Babinet C, Kemler R, Jacob F. Proc Natl Acad Sci USA. 1980;77:4113–4117. doi: 10.1073/pnas.77.7.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C, Wong WH. Genome Biol. 2001;2:0032.1–0032.11. [Google Scholar]
- 38.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.