Abstract
Using a combination of techniques we developed, we infected zebrafish embryos using pseudotyped retroviruses and mapped the genomic locations of the proviral integrations in the F1 offspring of the infected fish. From F1 fish, we obtained 2,045 sequences representing 933 unique retroviral integrations. A total of 599 were mappable to the current genomic assembly (Zv6), and 233 of the integrations landed within genes. By inbreeding fish carrying proviral integrations in 25 different genes, we were able to demonstrate that in ≈50% of the gene “hits,” the mRNA transcript levels were reduced by ≥70%, with the highest probability for mutation occurring if the integration was in an exon or first intron. Based on these data, the mutagenic frequency for the retrovirus is nearly one in five integrations. In addition, a strong mutagenic effect is seen when murine leukemia virus integrates specifically in the first intron of genes but not in other introns. Three of 19 gene inactivation events had embryonic defects. Using the strategy we outlined, it is possible to identify 1 mutagenic event for every 30 sequencing reactions done on the F1 fish. This is a 20- to 30-fold increase in efficiency when compared with the current resequencing approach [targeting induced local lesions in genomes (TILLING)] used in zebrafish for identifying mutations in genes. Combining this increase in efficiency with cryopreservation of sperm samples from the F1 fish, it is now possible to create a stable resource that contains mutations in every known zebrafish gene.
Keywords: genetics, retrovirus
Zebrafish (Danio rerio) has become a powerful model organism for studying vertebrate development. One of the primary reasons for the popularity of zebrafish is that they are particularly amenable to forward genetic studies to isolate mutations affecting early development. Most forward genetic studies have been conducted by using a chemical mutagen, the methylating agent ethylnitrosourea (ENU) (1). The mutated genes must then be identified by positional cloning (2, 3). However, forward genetic screens are fundamentally limited in their effectiveness by issues such as redundancy and the need to have a measurable phenotype. It is therefore also desirable to obtain mutations specifically in genes of interest and evaluate the effects of these mutations (i.e., “reverse” genetics). Unlike research using mice, a homologous recombination-based targeted gene knockout in zebrafish is still unavailable. As an alternative, a technique known as targeting induced local lesions in genomes (TILLING) that identifies fish harboring point mutations in a gene of interest within a population of ENU-mutagenized fish has been recently developed (4–6). However, this technique is labor-intensive and requires significant DNA sequencing capacity for each identified mutation (7, 8).
As an alternative to ENU, another mutagen that has been used for zebrafish genetic screens is the murine leukemia virus (MLV) (9–11). A great advantage of retroviral mutagenesis over ENU is that it allows the rapid identification of the mutated gene because of the presence of a proviral molecular “tag” at the site of insertion (11–13). Thus, saturation mutagenesis using the retrovirus as the mutagen followed by the rapid identification of the proviral insertion sites in the genome could be an alternative reverse genetics approach. Similar in principle to the Arabidopsis T-DNAExpress (http://signal.salk.edu/cgi-bin/tdnaexpress) or to gene-trap efforts in mouse embryonic stem cells (14–17), DNA fragments flanking the insertional mutagen (in our case, an MLV based retrovirus) are isolated, sequenced, and indexed to cryopreserved sperm samples (Fig. 1). Once saturation is reached, any desired mutant line could readily be generated through in vitro fertilization of the frozen sperm sample containing the integration of interest, and the F2 fish would be available for inbreeding in 3–4 months.
Fig. 1.
Construction of a zebrafish retroviral insertional mutant library. Pipeline for constructing the zebrafish insertional mutant library: (i) infect zebrafish embryos with pseudotyped MLV virus at the 1,000- to 2,000-cell stage; (ii) raise the injected founder fish with high infection rates, determined by qPCR; (iii) inbreed or outcross founders (depending on the infection rates of the founder fish) and raise the F1 fish; (iv) cryopreserve sperm samples from the F1 fish and perform LM-PCR followed by shotgun cloning, sequencing, and mapping the integrations in the corresponding sperm samples.
Results
To assess whether retroviral mutagenesis is amenable as a tool for saturation mutagenesis, we tested three aspects of mutagenic efficiency of this technology: (i) the efficiency of the retrovirus infecting the injected fish (founders) and the subsequent germ-line transmission of the proviral insertions to the F1 fish; (ii) the efficiency of proviral insertions disrupting gene expression; and (iii) the overall rate of production, which determines the workforce required to mutate every gene in a timely, cost-efficient manner.
Improved Infection Rates.
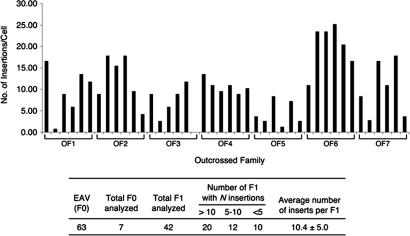
Similar to previous screening efforts using retroviral mutagenesis (12, 18, 19), we chose an MLV-based retrovirus pseudotyped with the vesicular stomatitis virus glycoprotein (20–22). The infection efficiency was assessed by quantitative PCR (qPCR), which determines the ratios of proviral DNA to a genomic reference. The average copy number of proviral integrations per cell in each batch of injected fish was determined at 2 days after injection and is called the embryo assay value (EAV). In a previous effort (21), the average EAV of injected founder fish was 14.3 integrations per cell. We improved the transfection and injection techniques (see Methods), so now the EAV of our injected founder fish increased significantly, ranging between 27 and 95, with an average value of 46.4 ± 24.1. This result was determined from 21 batches of injected founders using 10 independent virus preparations. This indicates an average 3-fold increase in the infection rate of the injected fish over what was previously achievable (12, 21). We then further assessed the germ-line transmission rate of our injected founder fish. We chose a batch of founders with an average EAV of 63 integrations per cell, raised them to sexual maturity, and outcrossed males with WT females to generate F1 families. We randomly chose seven F1 families, selected six fish from each family, and performed qPCR to determine the copy number of proviral insertions in each F1 fish. We found that the average number of proviral insertions per F1 fish is 10.4 ± 5.0 (Fig. 2). Because of the germ-line mosaicism of founder fish (viral infection begins at the 1,000- to 2,000-cell stage), there is an expected significant variation in the germ-line transmission rates of proviral insertions from founder to founder, as well as between the F1 siblings from each founder (Fig. 2). However, from a batch of injected founders with a high EAV, the chance of randomly selecting F1 fish with multiple proviral insertions is high (i.e., as Fig. 2 shows, 32 of 42, or 76% of randomly selected F1 fish harbor more than five proviral insertions). These results indicate that, with the current protocols, we can randomly select F1 fish and have a high confidence that every fish will have multiple proviral integrations.
Fig. 2.
Germ-line transmission rates tested in selected founders. Seven founders with an average EAV of 63 integrations per cell were randomly selected and outcrossed with WT fish to generate F1 families, OF1–OF7. Six F1 fish were randomly selected from each family, and the copy number of proviral integrations in each F1 fish was determined by quantitative PCR. Variations in germ-line transmission of proviral insertions can be seen between different founders (e.g., all selected F1 fish from family OF6 have >10 copies of proviral insertions per cell, whereas from OF5, only two F1 fish have more than five copies of proviral insertions per cell) and between the F1 siblings of each founder (e.g., the copy numbers of proviral insertions in selected F1 fish from OF1 range between 0.7 and 16.6 copies per cell). Overall, 32 of 42, or 76% of randomly selected F1 fish harbor more than five proviral insertions per cell with an average value of 10.4 ± 5.0 copies per cell.
Efficiently Mapping Unique Retroviral Integrations.
To identify unique proviral integrations from the F1 generation efficiently, the challenge becomes selecting the appropriate number of F1 fish from each founder family to maximize the number of unique integrations while minimizing any redundancy that might occur from multiple F1 fish carrying the same integration. In principle, it is possible to individually isolate the flanking DNA from each integration in each fish before the sequencing is performed, but in terms of workflow, the advantages of such an approach are far outweighed by the increase in required labor. A more practical and streamlined approach would be to determine empirically a number of randomly selected F1 fish and a number of sequences obtained per F1 fish that would yield a high number of unique integrations. This approach would significantly reduce labor, allowing for a very small number of steps in the process: (i) inject founder fish, (ii) outcross to WT fish, (iii) raise a small number of male F1 fish from each cross, (iv) isolate and sequence proviral integration sites, and (v) cryopreserve the sperm (Fig. 1).
To empirically establish a guideline for this “random selection” approach, we selected 32 high-quality founders (i.e., EAV >25), outcrossed them with WT fish to generate the F1 generation, and then randomly selected six F1 fish per founder family for linker-mediated PCR (LM-PCR) analysis. After LM-PCR, a screen was performed to eliminate those F1 fish showing few or no PCR products (by simply running a portion of the LM-PCR products on gels) before the shotgun cloning. We found that the number of sequences that would generate unique integration sites was directly proportional to the EAV of founders. We could increase the number of sequences per F1 from 4 to 12 according to the increases in EAV of the founders while maintaining ≈45% of sequences as unique sequences (Table 1). Similar results were obtained from founders with lower infection rates if we inbred two founders instead of outcrossing to WT (Table 1). Thus, by selecting one to six F1 fish (average 3.5) per founder and using founders' EAV as the guideline to determine the number of sequences per F1, we can consistently obtain ≈50% of sequences identifying unique integration sites or on average 2–10 unique sequences per founder depending on their “quality” (i.e., infection rate) (Table 1).
Table 1.
Test of the random selection approach
| Infection rate of F0 | EAV 5–10 | EAV 25–35 | EAV 35–70 | EAV >70 |
|---|---|---|---|---|
| Number of F0 | 54 | 9 | 16 | 7 |
| Type of cross | Inbreed | Outcross | Outcross | Outcross |
| Number of F1 per F0 (before LM-PCR) | 6 | 6 | 6 | 6 |
| Number of F1 per F0 (after LM-PCR)* | 1–6 | 1–6 | 1–6 | 1–6 |
| Total number of F1 cloned | 108 | 35 | 35 | 23 |
| Number of sequences per F1 | 4 | 4 | 8 | 12 |
| Number of analyzed sequences† | 227 | 85 | 183 | 151 |
| Unique sequences | 126 (56%) | 39 (46%) | 83 (45%) | 68 (45%) |
| Unique sequences per F0 | 2.3 | 4.3 | 5.2 | 9.7 |
*F1 with few or no LM-PCR products were excluded from cloning.
†Short (<18-bp), linker-only, and ambiguous sequences were excluded.
Retroviral Integrations in Genes Effectively Reduce mRNA Levels.
The second critical aspect of this approach that needed to be determined was how efficiently the proviral insertions disrupted gene expression. To establish the distribution profile of proviral integration sites in the zebrafish genome, ≈300 founder families, a total of 854 F1 fish, were subjected to high-throughput cloning and sequence analysis using the “random selection” approach. Of 2,045 total sequence reads, we obtained 933 unique sequences meeting our validity criteria (see Methods). Of those 933 reads, we were able to map 599 different integration sites in the zebrafish genome based on the latest genome assembly (Zv6) [Fig. 3A and supporting information (SI) Table 3]. Those mapped integrations distribute roughly evenly across all chromosomes (Fig. 3B). Thirty-nine percent of mapped integrations (233/599) landed in Ensembl annotated genes (Fig. 3A), many corresponding to previously uncharacterized genes and ESTs. Eleven integrations landed in regions with two overlapping genes resulting in 244 “gene hits” total. Ninety-two percent (225/244) of gene hits were in introns, and 19 integrations landed in exons. Overwhelmingly, 59% (132/225) of integrations in introns were in the first intron (Fig. 3C). Twenty-five percent of mapped integrations (150/599) landed within 3 kb of genes with 4-fold more integrations landing at the upstream side of genes compared with the downstream side (20% upstream vs. 5% downstream) (Fig. 3A). Together, our data show that 64% (383/599) of mapped integrations landed either in genes or within 3 kb upstream or downstream of genes with a strong preference (65%, 250/383) toward the first intron and the upstream region (<3 kb) of genes. This distribution of integrations is consistent with our previous studies of integration preferences for MLV (13).
Fig. 3.
Summary of 933 integrations. (A) BLAST analysis of 933 proviral integrations; 599 of 933 sequences could be mapped in the zebrafish genome based on the latest genome assembly Zv6. (B) Distribution of 599 mapped proviral integrations across chromosomes. (C) Distribution of 225 proviral integrations landed in introns; 59% (132/225) of integrations in introns landed in the first intron.
Next we examined how often gene expression was affected by retroviral integration. We selected 25 integrations that landed within genes in either an intron or an exon or in the near upstream proximity (mostly within 1 kb of the transcriptional start) or downstream proximity. We either identified several F1 fish carrying the same integration or outcrossed the F1 fish and raised an F2 generation to inbreed all 25 integrations. We established the genotype for 24 embryos from each inbreed and then, by quantitative RT-PCR, compared the RNA transcript levels between integration homozygotes (−/−) and their siblings with WT background (+/+). Of 25 gene hits tested, 11 show significant reduction (>70%) in RNA levels, and 8 show >90% reduction (Fig. 4). It can clearly be seen that there is a strong trend for integrations in the first intron to be mutagenic; 8 of 10 cases with integrations in the first intron showed >70% reduction in mRNA levels; 6 of 10 showed >90% reduction. The mechanism causing the reduction in detected mRNA levels is not clear. The distance between the integration and splicing sites might be a factor that contributes to the knockdown as all eight first intron hits that result in reduced mRNA levels have integrations that land within 400 bp of the splicing sites. With our RT-PCR strategy, with primers typically in the exons on either side of the proviral integration site, we cannot differentiate between destabilized mRNAs, truncated mRNAs, or misspliced transcripts. Regardless, the functional mutagenicity of the approach is quite strong. Integrations landing in the putative promoter region (presumably within 1 kb upstream of genes) may also be mutagenic; six of seven integrations in this putative promoter region caused at least moderate gene knockdowns with two of them causing >70% reduction (Table 2). A summary of the integrations suggests that the retrovirus is remarkably mutagenic. Roughly 20% (exon hits plus 80% of first intron hits) of the integrations will result in the mRNA level of a gene being reduced by >70%.
Fig. 4.
Quantitative RT-PCR analysis of RNA transcript levels in integration homozygotes. (A) Twenty-five integrations landing within genes in either an intron or an exon or in the near upstream or downstream proximity were inbred. Total RNA from 24 embryos of each inbreed was isolated and the genotype was determined. The RNA transcript levels between three integration homozygotes (−/−) and three siblings with the WT background (+/+) were then compared by qRT-PCR. Of 25 gene hits tested, 11 of 25 show significant reduction (>70%) in RNA levels; 8 of 25 show >90% reduction. There is a strong trend for integrations in the first intron to be mutagenic; 8 of 10 cases with integrations in the first intron showed >70% reduction in gene expression; 6 of 10 showed >90% reduction.
Table 2.
Detailed summary of 25 tested integrations shown in Fig. 4
| Ensembl gene | Gene size, bp | Predicted product | Position of integration | Orientation (provirus vs. gene) | Visual phenotype | Relative expression (−/− vs. +/+), % |
|---|---|---|---|---|---|---|
| ENSDARG00000060820 | 5,766 | TG | Exon 3 | + | − | 0.3 ± 0.3 |
| ENSDARESTG00000015436 | 14,955 | XP_686562 | 42 bp upstream | + | − | 46.7 ± 15.4 |
| ENSDARG00000052482 | 104,770 | RhoGEF11 | 151 bp upstream | + | − | 0.1 ± 0.1 |
| ENSDARG00000002467 | 6,568 | RDH14 | 261 bp upstream | + | − | 20.1 ± 9.9 |
| ENSDARG00000058995 | 12,032 | JMJD4 | 296 bp upstream | + | − | 35.0 ± 6.0 |
| ENSDARG00000008064 | 28,144 | zgc:76877 | 448 bp upstream | + | − | 92.1 ± 24.1 |
| ENSDARG00000041338 | 5,915 | mrpl15 | 660 bp upstream | − | − | 52.8 ± 14.3 |
| ENSDARG00000010462 | 2,502 | Sp9 | 896 bp upstream | + | − | 36.8 ± 11.1 |
| ENSDARG00000045911 | 21,017 | TULP4 | 2.1 kb upstream | + | − | 146.0 ± 39.1 |
| ENSDARG00000036388 | 12,396 | slc39a7 | Intron 1 (7 bp to intron 1/exon 2 junction) | + | + | 1.4 ± 0.8 |
| ENSDARESTG00000017041 | 3,687 | zfp294 | Intron 1 (46 bp to exon 1/intron 1 junction) | − | − | 1.5 ± 0.7 |
| ENSDARG00000045913 | 5,531 | mrps10 | Intron 1 (77 bp to exon 1/intron 1 junction) | − | − | 9.6 ± 4.2 |
| ENSDARG00000060651 | 6,112 | MAC2BP | Intron 1 (94 bp to exon 1/intron 1 junction) | − | − | 2.3 ± 1.9 |
| ENSDARG00000015889 | 11,535 | zc3h15 | Intron 1 (162 bp to exon 1/intron 1 junction) | + | − | 0.2 ± 0.1 |
| ENSDARESTG00000017933 | 7,904 | PRP39 | Intron 1 (271 bp to exon 1/intron 1 junction) | − | − | 28.0 ± 3.6 |
| ENSDARG00000023536 | 64,841 | nnt | Intron 1 (301 bp to exon 1/intron 1 junction) | + | − | 20.4 ± 2.4 |
| ENSDARESTG00000008690 | 73,527 | CCR4AF2 | Intron 1 (352 bp to exon 1/intron 1 junction) | + | − | 9.1 ± 5.8 |
| ENSDARG00000041337 | 9,219 | lypla1 | Intron 1 (7 bp to exon 1/intron 1 junction) | + | − | 59.9 ± 18.5 |
| ENSDARG00000003827 | 14,102 | zgc:55475 | Intron 1 (1,341 bp to exon 1/intron 1 junction) | + | − | 82.3 ± 28.0 |
| ENSDARG00000029930 | 31,106 | pea3 | Intron 2 (1,078 bp to exon 2/intron 2) | + | − | 96.9 ± 18.2 |
| ENSDARG00000014147 | 19,614 | axin2 | Intron 3 (693 bp to exon 2/intron 3 junction) | − | − | 60.5 ± 20.7 |
| ENSDARESTG00000001264 | 30,270 | zbp | Intron 3 (13,780 bp to exon 3/intron 3 junction) | − | − | 93.2 ± 20.6 |
| ENSDARG00000037180 | 93,461 | pcca | Intron 7 (329 bp to exon 7/intron 7 junction) | − | − | 87.4 ± 30.3 |
| ENSDARG00000037429 | 91,521 | tll1 | Intron 12 (85 bp to exon 12/intron 12 junction) | + | − | 116.9 ± 25.5 |
| ENSDARG00000033201 | 33,508 | CRIP2 | 679 bp downstream | + | − | 88.3 ± 42.5 |
In terms of visual embryonic phenotypes, we analyzed 19 lines that had significant knockdown of mRNA expression levels and identified three observable embryonic developmental defects (SI Fig. 5). This underscores one of the benefits of a reverse genetic approach, generation of a loss-of-function mutant in a gene of interest without relying on phenotypic screening; those mutants that do not show a detectable embryonic phenotype potentially could be a valuable resource for screening the late onset and/or adult phenotypes, identifying mutations that cause subtle changes in physiology or behavior, overcoming limitations caused by gene duplications by generating double mutations, or making systematic functional genomics approaches possible.
Discussion
The actual mutagenic rate of retroviral integrations has not been previously determined. In previous studies using retroviruses as a mutagen, there was some form of inherently biased selection placed on the approach before the integrations were examined for mutagenic rate. For example, the large-scale zebrafish retroviral mutagenesis effort at Massachusetts Institute of Technology required an observable embryonic phenotype to establish the mutation rate (12, 19). Here we demonstrate that ≈20% of all retroviral integrations will result in a gene disruption of >70% of the mRNA level when the integration is homozygosed. This mutation rate is high enough that retroviral mapping can realistically be used to generate a mutant resource.
An additional surprising discovery from the data is the strong effect retroviral integrations have on mRNA levels appears to be primarily limited to integrations in the first intron. Other introns typically had no or little effect on mRNA levels. The mechanism and significance of this effect are currently unclear, but the data provide strong evidence that MLV sequences in some way behave differently when in the first intron. The RT-PCR strategy we used was chosen for sensitivity but cannot differentiate from a variety of different mutagenic scenarios including: mRNA destabilization, premature truncation, or exon skipping, but whichever effect (or combination of effects) is occurring, the end result is a predictable disruption in normal gene expression.
We have shown that MLV has a strong preference for integrating in the 5′ end of the gene (13), and the data we present here confirm this is also true for MLV in the zebrafish genome. This preference for the first intron appears to be independent of gene or intron size and is more likely linked to the MLV preference for the transcriptional start site. It is interesting to consider that these two phenomena (5′ bias and first-intron effects) are linked in terms of the life cycle of MLV.
To achieve saturation mutagenesis, it is critical to have an efficient overall rate of production. Recent efforts in our laboratories have demonstrated that a steady, but modest effort can generate 200–500 injected founder fish per week by two trained individuals. More aggressive schedules could double that rate. Based on the average infection rate of 46 integrations per cell for founders, and the pilot data projecting the gene hit and gene disruption or knockdown rates, in 2 years, we would be able to generate enough mapped integrations to produce >20,000 gene disruptions (>90% reduction in gene expression) or ≈30,000 strong gene knockdowns (>70% reduction).
Projections are as follows: 200–500 founders per week by two trained individuals = 4,000 founders (2–3 months); 4,000 founders × four F1 males per founder = 16,000 F1 males; 16,000 F1 males × eight sequences per F1 = 128,000 reads; 128,000 reads‖× 0.55 (% analyzable reads) = 70,400 good reads; 70,400 good reads × 0.30 (% unique and mappable) = 21,120 mapped insertions; 21,120 mapped insertions × 0.4 (% gene hits)** = 8,448 gene hits; 8,448 gene hits × 0.55 (% the first intron hits) = 4,646 the first-intron hits; 4,646 the first-intron hits × 0.80 (% knockdowns) = 3,717 >70% knockdowns or 4,646 the first-intron hits × 0.60 (% disruptions) = 2,788 >90% knockdowns; 2 years → 29,736 strong knockdowns or 22,304 disruptions (from the first-intron hits only).
Assuming even distribution of integrations, this would represent knockdowns in most zebrafish genes. This nonphenotype-based genome-wide gene knockdown approach is particularly useful for studying genes that function in later stages of vertebrate animal life cycle and/or involve functional redundancy. In addition, the approach described here generates zebrafish with insertions that block or knockdown mRNA transcription in F1 generation. This greatly improves the number of fish that can be recovered for future studies.
It has been demonstrated that MLV does not randomly integrate but rather has a bias for integrating in the 5′ end of genes, therefore one important question is how many genes can actually be mutagenized using this approach. Given the relatively small number of integrations reported here and in the literature, this is difficult to predict. It is unlikely that any single approach can mutate all of the genes in the genome, but our data in this paper show that of 233 gene hits, six were hit twice, and two were hit three times. The number of multiple hits is slightly higher than would be expected for random integration but not inconsistent for a retrovirus with a known predisposition for landing in genes. Similarly, our data set of MLV integrations in human cells suggested there are some genomic regions that are more likely to receive integrations (24). It has not been determined exactly what aspect of these genomic regions makes them preferred targets, but one possibility is that MLV has been shown to have a weak bias for DNase hypersensitive sites (25). Gene expression also has a modest effect on integration preference (13). These effects are relatively small, and distribution of ≈80% of the integrations are indistinguishable from random integrations using current analyses. Evidence from the Hopkins Laboratory screen (Massachusetts Institute of Technology, Cambridge, MA) (19) where 390 unique mutations were identified from 525 isolated mutations (from ≈1,400 possible embryonic lethal mutations) suggests that the large majority of zebrafish genes can be mutated using this approach. As mapping of integrations proceeds, it will become obvious when sequencing more integrations will no longer be cost-effective in terms of the number of sequences required to obtain a new gene hit.
In terms of sequencing costs, this technique compares very favorably with the current state of the art for zebrafish reverse genetics, i.e., TILLING, which uses a resequencing of exons approach to detect mutations in specific genes. TILLING has many advantages including the ability to identify single base changes or small deletions, the possibility of identifying an allelic series, and the relative simplicity of using ENU as a mutagen. However, in TILLING, 5,000–10,000 sequencing reactions are done to identify mutations in a specific gene. Typically it takes ≈1,000 sequences to identify a sequence change that represents a likely mutation. From this work it takes ≈30–40 sequences to identify an integration with a high potential for causing mutation. This is a ≈30-fold increase in sequencing cost efficiency making a systematic large-scale effort to mutate most zebrafish genes economically feasible. Thus a retroviral integration resource nicely complements the TILLING approach and provides a quick first test for mutations in a gene. Further reductions in sequencing cost using emerging low-cost sequencing technologies could greatly increase the number of integrations mapped making saturation of the genome possible.
In conclusion, here we report the proof of principle for using retroviral mutagenesis to establish a permanent library of cryopreserved zebrafish gene disruptions. Once complete, this resource would be a convenient complement to TILLING for targeted gene disruptions in zebrafish. The level of coverage in terms of gene disruptions is directly related to the total number of integrations mapped. In principle, saturation of the genome is possible using this approach with a relatively small commitment in scientific resources. Such a resource will allow for systematic or functional genomics approaches that could not be accomplished by traditional genetic screening.
A commercial effort similar to our approach is being used by Znomics, Inc. (Portland, OR). The major difference between the two approaches is our use of F1s to map integrations instead of the original infected founder fish, significantly simplifying recovery but reducing the number of integrations that can be effectively mapped.
Methods
Retrovirus Production.
GT/186 cells, a 293gp/bsr-derived retrovirus producer cell line (21, 23), were seeded in 12 poly-l-lysine-coated 600-ml flasks at 40% confluence. The next day, cells in each flask were transfected with pCMV-G plasmid (22) using Lipofectamine transfection reagent for 8 h according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). For each flask, 8 μg of pCMV-G and 120 μl of Lipofectamine reagent were used. After transfection, the medium was replaced with DMEM supplemented with 10% FBS, penicillin, and streptomycin. Media collected at 24- and 48-h posttransfection were filter-sterilized and concentrated by ultracentrifugation at 27,000 rpm for 1.5 h at 4°C in a Beckman SW28 rotor. Viral pellets were resuspended in 25 μl of PBS and stored at 4°C before use in injection.
Generation of Founder Fish.
Synchronized embryos for injection were obtained from a lethal-free zebrafish line TAB-5 as described (18). Approximately 50 nl of the concentrated viral stock containing 8 μg/ml of polybrene and a trace amount of phenol red were injected into five to seven locations among the blastomeres of blastula-stage embryos (≈1,000–2,000 cell stage). Each embryo received two rounds of injection in a period of ≈15 min. Injected embryos were maintained in Holtfreiter's solution (60 mM NaCl/0.7 mM KCl/1 mM Hepes, pH 7.0/0.9 mM CaCl2) at 32°C overnight after a 90-min heat-shock period at 37°C. The next day, the injected embryos were transferred to dishes of filtered system water and raised under normal protocol.
Embryo Assay.
To ensure the injected founder embryos were efficiently infected, we determined the copy number of provirus in several injected embryos from each batch of injected founders at 2 days postinjection using a multiplex qPCR-based assay (designated as embryo assay) described by Amsterdam et al. (18). The number of proviral insertions per cell was computed by measuring the amplification rate of the SFG locus, which is specific to proviral DNA, and comparing the ratios of threshold values between founder embryo DNA and DNA with known copy numbers of proviral insertions; the results are normalized to the control rag2 locus, which is simultaneously measured. The average copy number of provirus in each batch of injected fish is called the EAV.
LM-PCR.
For LM-PCR, we used the method of Wu et al. (13). In brief, genomic DNA was digested with MseI and PstI. MseI cuts genomic DNA frequently. PstI cuts within the proviral sequence and is used to prevent amplification of an internal viral fragment from the 5′ LTR. The fragments were then ligated to a linker at the MseI restriction site. The first PCR was performed by using primers specific to the 3′ LTR and the linker to amplify the genomic DNA between these regions. The PCR products were diluted 1:50, and a nested PCR was performed to increase sensitivity and to reduce nonspecific amplification. The nested PCR products were directly shotgun cloned into a TOPO vector using the TOPO TA Cloning Kit for Sequencing (Invitrogen) and sequenced.
Mapping Integration Sites.
To map sequences to zebrafish genome, BLAST searches were performed on the Ensembl genome server (www.ensembl.org/Danio_rerio/blastview). All analysis used the annotation database specific to the zebrafish assembly version 6 (Zv6) released in March 2006. A sequence was considered to be from a genuine integration event if (i) it contained the 3′LTR sequence from the nested primer to the end of 3′LTR (CA) (in fact, 99% of verified sequences contained both the 3′LTR and the linker sequences), and (ii) it showed ≥95% identity to the genomic sequence over the high-quality sequence region.
Genotyping.
Fish were genotyped by PCR analysis of tail biopsy DNA using primers listed in SI Table 4. To identify the integration-positive fish, a universal primer complementary to the 5′ end of the proviral 3′ LTR and a gene-specific primer complementary to the genomic sequence adjacent to the 3′ end of the integration site were used; this combination will amplify a ≈600-bp amplicon from all integrations regardless of their locations in the genome. To distinguish between homozygous and heterozygous fish, a third gene-specific primer complementary to the same locus without integration, locating ≈300 bp upstream of the first gene-specific primer site, was added into the above primer combination in PCR; a single 600-bp, a single 300-bp, and both 600-bp/300-bp amplicons on gels indicate the integration homozygous, WT, and heterozygous fish, respectively.
Design of Quantitative RT-PCR Primers.
RT-PCR primers were designed to fulfill at least one of the following criteria to avoid the amplification of carryover genomic DNA in the total RNA preparation: (i) both primers are targeted to exons flanking one or more introns with a sum intronic sequence larger than 500 bp; (ii) one of the primers is targeted to the boundary of two consecutive exons so that the primer will specifically recognize the cDNA. As to the selection of exons the primers are targeted to, in the case with the integration landing in an intron (either the first intron or later introns), if possible, we would place the primers on the exons flanking the intron with the proviral integration. Thus, the effect of integration on the flanking exons could be assessed directly (if targeting to exons far downstream to the proviral integration, aberrant events near the integration such as skip splicing may not be detected). Because the virus vector contains a gene-trap cassette, designing primers targeting to the exons flanking the integrant intron also enables us to detect possible gene-trap events. In the case with the integration landing upstream or downstream of genes, we would place the primers to the upstream exons (mostly exon 1 and exon 2). The nucleotide sequences of primers, the locations of primers relative to the integration site, and the sizes of amplicons are detailed in SI Table 5.
Quantitative RT-PCR.
Total RNA was isolated from 6- to 8-day-old embryos using TRIzol reagent (Invitrogen). Reverse transcription and real-time PCR were performed by using the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen) according to the manufacturer's instructions, with 30 ng of total RNA and a gene-specific primer pair. Amplifications were performed by using iCycler (Bio-Rad, Hercules, CA). Typical conditions were as follows: 60°C for 15 min, 95°C for 5 min, then 95°C for 15 s and 60°C for 30 s (40 cycles). All reactions were performed in triplicates and normalized to the expression of bactin1 mRNA. The relative changes of gene expression between the homozygous mutants and the wild-type embryos were calculated by using comparative quantification as follows: ΔΔCt (ΔCt−/−-ΔCt+/+), where Ct is the cycle number at which amplification rises above the background threshold; ΔCt is the change in Ct between the targeted gene and bactin1; −/− is the homozygous mutant sample; and +/+ is the WT sample. Gene expression is then calculated as 2−ΔΔCt.
Cryopreservation of Sperm.
The sperm-freezing protocol was essentially same as described (8). To save space, sperm from one fish line were collected in multiple capillary glass tubes and stored in a single cryotube. To recover fish from frozen sperm samples, one capillary was used for each in vitro fertilization.
Supplementary Material
Acknowledgments
We thank Jiao Zhang, Yan Zhuang, Feifei Zhang, and Qian Zhang for technical assistance; Qingchun Cai, Yongfei Yang, and Weida Li for initial technical support; Yingdi Jia, Jingliang Chen, and Houhua Cui for maintaining zebrafish and fish facility; and Qichang Fan and Ruowen Ge for helpful discussion. This work was partially supported by the Natural National Science Foundation of China (Grants 30421004 and 30620120101), as well as the 973 Program from Ministry of Science and Technology of PR China (Grants 2005CB522504 and 2006CB943801). B.Z. was supported by New Century Excellent Talents in University and Excellent Young Teachers Program as well as the Program for Promoting Scientific Cooperation and Higher Education within American and Oceanian Regions, Ministry of Education of PR China. This work was also supported in part by funding from National Institutes of Health (Grant RR13227, to S.L.) and in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Abbreviations
- ENU
ethylnitrosourea
- TILLING
targeting induced local lesions in genomes
- MLV
murine leukemia virus
- qPCR
quantitative PCR
- EAV
embryo assay value
- LM-PCR
linker-mediated PCR.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF681140–EF681618).
This article contains supporting information online at www.pnas.org/cgi/content/full/0705502104/DC1.
Short (<18 bp) linker-only, and ambiguous sequences are excluded.
The percentage is expected to be close to 40% because of multiple genes affected by a single integration.
References
- 1.Solnica-Krezel L, Schier AF, Driever W. Genetics. 1994;136:1401–1420. doi: 10.1093/genetics/136.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talbot WS, Schier AF. Methods Cell Biol. 1999;60:259–286. doi: 10.1016/s0091-679x(08)61905-6. [DOI] [PubMed] [Google Scholar]
- 3.Bahary N, Davidson A, Ransom D, Shepard J, Stern H, Trede N, Zhou Y, Barut B, Zon LI. Methods Cell Biol. 2004;77:305–329. doi: 10.1016/s0091-679x(04)77017-x. [DOI] [PubMed] [Google Scholar]
- 4.Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RH, Cuppen E. Genome Res. 2003;13:2700–2707. doi: 10.1101/gr.1725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stemple DL. Nat Rev Genet. 2004;5:145–150. doi: 10.1038/nrg1273. [DOI] [PubMed] [Google Scholar]
- 6.Wienholds E, Plasterk RH. Methods Cell Biol. 2004;77:69–90. doi: 10.1016/s0091-679x(04)77004-1. [DOI] [PubMed] [Google Scholar]
- 7.Draper BW, McCallum CM, Stout JL, Slade AJ, Moens CB. Methods Cell Biol. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- 8.Sood R, English MA, Jones M, Mullikin J, Wang DM, Anderson M, Wu D, Chandrasekharappa SC, Yu J, Zhang J, Paul Liu P. Methods. 2006;39:220–227. doi: 10.1016/j.ymeth.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Lin S, Gaiano N, Culp P, Burns JC, Friedmann T, Yee JK, Hopkins N. Science. 1994;265:666–669. doi: 10.1126/science.8036514. [DOI] [PubMed] [Google Scholar]
- 10.Gaiano N, Allende M, Amsterdam A, Kawakami K, Hopkins N. Proc Natl Acad Sci USA. 1996;93:7777–7782. doi: 10.1073/pnas.93.15.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Nature. 1996;383:829–832. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 12.Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, et al. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, Li Y, Crise B, Burgess SM. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 14.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 15.Skarnes WC, von Melchner H, Wurst W, Hicks G, Nord AS, Cox T, Young SG, Ruiz P, Soriano P, Tessier-Lavigne M, et al. Nat Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nord AS, Chang PJ, Conklin BR, Cox AV, Harper CA, Hicks GG, Huang CC, Johns SJ, Kawamoto M, Liu S, et al. Nucleic Acids Res. 2006;34:D642–8. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, et al. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Proc Natl Acad Sci USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naviaux RK, Costanzi E, Haas M, Verma IM. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W, Burgess S, Golling G, Amsterdam A, Hopkins N. J Virol. 2002;76:2192–2198. doi: 10.1128/jvi.76.5.2192-2198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi H, Takahashi M, Gage FH, Verma IM. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Luke BT, Burgess SM. Virology. 2006;344:292–295. doi: 10.1016/j.virol.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 25.Berry C, Hannenhalli S, Leipzig J, Bushman FD. PLOS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.