Abstract
The vitamin D receptor (VDR) mediates the biological actions of 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) through its capacity to recruit coregulatory proteins. This interaction is mediated via a coregulatory LxxLL motif. We screened a combinatorial (x)7LxxLL(x)7 phage library with purified VDR to identify peptides that displayed high affinity and selectivity for VDR. These peptides contained the consensus sequence Lx E/H x H/F P L/M/I LxxLL and exhibited significant sequence similarity to the active LxxLL box found in DRIP205. Nearly all LxxLL peptides interacted in a ligand-dependent manner directly with human VDR. However, a pattern of selectivity of the peptides for other members of the nuclear receptor family was also observed. Interestingly, the interaction between the VDR and many of the peptides was differentially sensitive to a broad assortment of VDR ligands. Finally, several of these peptides were shown to inhibit activation of a vitamin 1,25(OH)2D3-sensitive reporter gene. These studies suggest that the LxxLL motif can interact directly with the VDR and that this interaction is regulated by chemically diverse vitamin D ligands.
Keywords: Vitamin D receptor, LxxLL motif, phage display, DRIP205, antagonist
INTRODUCTION
The nuclear receptor superfamily is a collection of sequence-related transcription factors that regulate gene transcription in a ligand-dependent manner [1, 2]. The vitamin D receptor (VDR) is a member of the nuclear receptor superfamily that regulates the biological actions of 1,25-dihydroxyvitamin D3 (1,25(OH)2D3), which is the hormonal form of vitamin D3. 1,25(OH)2D3 binds with high affinity to the VDR in the nucleus of target cells forming a ligand-receptor complex critical for regulating bone mineralization and remodeling through an effect on calcium and phosphate homeostasis [3, 4]. It has also been shown that the vitamin D-VDR system has other noncalcemic effects such as a role in cell differentiation and in regulation of the immune system [5].
Along with other members of the nuclear receptor superfamily, the VDR binds DNA upon ligand activation, predominately as a heterodimer with the retinoid X receptor (RXR) [6]. Once the ligand-activated RXR/VDR heterodimer binds to a vitamin D3 response element (VDRE) on a target gene, the complex recruits additional coregulators that are essential for transcription such as the vitamin D receptor interacting proteins (DRIP/mediator) and the steroid receptor coactivator/p160 (SRC/p160) family of proteins [7, 8].
The interaction between nuclear receptors and coactivators is mediated by an LxxLL motif (where L is leucine and x is any amino acid) contained within the coactivator protein that is necessary and sufficient for the binding of these proteins to the receptor and enhancing its transcriptional activity [9]. The enhancement of transcriptional activity is dependent upon the integrity of the motif along with key residues in helix 12 (AF-2) of the nuclear receptor ligand-binding domain [9, 10]. In addition, studies have shown receptor selectivity for coactivators by detailing the importance of sequences N and C-terminal to the LxxLL motif [9, 11-13]. Furthermore, there is evidence for ligand-specific recruitment of coactivators. A previous study showed the VDR to have an increased ability to recruit an LxxLL-containing coregulatory protein when liganded with different vitamin D3 analogs [14]. Thus far a variety of studies have shown the importance of receptor and ligand identity, promoter context, and tissue specificity to overall coregulator function [7, 15]; however, the ability of individual receptors to recruit various coregulators in a specific manner is still unclear.
In order to better understand the interaction between nuclear receptors and coregulators, a combinatorial phage display approach can be used to probe ligand-induced conformational changes in the VDR and the nature of the VDR-coregulator interaction. Previously, this technique was used by Chang et al. to study the interaction between estrogen receptor (ER) α and LxxLL motifs, leading to the discovery of peptide antagonists of ERα and ERβ [12, 13]. We therefore used this screening process to identify VDR-selective LxxLL-containing peptides. In addition, a two-hybrid analysis of the interaction of the VDR with selected peptides in the presence of VDR agonists revealed ligand-specific differences in the interactions. Several of these peptides were also capable of inhibiting 1,25(OH)2D3-dependent upregulation of an osteocalcin-luciferase reporter gene.
MATERIALS AND METHODS
Compounds
1,25(OH)2D3 was obtained from Solvay (da WEESP, Netherlands). ZK159222 and ZK168281 was provided by Dr. Andreas Steinmeyer (Schering AG, Germany). 2-methylene-19-nor-(20S)-1,25-dihydroxyvitamin D3 (2MD), 2-methylene-19-nor-1α-hydroxyhomopregnacalciferol (2MP), and 2-methylene-19-nor-20(S)-1α-hydroxy-bishomopregnacalciferol (2MbisP) were obtained from Deltanoid Pharmaceuticals, Inc. (Madison, WI). KH1060, GS1790, EB1089, and MC1288 were a gift from Dr Lise Binderup (Leo Pharmaceuticals, Denmark). The non-steroid analog XC1734 was provided by Dr. R. Heyman (X-Ceptor Therapuetics, San Diego). Lithocholic acid (LCA), 9-cis-retinoic acid (9cisRA), dexamethasone, and thyroid hormone (T3) were all obtained from Sigma Chemical Co (St. Louis, MO).
Protein Purification
Full length human VDR was cloned into pET-29b vector obtained from Novagen (Darmstadt, Germany) and expressed with a C-terminal 6xHis tag and produced in BL21(DE3) codon Plus RIL cells obtained from Stratagene (San Diego, CA). Soluble hVDR protein was then purified to homogeneity using sequential Ni-NTA and SP-Sepharose column chromatography as previously described [16].
Phage Display Libraries
A peptide library in the format of (x)7LxxLL(x)7, where x is any amino acid and L is leucine, was screened using purified VDR . The construction of the library has been described previously [12]. Briefly, the top-strand oligonucleotide 5'-AGTGTGTGCCTCGAGA(NNK)7CTG(NNK)2CTGCTG(NNK)7TCTAGACTGTGCAGT-3' (N = A, C, G, or T; K = C or T) was gel purified, annealed to its complementary-strand oligonucleotide 5'-ACTGCACAGTCTAGA-3', and extended with Klenow polymerase in the presence of dNTPs to generate double-stranded DNA. The DNA was restriction-digested, ligated into Escherichia coli JS-5 cells, and amplified on 2YT plates to generate the library. The library contained a complexity of 1.5×108 different peptide sequences.
Phage Display
hVDR protein (5 pmoles/well ) treated with either 1,25(OH)2D3 or ZK159222 was immobilized to a 96-well cell culture plate using NaHCO3 (pH 8.5). One well was plated for each ligand and the plate was left to incubate overnight at 4°C. The wells were blocked with 2% milk and washed prior to incubating the protein for three hours with pre-cleared phage library treated with ligand. After nonbinding phage were removed by washing, the bound phage were eluted and amplified in E. coli DH5αF' cells for five hours. The cells were centrifuged and the supernatant collected for use in subsequent rounds of screening [12, 13]. Four rounds of panning were performed where the enrichment of VDR-binding phage was tested by enzyme-linked immunosorbent assay (ELISA).
ELISA
An ELISA was carried out in a manner similar to that of the phage display screening. First, hVDR was treated and plated as described above along with a BSA (or milk) control. After the protein-coated wells were blocked and washed, pre-cleared phage stock treated with hormone was added to each well. Nonbinding phage were removed by washing, which was followed by an incubation with a 1:5000 dilution of HRP-conjugated anti-M13 antibody. Bound antibody was detected using 2',2'-azino-bis-3-ethylbenzthiazoline-6-sulfonic acid (ABTS) containing 0.05% H2O2 where the color change was measured at 405 nm using a plate reader.
Transient Transfections
MC3T3-E1 were seeded into 24-well plates at a concentration of 5.0 × 104 cells/well in α-MEM (Mediatech, Herndon, VA) containing 10% FBS (Hyclone, Logan, UT) and were transfected 24 hrs later with Lipofectamine PLUS (Invitrogen, Carlsbad, CA) in serum and antibiotic-free medium. Individual wells were transfected with 50 ng of pCH110-βgal, 250 ng of a luciferase reporter vector, and 50 ng of a pMsx-LxxLL peptide or pMsx control plasmid (unless otherwise indicated). In the two-hybrid assays, 50 ng of a receptor-VP16 plasmid was transfected as well. After transfection, the cells were cultured in medium supplemented with 20% FBS with or without ligand. Cells were harvested 24 hrs after stimulation and the lysates assayed for luciferase and β-galactosidase activities as previously described [16]. Luciferase activity was normalized to β-galactosidase activity in all cases.
Plasmids
pVP16-hVDR, pVP16-estrogen receptor α (pVP16-ERα), pVP16-retinoic acid receptor α (pVP16-RARα), pVP16-retinoic X receptor α (pVP16-RXRα), pVP16-glucocorticoid receptor (pVP16-GR), pVP16-thyroid receptor β (pVP16-TRβ), pVP16-liver X receptor (pVP16-LXR), pVP16-farnesoid X receptor (pVP16-FXR), pGal4(5x)Luc3, RSV-βgal, and phOC(3900)-Luc gene have been previously reported [13]. pM-SRC-1 prepared from the pM vector containing the Gal4 DNA binding domain (Gal4DBD, residues 1-147) was also previously described [12, 13]. All of the Gal4DBD-LxxLL peptide fusions were constructed as previously described [12]. Briefly, DNA sequence coding the (x)7LxxLL(x)7 peptides were excised from the mBAX vector by restriction digest and subcloned into the pMsx vector (derived from the pM vector [Clontech] with a linker sequence to generate in-frame SalI and XbaI sites for cloning).
RESULTS
Affinity selection of ligand-dependent VDR binding peptides using phage display screening
Upon ligand binding, the receptor is believed to undergo a conformational change that mediates the formation of a heterodimeric complex with RXR. This heterodimeric complex is capable of recruiting a variety of coregulator proteins necessary for transcriptional regulation of vitamin D3-target genes through an LxxLL motif present in the coactivator structure [7, 9]. Previous studies have utilized a combinatorial phage display approach to explore the importance of flanking sequences to overall LxxLL motif-receptor interaction [12, 13]. This approach is useful given that sequences obtained from this type of screen often reflect sequences found in nature [17, 18] and have the potential to act as peptide antagonists for nuclear receptor transactivation [12, 13]. In our own previous studies, we have shown that small 19-amino acid (19-mer) LxxLL-containing peptides originally identified in a phage display screen using purified ERα and ERβ can also bind both VDR and RXR and inhibit vitamin D3 response [19, 20].
In this report, we screened the (x)7LxxLL(x)7 library directly with purified human VDR to identify peptide sequences that displayed higher affinity and selectivity for the VDR. The screen was carried out using bacterial expressed hVDR protein as described in Materials and Methods. Briefly, hVDR protein immobilized to a 96-well plate was incubated with pre-cleared phage library treated with ligand. After nonbound phage were removed by washing, the bound phage were eluted and amplified for use in subsequent rounds of screening. Four rounds of panning were performed and the enrichment of VDR-binding phage was tested by enzyme-linked immunosorbent assay (ELISA). The amino acid sequence of clones confirmed to bind VDR in the ELISA assay were deduced following DNA sequencing and summarized in Table 1. All but one of the agonist-derived peptides showed strong sequence conservation at several positions N-terminal to the LxxLL motif. The general trend was as follows: a hydrophobic residue positioned at −1 and −7 (I, L, or M), a proline (P) residue at −2, an aromatic residue (H or F) at −3, and a glutamate (E) in most of the agonist-derived peptides at −5. The strong sequence conservation at positions −1 through −3 of the LxxLL peptides identified here also coincides with the N-terminal flanking sequence of the active DRIP205 LxxLL motif, which lends validity to our screening process.
Table 1.
Sequences of the LxxLL peptides identified by phage display screening of the (x)7LxxLL(x)7 library using purified hVDR in the presence of VDR agonist or antagonist. Leucine residues are highlighted in gray. Boxes indicate residues N-terminal to the LxxLL motif that display high sequence similarity. Asterisks indicate LxxLL peptides confirmed to bind mammalian hVDR in the two-hybrid assay (see Fig 1). The sequence of the known LxxLL motif in the DRIP205 protein is included for sequence comparison.
| DRIP205 | N | L | M | N | L | L | K | D | N | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Agonist | 1 | D | V | P | W | Y | G | G | L | T | E | L | L | G | V | L | E | S | R | E | |
| 2 | T | M | L | T | S | L | L | Q | N | G | V | D | H | V | * | ||||||
| 3 | S | T | L | W | T | L | L | S | S | E | G | D | S | M | * | ||||||
| 4 | Q | R | L | W | D | L | L | D | L | P | S | P | T | S | * | ||||||
| 5 | G | S | L | M | Q | L | L | T | E | N | V | G | T | H | * | ||||||
| 6 | D | Q | L | T | Q | L | L | R | S | Y | D | A | G | L | * | ||||||
| 7 | S | M | L | V | D | L | L | E | L | Q | P | R | Y | L | * | ||||||
| 8 | V | S | L | L | N | L | L | M | Q | D | L | L | D | G | * | ||||||
| Antagonist | 9 | D | A | P | T | S | L | S | L | Q | R | L | L | H | P | N | M | S | X | A | |
| 10 | D | A | V | P | P | P | S | L | S | D | L | L | T | R | Y | P | E | W | P | ||
| 11 | D | A | P | W | G | R | S | L | N | S | L | L | D | G | Y | P | V | S | S | ||
| 12 | H | A | A | E | L | A | N | L | P | R | L | L | A | S | Y | E | D | R | L | ||
| 13 | L | D | L | E | G | S | S | L | N | V | L | L | R | P | K | I | D | G | M | ||
Two-hybrid analysis confirms that agonist-derived LxxLL peptides can bind mammalian hVDR as well as various nuclear receptors with differential activity
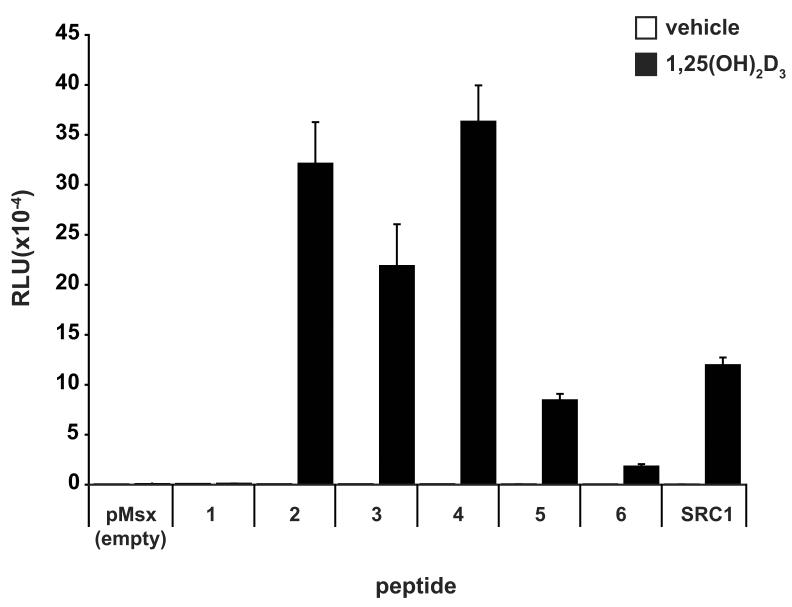
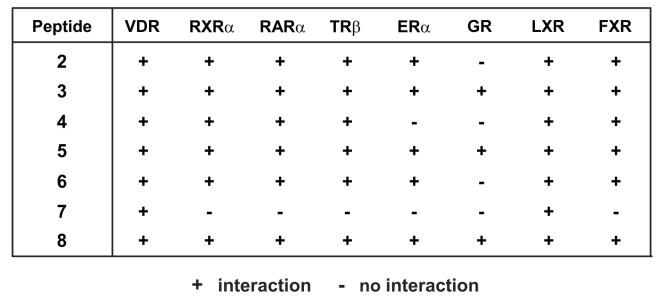
To confirm binding of the 19-mer LxxLL-containing peptides identified in our screen to mammalian expressed hVDR, we utilized a two-hybrid assay [12, 13, 19] in an MC3T3-E1 cell background with full-length hVDR expressed as a fusion protein containing the activation domain of VP16. Each peptide was produced as a fusion with the yeast Gal4 DNA binding domain and an interaction with pVP16-hVDR was assessed using the 5xGal4-Luc3 reporter gene. We included pM-SRC1 in our two-hybrid experiments as a positive control and the empty pMsx vector as a negative control. As seen in Fig. 1A and Fig. 1B (VDR column), the majority of the agonist-derived peptides were able to interact with VDR in the presence of 1,25(OH)2D3. Of the eight total agonist-derived peptides, only peptide #1 appeared unable to bind VDR. In addition, None of the antagonist-derived peptides, however, display sufficient affinity to interact directly with the VDR in the presence of either 1,25(OH)2D3 or ZK159222 in the two-hybrid assay (data not shown). It is possible that the fusion of these peptides to the Gal4 DBD reduced their affinity relative to that observed on the phage. As can be seen in the two-hybrid assay results shown in Fig. 2B, the agonist-derived peptides (2-8) showed differential activity in their binding to various nuclear receptors despite the strong sequence conservation in the region N-terminal to the LxxLL motif. In particular, peptide #7 displayed an increased selectivity given that it bound to only VDR and LXR. In contrast, peptides #3, 5, and 8 showed the greatest promiscuity. Therefore, it appears that small LxxLL-containing peptides can in fact display preferential receptor binding and are not all functionally equivalent, which is consistent with findings in previous studies [12, 13].
Fig. 1.
Two-hybrid analysis confirms binding of the agonist-derived LxxLL peptides to mammalian hVDR and indicates differential binding of each peptide to various nuclear receptors. A, Two-hybrid assay examining the interaction between LxxLL peptides (1 - 6) and pVP16-hVDR. MC3T3-E1 cells were co-transfected with pCH110-βgal, pGal4(5x)luc3, pVP16-hVDR, and a single pMsx-LxxLL peptide or the pMsx control plasmid as described in Materials and Methods. Cells were treated with vehicle or 1,25(OH)2D3 (10−7 M) for 24 hrs and then evaluated for both luciferase and β-gal activity. Each point represents the normalized RLU average ± SEM of a triplicate set of transfections. B, Summary of the two-hybrid analysis of LxxLL peptides (2-8) with various nuclear receptors. MC3T3-E1 cells were transfected and evaluated as described in (A). The chart summarizes the presence (+) or absence (−) of an interaction between each peptide and the indicated nuclear receptor.
Fig. 2.
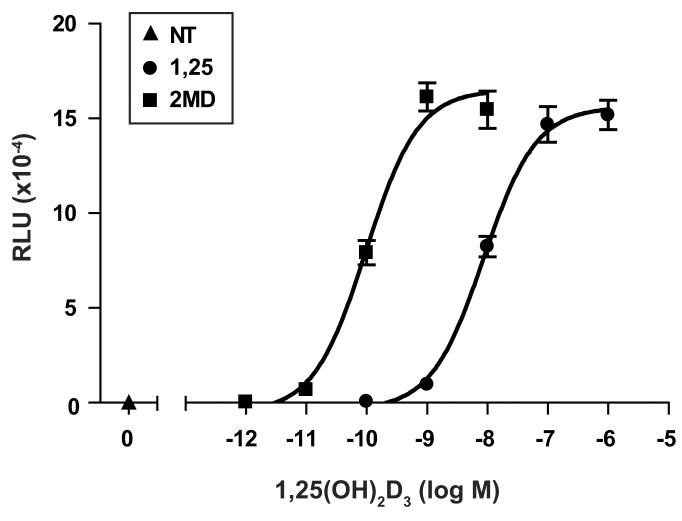
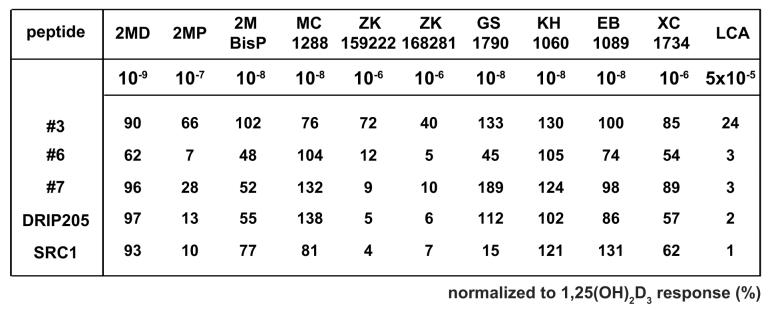
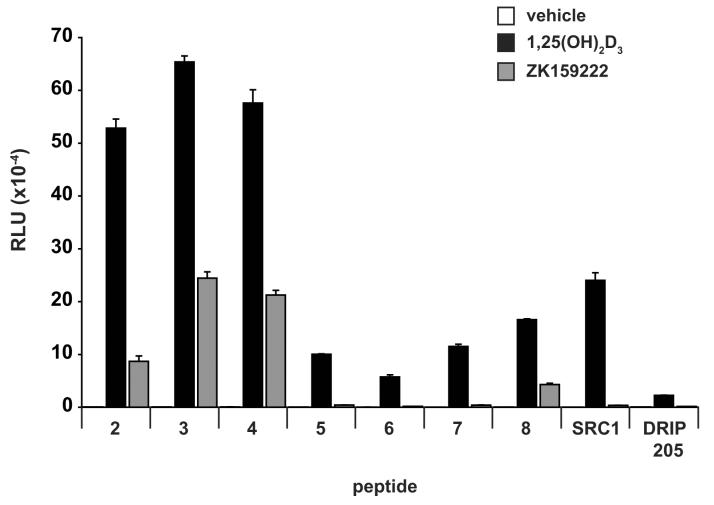
Two-hybrid analysis indicates potency, efficacy, and selectivity differences in the ability of agonist-derived LxxLL peptides to bind mammalian-expressed hVDR. A, Two-hybrid assay examining the interaction between LxxLL peptide #2 and pVP16-hVDR in the presence of 1,25(OH)2D3 versus 2MD. MC3T3-E1 cells were co-transfected with pCH110-βgal, pGal4(5x)luc3, pVP16-hVDR, and a single pMsx-LxxLL peptide as described in Materials and Methods. Cells were treated with vehicle, 1,25(OH)2D3 (10−10 M to 10−6 M) or 2MD (10−12 M to 10−8 M) for 24 hrs and evaluated for both luciferase and β-gal activity. Each point represents the normalized RLU average ± SEM of a triplicate set of transfections. B, Two-hybrid assay examining the interaction between LxxLL peptide #3 and pVP16-hVDR in the presence of various VDR ligands at maximal concentrations. MC3T3-E1 cells were transfected, treated with the indicated concentration of ligand for 24 hrs and evaluated as described in (A). Statistical analyses were carried out using one way ANOVA with Dunnett's multiple comparison post test (*, p<0.05; **, p<0.01 in comparison to 1,25(OH)2D3). C, Summary of the interaction between LxxLL peptides (3, 6, and 7), SRC1, or DRIP205 and VDR-VP16 in the presence of various VDR ligands. MC3T3-E1 cells were transfected, treated with the indicated concentration of ligand for 24 hrs and evaluated as described in (A). Each point represents the normalized RLU average of a triplicate set of transfections represented as a percentage of 1,25(OH)2D3 response. Raw data set for peptide #3 was shown in (B). D, Two-hybrid assay examining the interaction between LxxLL peptides (2-8), SRC1, or DRIP205 and pVP16-hVDR in the presence of 1,25(OH)2D3 versus ZK159222. MC3T3-E1 cells were transfected, treated with vehicle, 1,25(OH)2D3 (10−7 M), or ZK159222 (10−6 M) and evaluated as described in (A).
Two-hybrid analysis indicates potency, efficacy, and selectivity differences in the ability of the agonist-derived LxxLL peptides to bind VDR
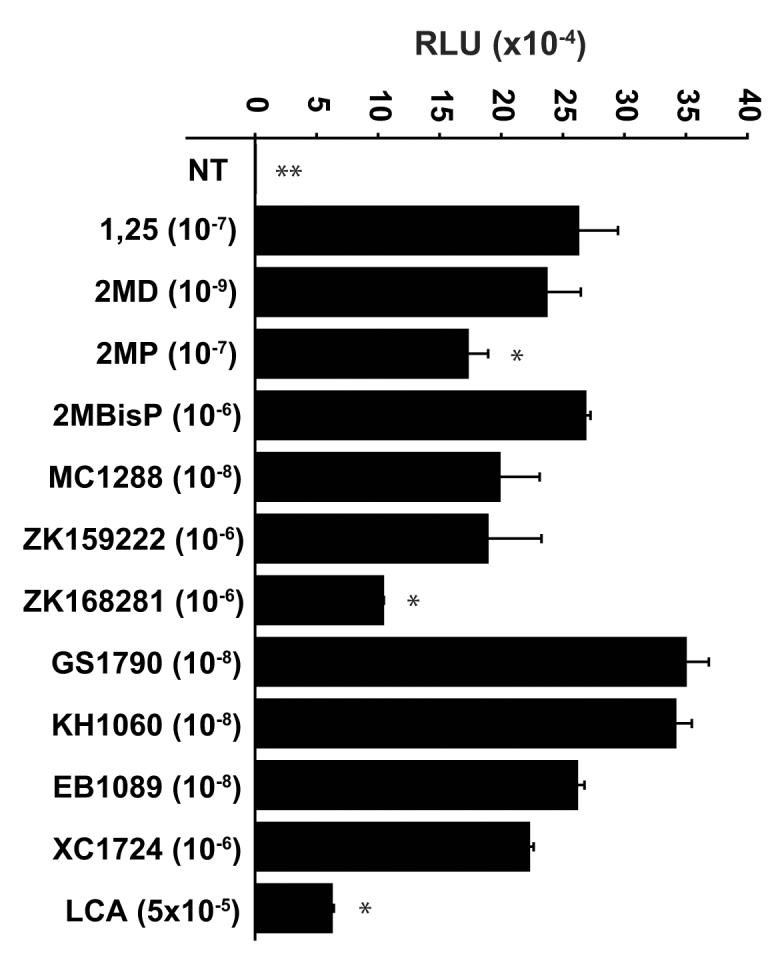
We next explored whether various VDR ligands, both agonists and antagonists, could promote differential binding of the LxxLL peptides to VDR. In the first of these studies, we asked if the highly potent vitamin D3 analog 2MD could promote an increased interaction between the LxxLL peptides identified in this study and VDR as compared to 1,25(OH)2D3. In our previous studies, 2MD manifested a two-log increased potency over 1,25(OH)2D3 in promoting VDR interaction with both SRC1 and DRIP205 [16]. This two-log potency difference between 2MD and 1,25(OH)2D3 was similarly observed in our analysis of the interaction between peptide #2 and VDR, as can be seen in Fig. 2A. While only the data for peptide #2 is shown in Fig 2A, the two-log increase in potency was observed with many of the agonist-derived LxxLL peptides (data not shown, but see also Fig 2B and 2C).
While there was a clear potency difference between 1,25(OH)2D3 and 2MD in promoting the LxxLL peptide-receptor interaction, there did not appear to be any difference in the efficacy of the two compounds. We then explored the possibility that other VDR ligands might be more efficacious than 1,25(OH)2D3 by performing additional two-hybrid analyses of several LxxLL-containing peptides with pVP16-hVDR in the presence of maximal concentrations of various VDR ligands. Maximal concentrations of each ligand were used in order to minimize any potency effects that might be observed due to differences in the affinity of the ligand for the receptor and/or for serum components such as vitamin D binding protein. A representative experiment which examined the interaction between peptide #3 and VDR can be seen in Fig 2B and a summary of the data for several peptides is documented in Fig 2C. As illustrated in Fig 2B, there was a statistically significant decrease in the ability of 2MP, ZK168281, and LCA to promote the interaction between peptide #3 and VDR. This result mirrors the general trend observed in Fig 2C where the shortened side chain analogs (2MP and 2MBisP), the antagonists (ZK159222 and ZK168281), and the secondary bile acid ligand (LCA) all seem to be less effective at promoting the peptide-receptor interaction. In a final experiment, we explored the differential binding of all eight of the agonist-derived peptides, SRC1 and DRIP205 in the presence of ZK159222 to assess the effect of antagonist binding on the LxxLL motif-VDR interaction in more detail. We continued to observe that the antagonist ZK159222 behaved as a weak agonist at best as compared to 1,25(OH)2D3. However, some peptides were unable to bind to the antagonist-liganded receptor, suggesting that ligand identity may be an important contributor to LxxLL-selectivity. In summary, differences in the ability of these peptides to bind to VDR in the presence of various ligands are apparent, which suggests that vitamin D3 analogs or VDR antagonists may be altering the conformation of the receptor in such a way as to influence potentially the recruitment of coactivator proteins.
Select agonist-derived LxxLL peptides can inhibit VDR-mediated transcription
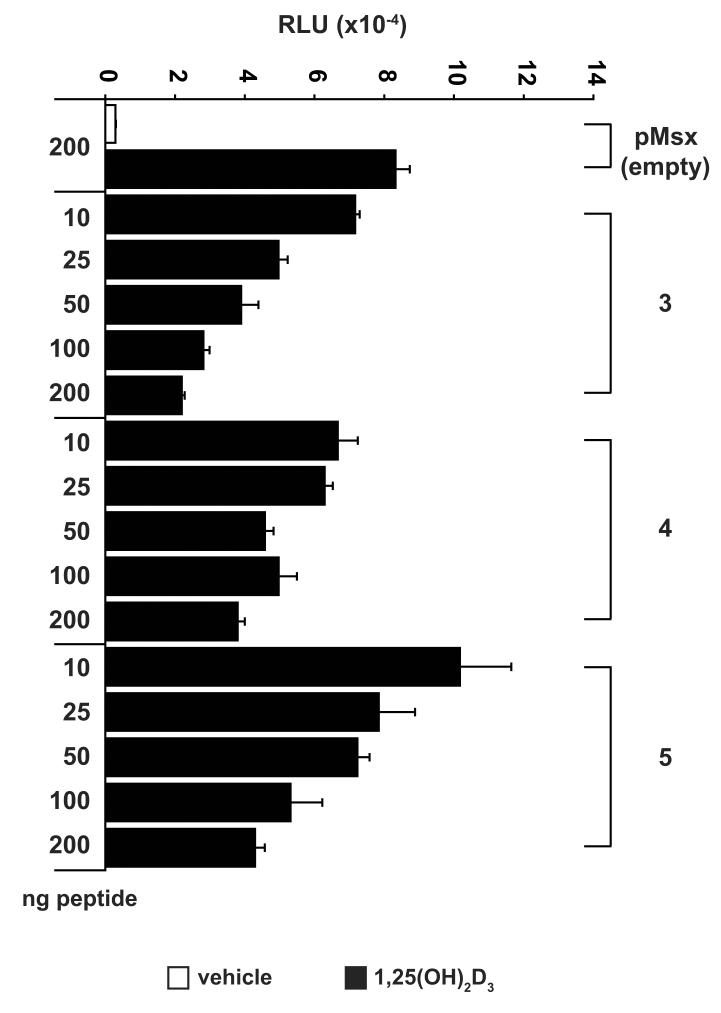
We next assessed whether the agonist-derived LxxLL peptides could modify a biological response in cell culture by determining whether they could block VDR-mediated transactivation of a transiently transfected osteocalcin promoter. If the peptides identified in our screen could mimic the interaction that occurs between the VDR and LxxLL-containing coregulatory proteins, they should be able to block transcription by impeding endogenous VDR-coactivator interaction. As can be seen in Fig. 3, the VDR-reactive LxxLL peptides #3, 4, and 5 were all capable of blocking the 1,25(OH)2D3-dependent upregulation of the osteocalcin promoter (observed in the presence of empty pMsx vector) in a dose dependent manner. This inhibition was specific to peptides #3, 4, and 5 as the other agonist-derived peptides (2, 6-8) were incapable of suppressing VDR activity (data not shown). Therefore, LxxLL-containing peptides with similar overall sequence identity can display differential biological effects on VDR activity.
Fig. 3.
Select agonist-derived LxxLL peptides can inhibit VDR-mediated transcription of an osteocalcin-luciferase reporter gene. MC3T3-E1 cells were co-transfected with pCH110-βgal, phOC(3900)-luc, and increasing amounts of a single pMsx-LxxLL peptide or the pMsx control plasmid as described in Materials and Methods. Cells were treated with vehicle or 1,25(OH)2D3 (10−7 M) for 24 hrs and evaluated for both luciferase and β-gal activity. Each point represents the normalized RLU average ± SEM of a triplicate set of transfections.
DISCUSSION
During 1,25(OH)2D3-mediated transcription, the VDR/RXR heterodimer recruits additional coregulatory proteins necessary for both chromatin modification and transcriptional activation [7, 8, 15, 21]. Nuclear receptor-coactivator interactions are mediated by LxxLL motifs located in the coactivator protein that interact with the AF-2 region of a nuclear receptor [9, 10]. In this study, we screened a (x)7LxxLL(x)7 phage library against VDR in the presence of 1,25(OH)2D3 or ZK159222, a selective antagonist of VDR-mediated transcription that competes with the natural ligand for binding to VDR [22-24]. Our agonist-derived class of LxxLL peptides showed strong sequence conservation across the region N-terminal to the LxxLL box giving the consensus sequence Lx E/H x H/F P L/M/I LxxLL, which was also consistent with the first three residues N-terminal to the DRIP205 LxxLL motif. Conversely, the sequences of the peptides identified in the presence of ZK159222 were restricted to the central LxxLL motif. The majority of the agonist-derived peptides interacted with mammalian VDR in a ligand-dependent manner while the antagonist-derived peptides showed no interaction with the receptor in a two-hybrid assay. In addition, two-hybrid analysis of the agonist-derived peptides with VDR in the presence of various VDR ligands indicated ligand potency, efficacy and selectivity differences in comparison to the natural hormone. Finally, several of the peptides were also shown to inhibit activation of a luciferase reporter gene fused to the vitamin D3-sensitive osteocalcin promoter.
The presence of a consensus sequence in the agonist-derived peptides identified in this study illustrates the importance of residues N-terminal to the LxxLL motif in mediating receptor-coactivator interaction. Interestingly, the conserved proline residue located at the -2 position in the LxxLL peptides and DRIP205 may be particularly important for mediating the LxxLL-VDR interaction. A similar phage display screen of ERα published by Chang et al. [12] identified three classes of LxxLL peptides that displayed differential binding to various nuclear receptors. Only the class of peptides that contained a conserved proline at the -2 position were able to interact with VDR in their studies [12]. The SRC1 LxxLL motif can still bind VDR, however, even though it lacks a proline at the −2 position. This suggests that residues N-terminal to the LxxLL motif can dictate receptor preference but not absolute selectivity. Lastly, while we did observe strong sequence similarity across the agonist-derived peptides, the sequence of the antagonist-derived peptides was restricted to a core LxxLL motif, which may explain their lack of VDR-binding in the two-hybrid assay. Similarly, the only agonist-derived peptide identified in our screen that was unable to bind VDR also lacked the consensus sequence. In summary, our results are consistent with previous studies that stress the importance of residues N-terminal to the LxxLL motif to overall receptor selectivity [9, 12, 13], which is in contrast to a recent study that suggested receptor recognition is dictated by residues C-terminal to the motif [11].
During our studies of the agonist-derived peptides and their interaction with VDR, we also found that these peptides displayed differential binding to various nuclear receptors. One peptide, in particular, showed a greater selectivity for VDR, given that it only interacted with VDR and LXR. Conversely, three other peptides were capable of binding all seven of the receptors identified and examined in this study. These findings are consistent with the idea that residues beyond the core LxxLL motif can dictate receptor preference, which is particularly interesting given the strong sequence similarity displayed in the N-terminal region of these peptides. Furthermore, the identification of a highly selective VDR-binding LxxLL peptide gives us the opportunity to use the sequence as a tool for targeting a known coactivator protein to the VDR through modification of the active LxxLL box to resemble the VDR-selective peptide. In fact, Gaillard et al. engineered the LxxLL motifs within the known coactivator PGC-1α in order to develop a highly selective coactivator for estrogen receptor-related receptor α (ERRα) [25]. This approach may prove useful for designing a VDR-selective coregulatory protein that could be used in a variety of different mechanistic studies.
While the sequence adjacent to the LxxLL motif may play a role in directing nuclear receptor-coactivator interaction, there is also evidence for ligand-specific effects on receptor conformation and coactivator recruitment. When the ligand-binding domain (LBD) of the estrogen receptor (ERα) was crystallized in the presence of an agonist and a peptide derived from the LXXLL motif region of a p160 family member, the peptide clearly bound the hydrophobic grove on the surface of the LBD. Conversely, when the receptor was crystallized in the presence of an antagonist, helix 12 blocked the coactivator binding pocket preventing an interaction with the peptide [26]. Our own studies suggest a difference in the conformation of the VDR in the presence of agonist versus antagonist given that select LxxLL peptides were unable to bind VDR in the presence of ZK159222. Furthermore, we found that certain VDR ligands (i.e. the shortened side-chain analogs, the antagonists, and LCA) were generally restricted in their ability to promote LxxLL-VDR interactions. While recent crystallography data of the rat VDR ligand binding domain complexed with the DRIP205 LxxLL peptide in the presence of 1,25(OH)2D3, 2MD, or 2MbisP showed no change in the coactivator binding pocket [27], the effects of various ligands on the structure of VDR in its entirety is still unknown.
In similar phage display screens of the estrogen receptor mentioned above, the investigators were successful in identifying peptides capable of antagonizing ERα versus ERβ-mediated transcription [12, 13, 28]. In this study, we were also interested in identifying LxxLL-containing peptides that could serve as tools for studying VDR-mediated gene transcription given their ability to compete with endogenous coactivator proteins and block VDR-mediated transcription. Accordingly, we were also successful in identifying select LxxLL peptides that strongly inhibited VDR-mediated transcription of a reporter gene. We were, however, unsuccessful in demonstrating that these peptides could block the upregulation of an endogenous 1,25(OH)2D3-sensitive gene. The discrepancy between the reporter assay and our endogenous mRNA studies may be due to several factors. First, we delivered the peptides using transient transfection Perhaps our method of delivery of the peptides by transient transfection wherein low transfection efficiency may have resulted in the production of the peptide complex in an insufficient percentage of cells to suppress endogenous response. Alternatively, the fusion of the peptide to the Gal4-DBD may hinder its ability to bind effectively at a natural chromatin binding site. Nevertheless, the fact that only select peptides were able to block reporter gene activity suggests that not all LxxLL peptides are functionally equivalent. It remains possible, however, that the sequence of these peptides could be useful in the subsequent design of a small molecule inhibitor of VDR-mediated transcription for both mechanistic studies and potentially therapeutic purposes.
In conclusion, we used a combinatorial phage display approach to identify a class of LxxLL-containing peptides with the consensus sequence Lx E/H x H/F P L/M/I LxxLL that bind to VDR in a mammalian two-hybrid assay. These peptides show differential binding to VDR in the presence of various VDR-ligands, which suggests that a conformational change in the receptor that influences coactivator recruitment may occur in the presence of different ligands. We also observed these peptides to have diverse binding patterns to different nuclear receptors with one peptide in particular being highly selective for VDR. This finding highlights the importance of residues N-terminal to the LxxLL motif in dictating receptor preference. Finally, we found that select peptides were capable of inhibiting activation a vitamin D3-sensitive reporter gene, which will allow us to use these peptides in future mechanistic studies of VDR-mediated gene regulation.
ACKNOWLEDGMENTS
We thank members of the Pike and McDonnell laboratories for their individual contributions to this work. This work was supported by National Institutes of Health Grant DK72281 (J.W.P.) and DK62434 (D.P.M.)
Abbreviations used
- VDR
vitamin D receptor
- 1,25(OH)2D3
1,25-dihydroxyvitamin D3
- RXR
retinoid X receptor
- VDRE
vitamin D3 response element
- DRIP
vitamin D receptor interacting proteins
- SRC1
steroid receptor coactivator 1
- AF-2
activation function 2
- ER
estrogen receptor
- 2MD
2-methylene-19-nor-(20S)-1,25-dihydroxyvitamin D3
- 2MP
2-methylene-19-nor-1α-hydroxyhomopregnacalciferol
- 2MbisP
2-methylene-19-nor-20(S)-1α-hydroxy-bishomopregnacalciferol
- LCA
lithocholic acid
- 9cisRA
9-cis-retinoic acid
- dex
dexamethasone
- T3
thyroid hormone
- HRP
horseradish peroxidase
- βgal
β-galactosidase
- FBS
fetal bovine serum
- RAR
retinoic acid receptor
- GR
glucocorticoid receptor
- FXR
farnesoid X receptor
- LXR
liver X receptor
- TR
thyroid receptor
- PGC-1
PPARγ coactivator 1
- ERR
estrogen receptor-related receptor
Footnotes
The authors state that they have nothing to declare
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haussler MR, Whitfield GK, Haussler CA, Hsieh JC, Thompson PD, Selznick SH, Dominguez CE, Jurutka PW. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res. 1998;13:325–349. doi: 10.1359/jbmr.1998.13.3.325. [DOI] [PubMed] [Google Scholar]
- 4.Jurutka PW, Whitfield GK, Hsieh JC, Thompson PD, Haussler CA, Haussler MR. Molecular nature of the vitamin D receptor and its role in regulation of gene expression. Rev Endocr Metab Disord. 2001;2:203–216. doi: 10.1023/a:1010062929140. [DOI] [PubMed] [Google Scholar]
- 5.DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. 2004;80:1689S–1696S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- 6.Glass CK. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 7.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 8.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 9.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 10.Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 11.McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, Inostroza J, Torchia J, Nolte RT, Assa-Munt N, Milburn MV, Glass CK, Rosenfeld MG. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C, Norris JD, Gron H, Paige LA, Hamilton PT, Kenan DJ, Fowlkes D, McDonnell DP. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta. Mol Cell Biol. 1999;19:8226–8239. doi: 10.1128/mcb.19.12.8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall JM, Chang CY, McDonnell DP. Development of peptide antagonists that target estrogen receptor beta-coactivator interactions. Mol Endocrinol. 2000;14:2010–2023. doi: 10.1210/mend.14.12.0561. [DOI] [PubMed] [Google Scholar]
- 14.Issa LL, Leong GM, Sutherland RL, Eisman JA. Vitamin D analogue-specific recruitment of vitamin D receptor coactivators. J Bone Miner Res. 2002;17:879–890. doi: 10.1359/jbmr.2002.17.5.879. [DOI] [PubMed] [Google Scholar]
- 15.McKenna NJ, O'Malley BW. Minireview: nuclear receptor coactivators--an update. Endocrinology. 2002;143:2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto H, Shevde NK, Warrier A, Plum LA, DeLuca HF, Pike JW. 2-Methylene-19-nor-(20S)-1,25-dihydroxyvitamin D3 potently stimulates gene-specific DNA binding of the vitamin D receptor in osteoblasts. J Biol Chem. 2003;278:31756–31765. doi: 10.1074/jbc.M304737200. [DOI] [PubMed] [Google Scholar]
- 17.Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 18.Pirozzi G, McConnell SJ, Uveges AJ, Carter JM, Sparks AB, Kay BK, Fowlkes DM. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 19.Pathrose P, Barmina O, Chang CY, McDonnell DP, Shevde NK, Pike JW. Inhibition of 1,25-dihydroxyvitamin D3-dependent transcription by synthetic LXXLL peptide antagonists that target the activation domains of the vitamin D and retinoid X receptors. J Bone Miner Res. 2002;17:2196–2205. doi: 10.1359/jbmr.2002.17.12.2196. [DOI] [PubMed] [Google Scholar]
- 20.Pike JW, Pathrose P, Barmina O, Chang CY, McDonnell DP, Yamamoto H, Shevde NK. Synthetic LXXLL peptide antagonize 1,25-dihydroxyvitamin D3-dependent transcription. J Cell Biochem. 2003;88:252–258. doi: 10.1002/jcb.10336. [DOI] [PubMed] [Google Scholar]
- 21.Rachez C, Freedman LP. Mechanisms of gene regulation by vitamin D(3) receptor: a network of coactivator interactions. Gene. 2000;246:9–21. doi: 10.1016/s0378-1119(00)00052-4. [DOI] [PubMed] [Google Scholar]
- 22.Herdick M, Steinmeyer A, Carlberg C. Antagonistic Action of a 25-Carboxylic Ester Analogue of 1alpha ,25-Dihydroxyvitamin D3 Is Mediated by a Lack of Ligand-induced Vitamin D Receptor Interaction with Coactivators. J. Biol. Chem. 2000;275:16506–16512. doi: 10.1074/jbc.M910000199. [DOI] [PubMed] [Google Scholar]
- 23.Tocchini-Valentini G, Rochel N, Wurtz JM, Moras D. Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification. J Med Chem. 2004;47:1956–1961. doi: 10.1021/jm0310582. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20:305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 25.Gaillard S, Grasfeder L, Haeffele C, Lobenhofer E, Chu T, Wolfinger R, Kazmin D, Koves T, Muoio D, Chang C, McDonnell DP. Engineering Receptor Selective Coactivators: An approach to define the molecular pathways regulated by receptor-coactivator pairs. Mol Cell. 2006 doi: 10.1016/j.molcel.2006.10.012. In press. [DOI] [PubMed] [Google Scholar]
- 26.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 27.Vanhooke JL, Benning MM, Bauer CB, Pike JW, DeLuca HF. Molecular structure of the rat vitamin D receptor ligand binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry. 2004;43:4101–4110. doi: 10.1021/bi036056y. [DOI] [PubMed] [Google Scholar]
- 28.Norris JD, Paige LA, Christensen DJ, Chang CY, Huacani MR, Fan D, Hamilton PT, Fowlkes DM, McDonnell DP. Peptide antagonists of the human estrogen receptor. Science. 1999;285:744–746. doi: 10.1126/science.285.5428.744. [DOI] [PubMed] [Google Scholar]