Abstract
In macrophages, enzymes that synthesize or hydrolyze phosphatidylinositol (3,4,5)-trisphosphate [PI(3,4,5)P3] regulate Fcγ receptor-mediated phagocytosis. Inhibition of phosphatidylinositol 3-kinase (PI3K) or overexpression of the lipid phosphatases phosphatase and tensin homologue (PTEN) and Src homology 2 domain-containing inositol phosphatase (SHIP-1), which hydrolyze PI(3,4,5)P3 to phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2], respectively, inhibit phagocytosis in macrophages. To examine how these enzymes regulate phagosome formation, the distributions of yellow fluorescent protein (YFP) chimeras of enzymes and pleckstrin homology (PH) domains specific for their substrates and products were analyzed quantitatively. PTEN-YFP did not localize to phagosomes, suggesting that PTEN regulates phagocytosis globally within the macrophage. SHIP1-YFP and p85-YFP were recruited to forming phagosomes. SHIP1-YFP sequestered to the leading edge and dissociated from phagocytic cups earlier than did p85-cyan fluorescent protein, indicating that SHIP-1 inhibitory activities are restricted to the early stages of phagocytosis. PH domain chimeras indicated that early during phagocytosis, PI(3,4,5)P3 was slightly more abundant than PI(3,4)P2 at the leading edge of the forming cup. These results support a model in which phagosomal PI3K generates PI(3,4,5)P3 necessary for later stages of phagocytosis, PTEN determines whether those late stages can occur, and SHIP-1 regulates when and where they occur by transiently suppressing PI(3,4,5)P3-dependent activities necessary for completion of phagocytosis.
INTRODUCTION
Phospholipid-modifying enzymes regulate Fc receptor-mediated phagocytosis. When the expression or activities of these enzymes are perturbed, phagocytosis is altered or inhibited. Type I phosphatidylinositol 3-kinase (PI3K) phosphorylates the 3′ position of the inositol headgroup of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] and phosphatidylinositol 4-phosphate, producing phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] and phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2], respectively (Auger et al., 1989; Hawkins et al., 1992). Type I PI3K is made up of a regulatory p85 subunit and a catalytic p110 subunit. p85 is recruited via its Src homology 2 (SH2) domains to phosphorylated immunoreceptor tyrosine-based activation motifs (ITAMs) present in ligated FcγRIIa; then, conformational changes allow it to bind and activate the catalytic subunit (Dhand et al., 1994; Chacko et al., 1996; Lowry et al., 1998). Treatment of macrophages with wortmannin or LY294002, compounds that inhibit the catalytic subunit of PI3K, results in formation of phagocytic cups that are unable to close (Araki et al., 1996; Cox et al., 1999). Thus, inhibition of PI3K activity allows phagocytosis to begin, but it inhibits it at later stages.
The lipid phosphatases phosphatase and tensin homologue (PTEN) and Src homology 2 (SH2) domain-containing inositol phosphatase (SHIP-1) counteract PI3K activity by dephosphorylating its product, PI(3,4,5)P3. PTEN lowers PI(3,4,5)P3 levels in the plasma membrane by dephosphorylating the 3′ position of the inositol ring, producing PI(4,5)P2 (Maehama and Dixon, 1998). Overexpression of PTEN in a heterologous COS7 signaling system inhibits FcR-mediated phagocytosis (Kim et al., 2002). Murine macrophages lacking PTEN exhibit enhanced rates of phagocytosis and increased levels of phosphorylated Akt, a serine/threonine kinase that regulates cell survival (Cao et al., 2004).
SHIP-1 dephosphorylates the 5′ position of PI(3,4,5)P3, yielding PI(3,4)P2 (Damen et al., 1996). Manipulation of SHIP-1 also alters FcR-mediated phagocytosis. SHIP-1 is recruited to both the ITAM- and immunoreceptor tyrosine-based inhibitory motif domains of FcγRIIa and FcγRIIb, respectively, via an N-terminal SH2 domain (Nakamura et al., 2002; Tridandapani et al., 2002a). The binding of SHIP-1 to FcγRIIa also requires the presence of the small adapter protein Shc (Tridandapani et al., 1999). The interaction between SHIP-1 and Shc blocks the formation of a Grb2–Shc adapter protein complex, resulting in a decrease in Ras signaling and decreased nuclear factor-κB–dependent signaling (Tridandapani et al., 2002b). Once recruited to the phagosome, SHIP-1 can dephosphorylate PI(3,4,5)P3, down-regulating PI3K activity. Gene deletion of SHIP-1, or inhibition through overexpression of dominant-negative constructs, accelerates phagocytosis (Cox et al., 2001; Nakamura et al., 2002). Conversely, overexpression of functional SHIP-1 inhibits phagocytosis of large particles (Cox et al., 2001).
As is evidenced by these studies involving enzyme manipulation, proper control of phosphoinositide dynamics on the phagosomal membrane is essential to FcR-mediated phagocytosis. Pleckstrin homology (PH) domain proxies, which bind to specific inositol headgroups of phosphoinositides, have been used to track phosphoinositide dynamics in cells during signaling events such as growth factor stimulation. The PH domain of PLCδ1 has been used to track PI(4,5)P2 dynamics during phagocytosis (Botelho et al., 2000). The PH domain of Akt has been used to follow PI(3,4,5)P3 and/or PI(3,4)P2, but the two phosphoinositides have not been examined separately during phagocytosis with probes specific for each (Marshall et al., 2001). Previous studies have shown that PI3K and SHIP-1 localize to the phagosome and PTEN does not (Marshall et al., 2001).
Although it is established that these lipid-modifying enzymes are necessary for phagocytosis, it is not known how they affect the process. They could regulate all-or-none “stop/go” decisions or they could modulate the complex sequence of activities necessary for phagosome formation. If the latter, then one might expect relevant lipid-modifying enzymes to be regulated during the course of phagosome formation. It is therefore necessary to examine the relative kinetics of the phosphoinositides as well as the lipid-modifying enzymes that control them.
This study examined the localization dynamics of SHIP-1, PI3K, PTEN, and their substrates and products during FcR-mediated phagocytosis. We expected that SHIP-1 facilitates signal maturation, catalyzing the transition from PI(3,4,5)P3 to PI(3,4)P2 during late stages of phagocytosis (Swanson and Hoppe, 2004). Instead, we found that PI3K and SHIP-1 were both recruited during initial cup formation, but later they exhibited differential localization on the phagosome, with SHIP-1 concentrated at the leading edge of the phagocytic cup. PTEN was excluded from the phagosome. An initial shallow gradient of PI(3,4,5)P3 toward the edges of the phagocytic cup was lost as SHIP-1 sequestered to the leading edge. These dynamics indicate that during FcR-mediated phagocytosis, SHIP-1 acts as a regulator phosphatase, which transiently suppresses PI(3,4,5)P3-dependent activities necessary for phagosome formation.
MATERIALS AND METHODS
Molecular Cloning and DNA Manipulation
Plasmids encoding monomeric versions (A207K) of cyan fluorescent protein (CFP) and citrine (yellow fluorescent protein, YFP) were used for all constructs (Zacharias et al., 2002). The PH domains of human phospholipase C (PLC)δ1 and Bruton's tyrosine kinase (Btk) were a gift from Tamas Balla (National Institutes of Health, Bethesda, MD) (Varnai and Balla, 1998; Varnai et al., 1999). The PLCδ1PH construct was polymerase chain reaction (PCR) amplified adding Xho1 and BamH1 restriction sites, and then it was subcloned into pmCitrine-N1 (Clontech, Mountain View, CA). The BtkPH construct was PCR amplified and subcloned into the pmCitrine-N1 vector between Xho1 and HindIII. The AktPH construct was a gift from Tobias Meyer (Stanford University, Palo Alto, CA) and was subcloned into pmCitrine-C1 between BamH1 and Xba1 (Clontech). Mutant PH domain constructs for PLCδ1 (R40L), Btk (R28C), and Tapp1 (R211L) were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Human Tapp1 constructs were obtained from MRC Protein Phosphorylation Unit (University of Dundee, Dundee, Scotland), and the C-terminal PH domain was subcloned into pmCitrine-C1 at the EcoR1–BamH1 site. Human p85 was obtained from William Gullick (University of Kent, Kent, United Kingdom) (Gillham et al., 1999) and subcloned into pmCitrine-C1 between EcoR1 and BamH1. Fusing p85 to the C-terminal of a fluorescent protein, such as green fluorescent protein (GFP), does not interfere with the ability of p85 to bind to the catalytic subunit, p110 (Gillham et al., 1999). Murine SHIP-1 was donated by Mark Coggeshall (Oklahoma Medical Research Foundation, Oklahoma City, OK) and subcloned using Xho1 and EcoR1 restriction sites into pmCitrine-C1 (Tridandapani et al., 1997). Human FcγRIIa, originally cloned by Jeffrey Ravetch (Rockefeller University, New York, NY), was donated by Susheela Tridandapani (Ohio State University) and subcloned into pmCFP-N1 between Kpn1 and EcoR1 (Brooks et al., 1989). Murine PTEN was obtained from the I.M.A.G.E. consortium from a murine cDNA library and cloned into the EcoR1–BamH1 site of pmCtirine-C1 (Strausberg et al., 2002). PTEN-YFP fusions have previously been used to study plasma membrane localization in living mammalian cells (Vazquez et al., 2006).
Tissue Culture and Transfection
RAW264.7 cells (RAWs), a murine macrophage-like cell line (American Type Culture, Manassas, VA) were cultured at 37°C with 5% CO2. RAWs were cultured in Advanced-DMEM with 2% heat inactivated fetal bovine serum, 4 mM l-glutamine, 20 U/ml penicillin, and 20 μg/ml streptomycin by using cell culture reagents (Invitrogen, Carlsbad, CA). RAWs were prepared for ratiometric microscopy by plating ∼2.5 × 105 cells per coverslip the day before imaging. After cells had attached to the coverslip (∼3 h), cells were transfected with plasmids encoding the fluorescent chimeras, by using FuGene-6 as described in the manufacturer's protocol (Roche Diagnostics, Indianapolis, IN). For microscopy, coverslips were assembled into Leiden chambers (Harvard Apparatus, Holliston, MA) at 37°C in Ringer's buffer (155 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 2 mM NaH2PO4, 10 mM glucose, and 10 mM HEPES at pH 7.2). IgG-opsonized sheep erythrocytes were prepared and added to the macrophages as described previously (Knapp and Swanson, 1990). Polystyrene rods were opsonized with rabbit IgG through adsorption as described previously (Champion and Mitragotri, 2006).
Ratiometric Microscopy
Ratiometric images were acquired on an inverted fluorescence microscope (Nikon TE300) with a 60× 1.4 Planapo objective, a mercury arc lamp as the source of epifluorescent light, and a cooled digital charge-coupled device camera (Quantix; Photometrics, Tucson, AZ). The microscope was equipped with trans and epifluorescence shutters, a temperature-controlled stage, filter wheels for both excitation and emission filters, and dichroic mirrors that allow detection of multiple fluorophores. All images were acquired and processed using MetaMorph version 6.2r6 (Molecular Devices, Sunnyvale, CA). Fluorescence excitation and emission wavelengths were selected via a JP4v2 filter set (Chroma Technology, Rockingham, VT) and Lambda 10–2 filter wheel controller (Sutter Instrument, Novato, CA).
RAW 264.7 macrophages expressing fluorescent proteins were observed undergoing phagocytosis after delivering ∼2 × 105 IgG-opsonized erythrocytes to the target area of the coverslip. On landing of an erythrocyte on the macrophage, YFP, CFP, and phase-contrast images were recorded every 30 s until completion of phagocytosis (∼15 min). The ratio image (RM) was then calculated, representing the molar ratio of YFP chimera to CFP at every pixel in the cell.
To generate molar ratio images based upon stoichiometric fluorescence resonance energy transfer (FRET) methods (Hoppe et al., 2002), chimeric YFP was expressed with soluble CFP, which served as a marker of cell thickness. The RM value was calculated assuming that there was no FRET between the fluorescent molecules:
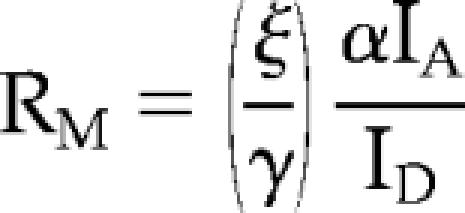 |
where IA corresponds to the YFP image (excitation 505 nm, emission 540 nm), and ID corresponds to the CFP image (excitation 435 nm, emission 490 nm). When two chimeras were expressed together, it was not possible to assume that there would be no FRET. Therefore, the ratio was calculated as follows:
 |
where IF corresponds to the FRET image (excitation 435 nm, emission 540 nm). The FRET coefficients α, β, γ, and ξ were calculated from calibrating the microscope with a series of images from cells transfected with CFP, citrine, or a linked CFP-citrine chimera with known FRET efficiency (Hoppe et al., 2002). Shading-correction images were taken by imaging solutions of CFP and YFP sandwiched between two coverslips.
To avoid effects of overexpression, macrophages not saturated in YFP fluorescence were selected for imaging. In addition, to ensure that the kinetics during phagocytosis were not affected, the time from the beginning of cup formation to phagosome closure was measured, and the average time of phagocytosis was calculated for each YFP chimera (Supplemental Figure 1). With the exception of PTEN-YFP, no probes inhibited similar rates of phagocytosis.
Particle Tracking
Recruitment of YFP chimeras to phagosomes was measured using the particle-tracking algorithm TRACKOBJ in MetaMorph. As described previously, a 5-μm region was drawn on the target erythrocyte, allowing it to be tracked as it was internalized into the macrophage (Hoppe and Swanson, 2004). For every frame in a stack of images comprising a movie, RM in the cell (RC) was computed as well as RM in the phagosome (RP). The recruitment index was calculated as RP/RC, with a baseline value of 1 indicating RM in the phagosome was equal to RM in the cell.
To align multiple phagocytic events from different time-lapse sequences, only phagocytic events that were <12 min in length from the time of erythrocyte attachment to closure were chosen for alignment and analysis. A number of recordings were excluded by this selection process. A circular region was drawn over the phase-contrast image, marking where the erythrocyte would contact the cell. When an increase in CFP fluorescence was detected in the region, indicating the beginning of pseudopod extension, that time point was designated time 0, the beginning of cup formation. Multiple phagocytic image series were then aligned for analysis based on that operational definition of time 0. The slope of the recruitment index was calculated as the change in the average RP/RC divided by the change in time.
Phagosomal Gradient Analysis
To measure the difference in ratio along the length of the phagosome, we analyzed time series' in which phagocytosis presented as a side-view in the microscope. Three 2-μm rectangular regions were drawn over the extending phagosome. R1 corresponded to the leading edge of the phagosome, R2 corresponded to the central area of the phagosome, and R3 corresponded to the rear of the phagosome. The average ratio in each region, R, for every frame in the stack was logged. To measure gradients of probes along the length of the phagosome, the ratios in regions R1, R2, or R3 were divided by the ratio in the cell, RC, for every time point. These values were averaged together over multiple phagocytic events and plotted as a function of time after the initiation of phagocytosis.
To study enzyme dynamics along extended phagosomes, IgG-opsonized polystyrene rods (a gift from Julie Champion, University of California, Santa Barbara) were added to macrophages, and coverslips were scanned for fluorescent, transfected macrophages internalizing the rods (Champion and Mitragotri, 2006). To measure the difference in ratio along the length of the phagosome, 2-μm regions were drawn over the phagosome, and the average ratio for each region (RR) was logged as well at the average ratio in the cell (RC). To normalize the gradients across multiple phagosomes, the ratio in each region was divided by the ratio for each cell (RR/RC). The normalized region ratios were then averaged over multiple phagosomes.
Statistical Analysis
Statistical tests of significance were applied to the gradient ratio R1/RC as compared with R2/RC or R3/RC, by using the Student's t test, assuming unequal variances.
RESULTS
Enzyme Dynamics
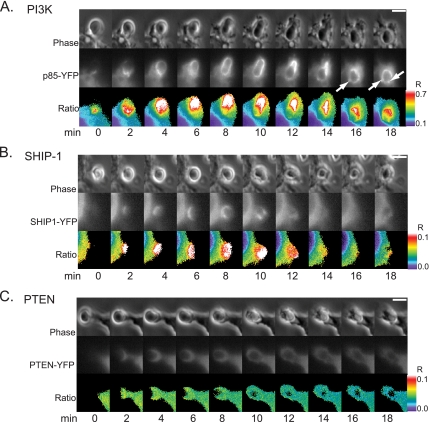
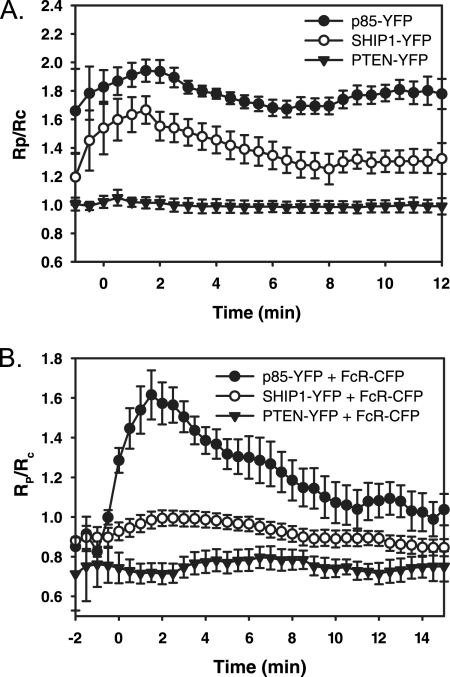
To localize lipid-modifying enzymes during phagocytosis, fluorescent fusions of YFP with PTEN, SHIP-1, and the p85 regulatory subunit of PI3K were coexpressed with CFP in RAW 264.7 macrophages. IgG-opsonized sheep erythrocytes were added to transfected macrophages and YFP chimera recruitment to the phagosome was measured using ratiometric fluorescence microscopy, calculating the molar ratio, RM in the absence of FRET (Figures 1 and 2). p85-YFP was cytosolic in the resting macrophage, redistributed to the phagosomal membrane upon initiation of phagocytosis and persisted until after closure. As the phagosome closed, punctate fluorescent structures occurred on the phagosomal membrane (Figure 1A; see Supplemental Movie 1). SHIP-1-YFP was also recruited to the phagosomal membrane from the cytosol upon initiation of phagocytosis. It was concentrated at the advancing edge of the phagocytic cup and dissociated from the phagosome membrane at the base of the cup (Figure 1B; see Supplemental Movie 2). Overexpression of PTEN-YFP inhibited phagocytosis in a manner similar to wortmannin treatment: phagocytic cups formed around the erythrocyte but failed to close (data not shown). Ratiometric data were obtained from five cells expressing PTEN-YFP that completed phagocytosis (Figure 2A). PTEN-YFP never localized to the phagosome (Figure 1C). Using a particle-tracking algorithm that measured association of chimeras with phagosomes, we found generally similar patterns of recruitment for p85-YFP and SHIP1-YFP (Figure 2A) with p85-YFP exhibiting higher levels of recruitment. Both YFP chimeras were associated with the phagosome until closure.
Figure 1.
Ratiometric imaging of p85-YFP, PTEN-YFP, and SHIP1-YFP relative to CFP during phagocytosis of IgG-opsonized erythrocytes. Phase-contrast (top), YFP (middle), and ratio (bottom) image time series of RAW macrophages during FcR-mediated phagocytosis. Color bars indicate the range of the calculated RM in the ratio time series (bar, 5 μm). (A) p85-YFP was present on the phagosome during phagocytosis. Arrows indicate the punctate structures that formed after closure. (B) SHIP1-YFP was present on the phagosomal membrane during phagocytosis. (C) PTEN-YFP remained distributed throughout the cytosol during phagocytosis.
Figure 2.
Localization dynamics of lipid-modifying enzymes during phagocytosis. (A) Plots of RP/RC indicating the dynamics of YFP-chimera localization to phagosomes, averaged for multiple phagocytic events. Error bars are SE of the mean (SEM). Tracking analysis indicated the patterns of p85-YFP (black circles; n = 11), SHIP1-YFP (white circles; n = 8) and PTEN-YFP (black triangles, n = 5) on phagosomes. (B) Tracking analysis of p85-YFP, SHIP1-YFP, or PTEN-YFP expressed with FcγRIIa-CFP during phagocytosis of IgG-opsonized erythrocytes. Plots of RP/RC showing the ratio between the YFP and CFP chimera averaged over multiple phagocytic events. Black circles show the ratio between p85-YFP and FcγRIIa-CFP (n = 9). White circles show the ratio between SHIP1-YFP and FcγRIIa-CFP (n = 18). Black triangles show the ratio between PTEN-YFP and FcγRIIa-CFP (n = 9). Error bars represent SEM.
The early association of SHIP-1 with the forming phagocytic cup, and its apparent loss from the phagosome before p85 suggested that SHIP-1 functions early in the process. To better understand the dynamics of p85, SHIP-1, and PTEN relative to FcR activation, their association and dissociation dynamics were normalized to distributions of fluorescent FcγRIIa. When expressed in RAW macrophages, FcγRIIa-GFP is initially present on resting plasma membrane; it concentrates on phagosomes during phagocytosis and remains there until after closure (Lee et al., 2005). SHIP1-YFP, p85-YFP, or PTEN-YFP was expressed in macrophages with FcγRIIa-CFP and the ratio between the chimeras was measured during phagocytosis (Figure 2B). On initiation of phagocytosis, the ratio of p85-YFP to FcγRIIa-CFP on the phagosome increased dramatically, indicating the increasing numbers of activated FcR recruiting p85 during formation of the phagocytic cup. This ratio peaked after 2 min, but it stayed elevated until after phagosome closure (10 min). In contrast, the ratio of SHIP1-YFP to FcγRIIa-CFP increased slightly on the phagosome and decreased to initial levels by 8 min, indicating a weak and transient association of SHIP-1 with FcγRIIa. The ratio change between chimeras was more dramatic with p85-YFP, consistent with the first ratiometric measurements showing that p85-YFP persisted on phagosomes longer than SHIP1-YFP (Figure 2A). After closure (8–11 min), the slope of the ratio between SHIP1-YFP and FcγRIIa-CFP did not change (slope = 0.00), whereas the ratio between p85-YFP and FcγRIIa-CFP continued to drop (slope = −0.05). In contrast, the ratio between PTEN-YFP and FcγRIIa-CFP stayed relatively constant, with a slight decrease during the initial stages of phagocytosis. This dip in the ratio suggests that PTEN-YFP may be excluded from phagosomal membranes early in the process (Figure 2B).
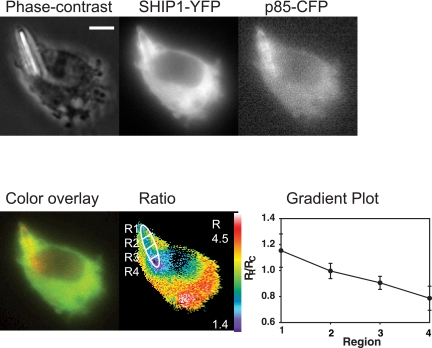
To examine the distribution of PI3K and SHIP-1 along the length of the phagosome, we coexpressed SHIP-1-YFP and p85-CFP and measured the molar ratio, RM, of the two enzymes using FRET stoichiometry (Figure 3A). Because both proteins are cytosolic in resting macrophages, the recruitment index RP/RC between the two enzymes started at a baseline of 1.0. During cup formation, the SHIP1-YFP/p85-CFP ratio decreased due to the increase of p85-CFP on the phagosome. The ratio remained low until after closure (7.5 min), indicating the persistence of p85-CFP on the phagosomal membrane. A plot of the ratio of the localization indices of SHIP1-YFP and FcγRIIa-CFP versus p85-YFP and FcγRIIa-CFP (from data in Figure 2B) showed a pattern similar to that seen when the two enzymes were expressed together (Figure 3A). The different magnitude of RP/RC values in these two curves may be attributable to the increased number of FcR in phagosomes of cells expressing FcγRIIa-CFP.
Figure 3.
Relative distributions of PI3K and SHIP-1 along the length of the phagosome. (A) Tracking analysis of SHIP1-YFP with p85-CFP during phagocytosis of IgG-opsonized erythrocytes (black circles; n = 8). RP/RC values >1.0 indicate a rise in SHIP1-YFP on the phagosome compared with p85-CFP. As a control, the localization index of SHIP1-YFP + FcγRIIa-CFP was divided by p85-YFP + FcγRIIa-CFP (from Figure 2B; white circles). (B) Macrophage phagosome depicted by phase contrast, p85-CFP, SHIP1-YFP, color overlay, and ratio (bar, 5 μm). The color bar indicates the range of the calculated RM in the ratio image. p85-CFP was pseudocolored red and SHIP1-YFP was pseudocolored green in the color overlay image. To quantify the gradient between SHIP1-YFP and p85-CFP, the length of the phagosome was divided into three regions: R1, the leading edge of the phagosome; R2, the middle area of the phagosome; and R3, the base of the phagosome. (C) Gradient plot representing the ratios of the leading edge of the phagosome (R1) and the center and basal areas (R2 and R3, respectively) divided by the ratio in the cell, RC. Black circles represent R1/RC in macrophages transfected with SHIP1-YFP and p85-CFP (SY-PC), white circles represent R2/RC, and black triangles represent R3/RC (n = 5). In macrophages expressing free YFP and CFP (Y-C), white triangles represent R1/RC, black squares represent R2/RC, and white triangles represent R3/RC (n = 5). Asterisks denote values in R1/RC that are significantly different from the values for R2/RC (p < 0.05).
The different kinetics of association and dissociation for p85 and SHIP-1 should create different relative enzyme distributions along the length of the phagosome. In cells expressing chimeras of both enzymes, SHIP1-YFP seemed concentrated at the front of the forming phagosomes, whereas p85-CFP was more evenly distributed over the phagosomal membrane (Figure 3B). To quantify this distribution for phagocytic events that were side-views, the length of the phagosome was divided into three rectangular bands. Region R1 was defined as the advancing edge of the phagocytic cup, R2 was the region just proximal to R1, and R3 was the region most distal to the edge. The average ratio value between SHIP-1-YFP and p85-CFP was calculated in each region, and these regions were tracked during phagocytosis (Figure 3B). To compare data in cells expressing different overall levels of YFP and CFP chimeras, each region ratio was divided by the ratio in the cell, RC. Differences in ratio values between region R1/RC and regions R2/RC or R3/RC indicate a gradient of enzyme distribution along the length of the phagosome. During the first 2 min of phagocytosis, the values for R1/RC, R2/RC, and R3/RC were all similar, indicating no gradient. After 2 min, however, R1/RC increased relative to R2/RC and R3/RC, indicating higher ratios of SHIP1-YFP to p85-CFP in the leading edge of the phagosome, compared with in the middle or base of the cup (Figure 3C). During cup closure (6–7 min), R1/RC and R2/RC were significantly different (p < 0.05). As a control, the gradients in phagosomes of macrophages expressing free YFP and CFP were measured. In that case, R2/RC and R3/RC did not differ from R1/RC (Figure 3C). Thus, the quantitative analyses indicate that SHIP-1 was sequestered to the front of the forming phagosome, perhaps due to transient association of the enzyme with ligated FcR.
This finding was further confirmed through examination of p85-CFP and SHIP1-YFP distribution on extended phagosomes (Figure 4). Previous work has shown that when fed IgG-opsonized ellipsoid particles, macrophages phagocytose the rods when the narrow end attaches to the plasma membrane (Champion and Mitragotri, 2006). To better assess the gradient between p85 and SHIP-1 along the length of phagosomes, we fed macrophages opsonized polystyrene rods and measured the ratio as the elongated particles were internalized (Figure 4). As the phagosome closed around the rod, SHIP1-YFP was virtually undetectable at the base of the phagosome but was strongly concentrated on the leading edge, whereas p85-CFP persisted along the length of the particle. The gradient of SHIP-1 was also evident in region ratio measurements in multiple extended phagosomes (Figure 4).
Figure 4.
Relative distributions of PI3K and SHIP-1 along extended phagosomes. Macrophage phagosome of a polystyrene rod depicted by phase contrast, SHIP1-YFP, p85-CFP, color overlay, and ratio (bar, 5 μm). The color bar indicates the range of the calculated RM in the ratio image. p85-CFP was pseudocolored red and SHIP1-YFP was pseudocolored green in the overlay image. To quantify the gradient between SHIP1-YFP and p85-CFP, the length of the phagosome was divided into four regions, as shown in the ratio image. The gradient plot represents the average RM for each region (R1–R4) divided by the average RM in the cell for multiple phagosomes, plotted as RR/RC (n = 4).
Phosphoinositide Dynamics
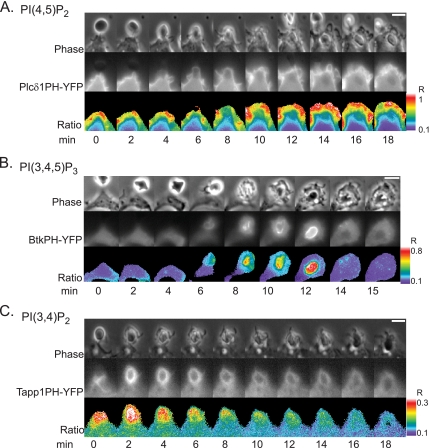
The spatial distributions of the lipid-modifying enzymes SHIP-1 and PI3K should influence the dynamics of their phosphoinositide products. To monitor phosphoinositides in the cell during FcR-mediated phagocytosis, we observed YFP chimeras of binding domains specific for certain phosphoinositides. The PH domain of PLCδ1 binds PI(4,5)P2 specifically (Lemmon et al., 1995). PLCδ1PH-YFP was concentrated on plasma membranes of unstimulated cells (Figure 5A). As phagocytosis began and the macrophage surrounded the opsonized erythrocyte, PLCδ1PH-YFP was concentrated around the advancing edge, and then detached from the membrane as the phagosome closed, as noted in previous studies (Botelho et al., 2000). Tracking analysis showed that the localization index for PLCδ1PH was initially elevated but steadily decreased, indicating loss of PI(4,5)P2 from the phagosome (Figure 6A). As a control for the specificity of the PH domain interaction with PI(4,5)P2, we expressed and measured a YFP chimera of a point mutant in the PH domain of PLCδ1, where arginine 40 was mutated to leucine (Varnai and Balla, 1998). This construct showed no recruitment to the phagosomal membrane, indicating that the nonmutated PLCδ1PH-YFP reported local increases in PI(4,5)P2 (Figure 6A). The PH domain of Btk specifically binds to PI(3,4,5)P3 (Rameh et al., 1997). BtkPH-YFP was present in the cytosol of resting macrophages but redistributed to the phagosome until closure (Figure 5B; see Supplemental Movie 3). Tracking showed that BtkPH-YFP localized to the phagosomal membrane from the initiation of phagocytosis until closure, when it redistributed back to the cytosol (Figure 6B). The point mutant for the Btk PH domain (R28C), which inhibits binding to PI(3,4,5)P3 (Rawlings et al., 1993; Thomas et al., 1993; Varnai et al., 1999) never localized to the phagosome (Figure 6B). The C-terminal PH domain of Tapp1 specifically binds to PI(3,4)P2 (Dowler et al., 2000). Tapp1PH-YFP localized to the phagosome upon initiation of phagocytosis, where it remained distributed around the phagosomal membrane until closure, reaching a maximal RP/RC of 1.33 (Figures 5C and 6C; see Supplemental Movie 4). Tracking showed a pattern of recruitment for Tapp1PH-YFP similar to that observed with the Btk PH domain. Control chimeras containing the arginine mutant (R211L), which abrogates binding to PI(3,4)P2 (Dowler et al., 2000), never localized to the phagosomal membrane (Figure 6C). The dynamics of BtkPH-YFP and Tapp1PH-YFP on phagosomes indicated that both PI(3,4,5)P3 and PI(3,4)P2 localized to the phagosomal membrane with similar kinetics, generally consistent with the localization dynamics of p85-YFP and SHIP1-YFP. Examination of the ratios in cells expressing BtkPH-YFP and Tapp1PH-CFP showed a drop during cup formation, indicating an increase in Tapp1PH-CFP relative to BtkPH-YFP on the phagosome. The ratio then rose, indicating a relative increase in BtkPH-YFP on the phagosome during later stages of phagocytosis that finally reached baseline levels at phagosome closure (Figure 6D). Gradient analysis indicated a slight gradient between BtkPH-YFP and Tapp1PH-CFP distributions, with BtkPH-YFP initially higher at the leading edge of the phagocytic cup (Figure 6E), although none of the differences were statistically significant. By 4 min, the stage of phagocytosis when SHIP1-YFP was at the leading edge of the phagosome, even the shallow gradients of 3′ phosphoinositides had disappeared. The early shallow gradient of 3′ phosphoinositides may be due to BtkPH-YFP labeling plasma membrane outside of the phagocytic cup, as indicated by top-views of phagocytosis (Figure 6F). Perhaps during early stages of phagocytic cup formation, PI(3,4,5)P3 diffuses away from ligated FcR more than PI(3,4)P2 does.
Figure 5.
Ratiometric imaging of PLCδ1PH-YFP, BtkPH-YFP, and Tapp1PH-YFP during phagocytosis. Phase contrast (top), YFP (middle), and ratio (bottom) image time series of RAW macrophages phagocytosing IgG-opsonized red blood cells. Color bars indicate the range of the calculated RM in the ratio time series (bar, 5 μm). (A) PLCδ1PH-YFP was present on resting membrane. (B) BtkPH-YFP was cytosolic in the resting macrophage, but during cup formation it localized to the leading edge of the phagosome. (C) Tapp1PH-YFP was cytosolic in the resting macrophage, but after cup formation it localized to the leading edge of the phagosome.
Figure 6.
Localization dynamics of fluorescent PH domains tracking specific phosphoinositides. (A–C) Plots of RP/RC indicating the dynamics of YFP-chimera localization to phagosomes, averaged over multiple events. Black circles represent the wild-type PH domain, white circles represent negative controls in which arginine has been mutated, abolishing the ability of the PH domain to interact with its target. Error bars represent SEM. (A) Tracking indicated the disappearance of PLCδ1PH-YFP from the phagosomal membrane after the initiation of cup formation (wt n = 8, mut n = 9). A recruitment index <1.0 indicated there was more YFP chimera in other areas of the cell than on the phagosomal membrane. (B) Tracking showed the increase of BtkPH-YFP on the phagosomal membrane at the time of cup formation (wt n = 8, mut n = 12). (C) Tracking showed the increase of Tapp1PH-YFP on the phagosome at the time of cup formation (wt n = 9, mut n = 12). (D) Tracking showed the ratio between BtkPH-YFP and TappPH-CFP (n = 15). (E) To quantify the gradient between BtkPH-YFP and TappPH-CFP, the length of the phagosome was divided into three regions: R1, the leading edge of the phagosome, R2 the middle area of the phagosome, and R3, the base of the phagosome. The average ratio in each region was divided by the ratio in the cell, RC. Black circles represent R1/RC, white circles represent R2/RC, and black triangles represent R3/RC (n = 5). (F) The differences in formation along the phagosomal membrane of PI(3,4,5)P3 and PI(3,4)P2 are shown by the localization of BtkPH-YFP and TappPH-CFP to forming phagosomes. Phase contrast, YFP, CFP, and ratio images are shown (bar, 5 μm).
Thus, the dynamics of fluorescent PH domains indicated that 3′ phosphoinositides on the inner leaflet of membranes in phagocytic cups formed sharp concentration gradients with the contiguous plasma membrane and transient shallow gradients along the length of the cup. The shallow gradients diminished coincident with the appearance of SHIP-1 gradients.
DISCUSSION
The patterns of enzyme and phosphoinositide distributions during phagocytosis, summarized in Figure 7, indicated that the distributions and concentrations of 3′ phosphoinositides in forming phagocytic cups are regulated by differential association of lipid-modifying enzymes. PTEN never localized to the phagosomal membrane. Both p85 and SHIP-1 were recruited to the phagosome upon initiation of phagocytosis. However, although p85 remained on the phagosome, SHIP-1 became restricted to the leading edge. The sequestration of SHIP-1 was more dramatic during phagocytosis of rod-like particles. The differential localization of the two enzymes along the length of the phagosome indicated a spatial control of 3′ phosphoinoisitide dynamics in phagosomal cups. The differential dynamics of the three enzymes were further indicated by normalizing their distributions relative to FcγRIIa-CFP. p85 recruitment to phagosomal FcR increased and decreased more sharply than SHIP-1, whereas the distribution of PTEN was unchanged.
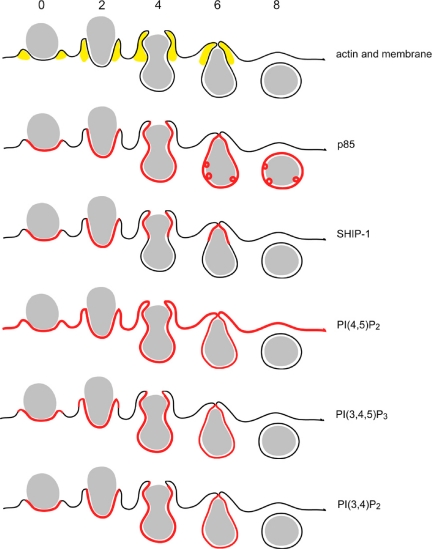
Figure 7.
Model for the distribution of lipid-modifying enzymes and their target phosphoinositides. The timing of actin localization is shown as a point of reference. Actin is recruited to the forming phagosome localizing from the base of the phagocytic cup along the sides of the phagosome (Hoppe and Swanson, 2004). After closure, it redistributes to the cytosol. PTEN is excluded from the phagosome, never localizing to the phagosomal membrane (data not shown). p85 is recruited immediately upon the initiation of phagocytosis. It remains distributed evenly along the length of the phagosome until closure, where it forms punctate structures persisting on the phagosome. SHIP-1 is also recruited at the beginning of phagocytosis and as the cup forms over the erythrocyte, it is sequestered to the leading edge of the phagosome. At closure, SHIP-1 leaves the phagosomal membrane. PI(4,5)P2 is present initially in resting plasma membrane. Through the course of phagocytosis, it distributes away from the phagosomal membrane. PI(3,4,5)P3 forms on the phagosome membrane with the initiation of phagocytosis. During the beginning of cup formation it concentrates on the forming edges of the phagosomal cup and then distributes more evenly over the length of the phagosome as the cup closes over the erythrocyte. PI(3,4,5)P3 is lost from the phagosomal membrane by the end of closure. PI(3,4)P2 is evenly distributed over the phagosomal membrane at the beginning of phagocytosis. By closure, PI(3,4)P2 is no longer detectable on the phagosome membrane.
The finding that PTEN did not localize to the phagosome prompts the question of how PTEN may regulate phagocytosis. Its lack of recruitment suggests that PTEN affects the magnitude of PI(3,4,5)P3-dependent signals on a global level, rather than at the site of phagocytosis. Overexpression of PTEN inhibited phagocytosis in a manner similar to wortmannin treatment: phagocytic cups formed but did not close. Thus, the role of PTEN in phagocytosis may be to regulate whether the late stages of phagocytosis necessary for ingestion of larger particles are allowed to occur at all. This may be related to roles for PI3K and PTEN in the regulation of cell size (Katso et al., 2001; Alvarez et al., 2003).
The products of both PI3K and SHIP-1 activities, PI(3,4,5)P3 and PI(3,4)P2, appeared on phagosomal membranes with similar overall kinetics. Both 3′ phosphoinositides were present during cup formation and persisted on the membrane until phagosome closure. Initially, a shallow gradient of these phosphoinositides could be detected along the length of the phagosome. Although both PI(3,4,5)P3 and PI(3,4)P2 were present during the beginning of cup formation, PI(3,4,5)P3 was slightly more concentrated than PI(3,4)P2 at the leading edge of the phagosome. This gradient was transient; within a few minutes, PI(3,4,5)P3 localized around the entirety of the phagosome, similar to the distribution of PI(3,4)P2. In contrast, PI(4,5)P2, whose levels are affected by many different enzymes, rapidly diminished on the phagosomal membrane. Given the continuity of the inner leaflet of membrane in the phagocytic cup with the inner leaflet of the plasma membrane, it is striking that such strong gradients of phosphoinositides exist at all. This indicates that the enzymes remodeling the membranes of the phagocytic cup are active locally and that lateral diffusion of the phosphoinositides out of the cup may be limited (Corbett-Nelson et al., 2006).
The gradients of 3′ phosphoinositides in the cup seem counterintuitive. We expected to observe gradients of substrate and product that would reflect the restricted distribution of SHIP-1 toward the distal margins of the phagocytic cup. Instead, PI(3,4,5)P3 was slightly elevated at the distal margin. However, this PI(3,4,5)P3 gradient was evident only during the early stages of phagocytosis, before the formation of detectable SHIP-1 gradients. We speculate that the higher ratios of BtkPH-CFP to TappPH-YFP at the distal margin of the early phagocytic cup occurred because PI(3,4,5)P3 is initially able to diffuse away from the ligated receptors to the membrane outside of the cup, whereas PI(3,4)P2 is restricted to the inner membrane where SHIP-1 is localized (Figures 6E and 7). As the distal margin of the forming phagosome matures into a discrete ring, the diffusion of PI(3,4,5)P3 away from ligated Fc receptors and across the lip of the cup may become more limited, restricting it to the inner membrane of the cup. Diffusion of phospholipids within this inner membrane domain would be sufficiently rapid that stable gradients of 3′ phosphoinositides along the length of the cup could not be maintained, despite the graded distribution of SHIP-1 within the cup. Rather, the changing ratio of SHIP-1 to PI3K in the growing phagosome may allow a gradual increase of PI(3,4,5)P3 concentrations within the entire membrane domain delimited by the lip at the distal margin.
Many other lipid-modifying enzymes participate in FcR-mediated phagocytosis, and some of these must surely affect the concentrations of PI(4,5)P2, PI(3,4,5)P3, and PI(3,4)P2. The phospholipases, PLA2, PLCγ1, and PLD all participate in phagocytosis. When their activities are inhibited, FcR-mediated phagocytosis is also inhibited (Lennartz and Brown, 1991; Kusner et al., 1999; Cheeseman et al., 2006). In addition, PKC depletion or inhibition halts IgG-opsonized particle ingestion in macrophages (Zheleznyak and Brown, 1992). SHIP-1 might not be the only 5-phosphatase that turns off PI3K signaling. Recent work has shown that SHIP-2, a widely expressed homolog of SHIP-1, is also recruited to the phagosome in FcR-mediated phagocytosis where it down-regulates phagocytosis (Ai et al., 2006). There are almost certainly additional enzymes, not yet identified, that are affecting the concentrations of PI(4,5)P2, PI(3,4,5)P3, and PI(3,4)P2. Nonetheless, the distributions reported here indicate PTEN, SHIP-1 and PI3K as primary determinants of the dynamics of these phosphoinositides in the phagosome.
The early and transient localization of SHIP-1 to the phagosome suggests a novel mechanism for regulating and coordinating progression through the distinct stages of FcR-mediated phagocytosis. Previous studies of the impact of PI3K inhibition have shown that PI(3,4,5)P3-independent signals are sufficient to initiate phagocytosis and to allow complete phagocytosis of particles <3 μm in diameter (Araki et al., 1996; Cox et al., 1999). As suggested previously (Swanson and Hoppe, 2004), diffusible phospholipid signaling molecules could coordinate the sequence of activities that follow FcR ligation. Consistent with this hypothesis, recent studies have identified a signal transition during phagocytosis, in which 3′ phosphoinositides mediate a transition from early to late stages of GTPase signaling (Beemiller et al., 2006). The transient localization of SHIP-1 could delay PI(3,4,5)P3-dependent inactivation of the early stage GTPases Arf6 or Cdc42. The gradual decrease in overall ratio of SHIP-1 and PI3K in the phagocytic cup could allow PI(3,4,5)P3 concentrations in the cup domain to increase, resulting in the signal transition and phagosomal closure.
Coggeshall et al. (2002) postulated two classes of inhibitory phosphatases affecting signal transduction, regulators and suppressors. Regulator phosphatases modulate a target signal, but they do not halt it entirely. Suppressor phosphatases function more specifically; upon activation they prevent a reaction. PTEN and SHIP-1 both inhibit PI3K signaling by dephosphorylating PI(3,4,5)P3. The ability of PTEN overexpression to halt phagocytosis without localizing to the phagosome suggests that it is a suppressor phosphatase. In contrast, the transient association of SHIP-1 with the phagosome indicates that it acts as a regulator phosphatase, such that local conversion of PI(3,4,5)P3 to PI(3,4)P2 helps regulate the sequence of stages during phagosome formation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI-35950 and AI-64668 (to J.A.S.). We are grateful for the helpful suggestions of Drs. Adam Hoppe, Peter Beemiller, Chris Ellson, Francesc Marti, and Susheela Tridandapani. We also thank Julie Champion and Samir Mitragotri (University of California Santa Barbara) for the generous donation of polystyrene rods.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0061) on April 18, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Ai J., Maturu A., Johnson W., Wang Y., Marsh C. B., Tridandapani S. The inositol phosphatase SHIP-2 down-regulates FcgammaR-mediated phagocytosis in murine macrophages independently of SHIP-1. Blood. 2006;107:813–820. doi: 10.1182/blood-2005-05-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez B., Garrido E., Garcia-Sanz J. A., Carrera A. C. Phosphoinositide 3-kinase activation regulates cell division time by coordinated control of cell mass and cell cycle progression rate. J. Biol. Chem. 2003;278:26466–26473. doi: 10.1074/jbc.M300663200. [DOI] [PubMed] [Google Scholar]
- Araki N., Johnson M. T., Swanson J. A. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J. Cell Biol. 1996;135:1249–1260. doi: 10.1083/jcb.135.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger K. R., Serunian L. A., Soltoff S. P., Libby P., Cantley L. C. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- Beemiller P., Hoppe A. D., Swanson J. A. A phosphatidylinositol- 3-kinase-dependent signal transition regulates ARF1 and ARF6 during Fcgamma receptor-mediated phagocytosis. PLoS Biol. 2006;4:e162. doi: 10.1371/journal.pbio.0040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho R. J., Teruel M., Dierckman R., Anderson R., Wells A., York J. D., Meyer T., Grinstein S. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J. Cell Biol. 2000;151:1353–1368. doi: 10.1083/jcb.151.7.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D. G., Qiu W. Q., Luster A. D., Ravetch J. V. Structure and expression of human IgG FcRII(CD32). Functional heterogeneity is encoded by the alternatively spliced products of multiple genes. J. Exp. Med. 1989;170:1369–1385. doi: 10.1084/jem.170.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Wei G., Fang H., Guo J., Weinstein M., Marsh C. B., Ostrowski M. C., Tridandapani S. The inositol 3-phosphatase PTEN negatively regulates Fc gamma receptor signaling, but supports Toll-like receptor 4 signaling in murine peritoneal macrophages. J. Immunol. 2004;172:4851–4857. doi: 10.4049/jimmunol.172.8.4851. [DOI] [PubMed] [Google Scholar]
- Chacko G. W., Brandt J. T., Coggeshall K. M., Anderson C. L. Phosphoinositide 3-kinase and p72syk noncovalently associate with the low affinity Fc gamma receptor on human platelets through an immunoreceptor tyrosine-based activation motif. Reconstitution with synthetic phosphopeptides. J. Biol. Chem. 1996;271:10775–10781. doi: 10.1074/jbc.271.18.10775. [DOI] [PubMed] [Google Scholar]
- Champion J. A., Mitragotri S. Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. USA. 2006;103:4930–4934. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman K. L., Ueyama T., Michaud T. M., Kashiwagi K., Wang D., Flax L. A., Shirai Y., Loegering D. J., Saito N., Lennartz M. R. Targeting of protein kinase C-epsilon during Fcgamma receptor-dependent phagocytosis requires the epsilonC1B domain and phospholipase C-gamma1. Mol. Biol. Cell. 2006;17:799–813. doi: 10.1091/mbc.E04-12-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggeshall K. M., Nakamura K., Phee H. How do inhibitory phosphatases work? Mol. Immunol. 2002;39:521–529. doi: 10.1016/s0161-5890(02)00209-2. [DOI] [PubMed] [Google Scholar]
- Corbett-Nelson E. F., Mason D., Marshall J. G., Collette Y., Grinstein S. Signaling-dependent immobilization of acylated proteins in the inner monolayer of the plasma membrane. J. Cell Biol. 2006;174:255–265. doi: 10.1083/jcb.200605044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D., Dale B. M., Kashiwada M., Helgason C. D., Greenberg S. A regulatory role for Src homology 2 domain-containing inositol 5′-phosphatase (SHIP) in phagocytosis mediated by Fc gamma receptors and complement receptor 3 (alpha(M)beta(2); CD11b/CD18) J. Exp. Med. 2001;193:61–71. doi: 10.1084/jem.193.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D., Tseng C. C., Bjekic G., Greenberg S. A requirement for phosphatidylinositol 3-kinase in pseudopod extension. J. Biol. Chem. 1999;274:1240–1247. doi: 10.1074/jbc.274.3.1240. [DOI] [PubMed] [Google Scholar]
- Damen J. E., Liu L., Rosten P., Humphries R. K., Jefferson A. B., Majerus P. W., Krystal G. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-triphosphate 5-phosphatase. Proc. Natl. Acad. Sci. USA. 1996;93:1689–1693. doi: 10.1073/pnas.93.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhand R., Hara K., Hiles I., Bax B., Gout I., Panayotou G., Fry M. J., Yonezawa K., Kasuga M., Waterfield M. D. PI 3-kinase: structural and functional analysis of intersubunit interactions. EMBO J. 1994;13:511–521. doi: 10.1002/j.1460-2075.1994.tb06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillham H., Golding M. C., Pepperkok R., Gullick W. J. Intracellular movement of green fluorescent protein-tagged phosphatidylinositol 3-kinase in response to growth factor receptor signaling. J. Cell Biol. 1999;146:869–880. doi: 10.1083/jcb.146.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins P. T., Jackson T. R., Stephens L. R. Platelet-derived growth factor stimulates synthesis of PtdIns(3,4,5)P3 by activating a PtdIns(4,5)P2 3-OH kinase. Nature. 1992;358:157–159. doi: 10.1038/358157a0. [DOI] [PubMed] [Google Scholar]
- Hoppe A., Christensen K., Swanson J. A. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys. J. 2002;83:3652–3664. doi: 10.1016/S0006-3495(02)75365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe A. D., Swanson J. A. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol. Biol. Cell. 2004;15:3509–3519. doi: 10.1091/mbc.E03-11-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katso R., Okkenhaug K., Ahmadi K., White S., Timms J., Waterfield M. D. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Peng X., De P. K., Geahlen R. L., Durden D. L. PTEN controls immunoreceptor (immunoreceptor tyrosine-based activation motif) signaling and the activation of Rac. Blood. 2002;99:694–697. doi: 10.1182/blood.v99.2.694. [DOI] [PubMed] [Google Scholar]
- Knapp P. E., Swanson J. A. Plasticity of the tubular lysosomal compartment in macrophages. J. Cell Sci. 1990;95:433–439. doi: 10.1242/jcs.95.3.433. [DOI] [PubMed] [Google Scholar]
- Kusner D. J., Hall C. F., Jackson S. Fc gamma receptor-mediated activation of phospholipase D regulates macrophage phagocytosis of IgG-opsonized particles. J. Immunol. 1999;162:2266–2274. [PubMed] [Google Scholar]
- Lee W. L., Kim M. K., Schreiber A. D., Grinstein S. Role of ubiquitin and proteasomes in phagosome maturation. Mol. Biol. Cell. 2005;16:2077–2090. doi: 10.1091/mbc.E04-06-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M. A., Ferguson K. M., O'Brien R., Sigler P. B., Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartz M. R., Brown E. J. Arachidonic acid is essential for IgG Fc receptor-mediated phagocytosis by human monocytes. J. Immunol. 1991;147:621–626. [PubMed] [Google Scholar]
- Lowry M. B., Duchemin A. M., Coggeshall K. M., Robinson J. M., Anderson C. L. Chimeric receptors composed of phosphoinositide 3-kinase domains and FCgamma receptor ligand-binding domains mediate phagocytosis in COS fibroblasts. J. Biol. Chem. 1998;273:24513–24520. doi: 10.1074/jbc.273.38.24513. [DOI] [PubMed] [Google Scholar]
- Maehama T., Dixon J. E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Marshall J. G., Booth J. W., Stambolic V., Mak T., Balla T., Schreiber A. D., Meyer T., Grinstein S. Restricted accumulation of phosphatidylinositol 3-kinase products in a plasmalemmal subdomain during Fc gamma receptor-mediated phagocytosis. J. Cell Biol. 2001;153:1369–1380. doi: 10.1083/jcb.153.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Malykhin A., Coggeshall K. M. The Src homology 2 domain-containing inositol 5-phosphatase negatively regulates Fcgamma receptor-mediated phagocytosis through immunoreceptor tyrosine-based activation motif-bearing phagocytic receptors. Blood. 2002;100:3374–3382. doi: 10.1182/blood-2002-03-0787. [DOI] [PubMed] [Google Scholar]
- Rameh L. E., et al. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J. Biol. Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- Rawlings D. J., et al. Mutation of unique region of Bruton's tyrosine kinase in immunodeficient XID mice. Science. 1993;261:358–361. doi: 10.1126/science.8332901. [DOI] [PubMed] [Google Scholar]
- Strausberg R. L., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. A., Hoppe A. D. The coordination of signaling during Fc receptor-mediated phagocytosis. J. Leukoc. Biol. 2004;76:1093–1103. doi: 10.1189/jlb.0804439. [DOI] [PubMed] [Google Scholar]
- Thomas J. D., Sideras P., Smith C. I., Vorechovsky I., Chapman V., Paul W. E. Colocalization of X-linked agammaglobulinemia and X-linked immunodeficiency genes. Science. 1993;261:355–358. doi: 10.1126/science.8332900. [DOI] [PubMed] [Google Scholar]
- Tridandapani S., Kelley T., Pradhan M., Cooney D., Justement L. B., Coggeshall K. M. Recruitment and phosphorylation of SH2-containing inositol phosphatase and Shc to the B-cell Fc gamma immunoreceptor tyrosine-based inhibition motif peptide motif. Mol. Cell. Biol. 1997;17:4305–4311. doi: 10.1128/mcb.17.8.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tridandapani S., Pradhan M., LaDine J. R., Garber S., Anderson C. L., Coggeshall K. M. Protein interactions of Src homology 2 (SH2) domain-containing inositol phosphatase (SHIP): association with Shc displaces SHIP from FcgammaRIIb in B cells. J. Immunol. 1999;162:1408–1414. [PubMed] [Google Scholar]
- Tridandapani S., Siefker K., Teillaud J. L., Carter J. E., Wewers M. D., Anderson C. L. Regulated expression and inhibitory function of Fcgamma RIIb in human monocytic cells. J. Biol. Chem. 2002a;277:5082–5089. doi: 10.1074/jbc.M110277200. [DOI] [PubMed] [Google Scholar]
- Tridandapani S., Wang Y., Marsh C. B., Anderson C. L. Src homology 2 domain-containing inositol polyphosphate phosphatase regulates NF-kappa B-mediated gene transcription by phagocytic Fc gamma Rs in human myeloid cells. J. Immunol. 2002b;169:4370–4378. doi: 10.4049/jimmunol.169.8.4370. [DOI] [PubMed] [Google Scholar]
- Varnai P., Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P., Rother K. I., Balla T. Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton's tyrosine kinase pleckstrin homology domain visualized in single living cells. J. Biol. Chem. 1999;274:10983–10989. doi: 10.1074/jbc.274.16.10983. [DOI] [PubMed] [Google Scholar]
- Vazquez F., Matsuoka S., Sellers W. R., Yanagida T., Ueda M., Devreotes P. N. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc. Natl. Acad. Sci. USA. 2006;103:3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zheleznyak A., Brown E. J. Immunoglobulin-mediated phagocytosis by human monocytes requires protein kinase C activation. Evidence for protein kinase C translocation to phagosomes. J. Biol. Chem. 1992;267:12042–12048. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.