Abstract
Cytotoxic necrotizing factor 1 (CNF1) is a protein toxin produced by some pathogenic strains of Escherichia coli that specifically activates Rho, Rac, and Cdc42 GTPases. We previously reported that this toxin prevents the ultraviolet-B–induced apoptosis in epithelial cells, with a mechanism that remained to be defined. In this work, we show that the proteasomal degradation of the Rho GTPase is necessary to achieve cell death protection, because inhibition of Rho degradation abolishes the prosurvival activity of CNF1. We hypothesize that Rho inactivation allows the activity of Rac to become dominant. This in turn leads to stimulation of the phosphoinositide 3-kinase/Akt/IκB kinase/nuclear factor-κB prosurvival pathway and to a remarkable modification in the architecture of the mitochondrial network, mainly consisting in the appearance of elongated and interconnected mitochondria. Importantly, we found that Bcl-2 silencing reduces the ability of CNF1 to protect cells against apoptosis and that it also prevents the CNF1-induced mitochondrial changes. It is worth noting that the ability of a bacterial toxin to induce such a remodeling of the mitochondrial network is herein reported for the first time. The possible pathophysiological relevance of this finding is discussed.
INTRODUCTION
Today, it is largely acknowledged that apoptosis, besides being an evolutionary conserved form of cell death that plays a pivotal role during development, morphogenesis, and cell homeostasis, is also critically implied in a constantly growing number of diseases (Fadeel and Orrenius, 2005). In fact, apoptosis can be regarded as a widespread strategy exploited by pathogenic bacteria to favor their own survival or spreading in the host (Fiorentini et al., 2003), often by producing protein toxins that mediate their long-range cross-talk with host cells. In this context, we have previously reported the ability of a protein toxin from Escherichia coli, namely the cytotoxic necrotizing factor 1 (CNF1), to prevent the ultraviolet-B (UVB)–induced apoptosis and to increase the expression of antiapoptotic Bcl-2 family proteins (Fiorentini et al., 1998). The precise mechanism by which CNF1 allows cells to survive, however, is not yet defined.
CNF1 is a protein toxin produced by some pathogenic strains of E. coli mainly involved in extraintestinal infections (Landraud et al., 2000). In eukaryotic cells, CNF1 binds to its receptor, reported to be the receptor of laminin (Kim et al., 2005), and it is endocytosed and released into the cytoplasm by an acidic-dependent mechanism (Contamin et al., 2000). Once in the cytoplasm, CNF1 exerts its enzymatic activity that is represented by deamidation of a pivotal glutamine residue of the guanosine triphosphate (GTP)-binding proteins Rho, Rac, and Cdc42 (glutamine 63 of Rho or glutamine 61 of Rac and Cdc42), giving rise to a glutamic acid (Flatau et al., 1997; Schmidt et al., 1997; Lerm et al., 1999). The glutamine residue modified by CNF1 lies in the switch 2 domain of Rho proteins, which is involved in GTP hydrolysis; thus, the modification exerted by CNF1 blocks the Rho GTPases in their GTP-bound activated state. Very high levels of activated Rho GTPases are recognized by cells that ubiquitinate and degrade them to more physiological levels (Doye et al., 2002). Although Rho, Rac and Cdc42 are mainly involved in the actin cytoskeleton organization (Hall, 1998), it is known that these proteins are also involved in a huge number of other cellular processes, such as gene transcription, cell proliferation, and survival. Thus, the consequence of CNF1-induced Rho activation is the induction of a number of actin-dependent phenomena, such as contractility, cell spreading (Fiorentini et al., 1988), assembly of focal adhesion plaques (Lacerda et al., 1997), and the induction of macropinocytosis (Falzano et al., 1993; Fiorentini et al., 2001), as well as the stimulation of new activities in cells, including the ability to counteract apoptosis (Fiorentini et al., 1998).
It is well known that, among the several molecules implied in the regulation of apoptotic cell death, an important role is played by the transcription factor nuclear factor-κB (NF-κB), whose activation has been reported to protect from various apoptotic stimuli in different cellular systems (for review, see Shishodia and Aggarwal, 2002). In this context, we recently demonstrated the ability of CNF1 to induce the activation of NF-κB (Boyer et al., 2004), highlighting a possible involvement of this transcription factor in the toxin-induced cell survival. NF-κB consists of five members that exist as both homo- and heterodimers, the best characterized form being a heterodimer composed of p50 and p65 subunits (for review, see Ghosh et al., 1998; Hayden and Ghosh, 2004). This heterodimer is sequestered in the cytoplasm by association with the inhibitory subunit IκBα (Beg et al., 1992). Signals leading to NF-κB activation trigger IκBα phosphorylation at two specific serine residues, allowing IκBα polyubiquitination and subsequent proteolytic degradation by the 26S proteasome. Thus, freed NF-κB is translocated into the nucleus for gene transactivation (Baldwin, 1996). Regarding CNF1, we have reported previously that activation of Rac and NF-κB by CNF1 is strictly linked via the recruitment of Rac, IκBα, and the Skp, Cullin, F-box–containing complex (responsible for the ubiquitination process) to the ruffling membranes (Boyer et al., 2004).
We undertook this study to analyze the molecular mechanism underlying the CNF1-induced protection from apoptosis. In the present article, we provide evidence that the antiapoptotic property of the toxin is dependent on the Rac-induced activation of the phosphoinositide 3-kinase (PI3K)/Akt/IKK/NF-κB prosurvival pathway and that it also relies on the Bcl-2–dependent remodeling of the mitochondrial network. Notably, the capacity of a bacterial toxin to promote the formation of a complex system of elongated and interconnected mitochondria is a hitherto uncharted novelty in the bacterial world.
MATERIALS AND METHODS
Cell Cultures, Treatments, and Inhibitors
Human epithelial HEp-2 cells were grown in minimal Eagle's medium supplemented with 10% fetal calf serum (Flow Laboratories, Rockville, MD), 5 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin.
CNF1 was obtained from the 392 ISS strain (kindly provided by V. Falbo, Department of Cell Biology and Neuroscience, Istituto Superiore di Sanità, Rome, Italy), and it was purified as described previously (Falzano et al., 1993). The plasmid coding for a nontoxic mutant of CNF1 that completely lacks the enzymatic activity (C866S) (Schmidt et al., 1998), was kindly provided by E. Lemichez (INSERM U627, Nice, France), and it was prepared as described previously (Flatau et al., 1997).
In all experiments, cells were seeded at a density of 2 × 104 cells/cm2, and 24 h after the seeding, cells were exposed for 1, 2, 4, 6, 12, 18, and 24 h to 10−10 M CNF1 or to 10−10 M CNF1 C866S.
UVB Exposure.
Control and treated HEp-2 cells were exposed to UVB irradiation in phosphate-buffered saline (PBS) by using a Philips TL 20 W/12 lamp as described previously (Malorni et al., 1995). Plastic Petri dishes containing cells were placed without covers at the vertical distance of 10 cm from the center of the tube of UVB. To eliminate UVC radiation, a Kodak filter (Kodacell TL 401) was placed on the Petri dishes during exposure. In these conditions, the UVB radiant flux density to the cells was 2.2 Wm−2, as verified by an Osram Centra UV meter. All the results reported herein were referred to cells still adhering to the substrate analyzed 24 h after UVB exposure.
Inhibitors.
For PI3K, IKK, and proteasome inhibition, cells were pre-incubated for 30 min with 100 ng/ml wortmannin (Alexis Biochemicals, San Diego, CA), 30 μM prostaglandin A1 (PGA1) (BioMol, San Diego, CA), and 2.5 μM lactacystin (BIOMOL Research Laboratories, Plymouth Meeting, PA), respectively, before being challenged with CNF1.
Transfection of HEp-2 Cells
Control and treated HEp-2 cells were transfected with 1 μg/35-mm Petri dish of plasmid DNA encoding myc-tagged dominant-positive form of the Rho GTPase (RhoV14) or DsRed-Mitochondria (Clontech, Palo Alto, CA), a plasmid DNA encoding DsRed-tagged protein that specifically recognizes and binds mitochondria. Lipofectamine (Invitrogen, Carlsbad, CA) was used to transfect cells according to the manufacturer's instructions. Eighteen hours after transfection, cells were exposed to CNF1 for 24 h. Efficiency of transfection was 30%.
To silence Rac1, Bcl-2, and Bcl-XL proteins, cells were transfected with specific small interfering RNAs (siRNAs) synthesized by M-Medical-Genenco (Florence, Italy). The following oligonucleotides were used: 5′-GGA GAU UGG UGC UGU AAA ATT-3′ or 5′-CCU UUG UAC GCU UUG CUC ATT-3′ (for Rac); 5′-AAC AUC GCC CUG UGG AUG ACU-3′ or 5′-CAG GAC CUC GCC GCU GCA GTT-3′ (for Bcl-2); and 5′-GGA GAU GCA GGU AUU GGU GTT-3′ (for Bcl-XL). Lipofectamine was used to transfect cells according to the manufacturer's instructions, and 36 h after silencing, cells were exposed to CNF1 for 24 h. To verify the actual occurrence of target protein silencing, cells were lysed and subjected to Western blot analysis, as described below.
Immunofluorescence and Confocal Microscopy
Control and CNF1-treated HEp-2 cells were processed as follows. For p65 immunostaining, cells were fixed in acetone/methanol (1:1, vol/vol) for 10 min at room temperature and air-dried. After 1 h of preincubation with PBS containing 10% of AB human serum, cells were incubated with an anti-p65 antibody (diluted 1:50; Santa Cruz Biotechnology, CA) for 1 h at room temperature. After three washing in PBS, cells were incubated for 1 h at room temperature with fluorescein isothiocyanate (FITC)-labeled anti-rabbit antibody. For apoptosis detection, cells were fixed in 3.7% paraformaldehyde and stained with Hoechst 33258 (Sigma-Aldrich, St. Louis, MO) as described previously (Fiorentini et al., 2001). Cells were mounted on glass coverslips and analyzed with an Olympus BX51 fluorescence microscope. For mitochondria immunostaining, cells were fixed in 3.7% paraformaldehyde in PBS for 30 min at room temperature, and then they were permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature. After washing in the same buffer, samples were incubated with an anti-mitochondria antibody (diluted 1:1000; BD Biosciences Transduction Laboratories, Lexington, KY) for 1 h at room temperature. After three washing in PBS, cells were incubated for 1 h at room temperature with a tetramethylrhodamine B isothiocyanate-labeled anti-mouse antibody. Cells transfected with DsRed-Mitochondria were fixed in 3.7% paraformaldehyde. Both cells immunostained with anti-mitochondria and transfected with DsRed-Mitochondria were analyzed with a Leica TCS SP2 spectral confocal microscope (Leica Microsystems, Wetzlar, Germany) equipped with Argon-HeliumNeon (Ar-HeNe) lasers. Images represent orthogonal maximum projection of a series of optical sections, obtained by calculating maximum intensity values. The excitation and emission wavelengths were 550 and 570 nm, respectively. Fluorescence emissions were collected after passage through a DD488/543 filter in a detection bandwidth of 555–620 nm. Images were processed by using the LCS (Leica Microsystems) software program.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total cellular RNA was extracted using the RNeasy kit (QIAGEN, Hilden, Germany). RNA was reverse transcribed into cDNA and amplified by PCR by using the Access RT-PCR System (Promega, Madison, WI) according to the manufacturer's instructions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Bcl-2, and Bcl-XL set of primers were synthesized by M-Medical-Genenco. The following couples of primers were used: Bcl-2: sense 5′-GCG TCA ACC GGG AGA TGT CGC CC-3′, antisense 5′-TTT CTT AAA CAG CCT GCA GCT TTG-3′; Bcl-XL: sense 5′-AGA GAA GGG GGT GGG AGG GTA-3′, antisense 5′-ATT GGT GAG TCG GAT CGC AGC-3′; and GAPDH: sense 5′-GTC TTC ACC ATG GAG AAG GTC-3′, antisense 5′-CAT GCC AGT GAG CTT CCC GTT CA-3′. To exclude false positive results due to contamination, in all experiments a control RT-PCR without RNA was conducted. For densitometry analysis, the reaction products were electrophoresed through a 1.8% agarose gel (Bio-Rad, Hercules, CA), and ethidium bromide-stained gels, scanned using the Imaging Densitometer GS-700 (Bio-Rad), were quantified by means of MultiAnalist software (Bio-Rad). The levels of mRNA were normalized to GAPDH mRNA levels.
Protein Extraction and Western Blot
Cells were lysed in boiled sample buffer 1× (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, and 100 mM dithiothreitol). Twenty-five micrograms of total protein extracts were resolved on 10 or 12% SDS-polyacrylamide gel electrophoresis (PAGE) and electrically transferred onto polyvinylidene difluoride membranes (Bio-Rad). Membranes were blocked with Tris-buffered saline-Tween 20 (TBS-T) (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.02% Tween 20) containing 5% skimmed milk (Bio-Rad) for 30 min at room temperature, and then they were incubated overnight at 4°C with primary antibodies diluted in TBS-T containing 2% milk. The following primary antibodies were used: mouse monoclonal anti-Rac1 (1:3500; BD Biosciences Transduction Laboratories), rabbit polyclonal anti-Bcl-XL (1:500; Santa Cruz Biotechnology, CA), mouse monoclonal anti-Bcl-2 (1:200; Santa Cruz Biotechnology, CA) and mouse monoclonal anti-α-tubulin (1:10,000; Sigma-Aldrich).
After extensive washing in TBS-T, immunocomplexes were detected with horseradish peroxidase-conjugated species-specific secondary antibodies (Jackson Laboratory, Bar Harbor, ME) followed by enhanced chemiluminescence reaction (Pierce Chemical, Rockford, IL).
Activated Rho GTPases Pull-Down and Akt Kinase Assay
Pull-down assays were performed as described previously (Travaglione et al., 2005). Cells were lysed in 1) 50 mM HEPES, pH 7.4, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS, 0.5 M NaCl, and 10 mM MgCl2, plus protease inhibitors (to detect Rho-GTP) or 2) 50 mM HEPES, pH 7.4, 0.1 M NaCl, 10 mM MgCl2, 5% glycerol, 1% NP-40, and 10 mM NaF, plus protease inhibitors (to detect Rac/Cdc42-GTP). The cleared lysates were incubated with 80 μg of glutathione S-transferase (GST)-Rhotekin (for Rho; Cytoskeleton, Denver, CO) and GST-PAK-CD (for Rac/Cdc42, prepared as described previously; Travaglione et al., 2005) fusion proteins, bound to glutathione-coupled Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) for 40 min at 4°C. Beads were washed three times in 1) 50 mM HEPES, pH 7.4, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS, 0.25 M NaCl, 5 mM MgCl2, plus protease inhibitors (for Rho) or 2) 50 mM HEPES, pH 7.4, 0.1 M NaCl, 10 mM MgCl2, 5% glycerol, 0.5% NP-40, and 10 mM NaF, plus protease inhibitors (for Rac/Cdc42). The bound proteins were eluted in sample buffer before being subjected to SDS-PAGE and immunoblotting with the following specific antibodies: mouse monoclonal anti-RhoA (1:500; Cytoskeleton), mouse monoclonal anti-Rac1 (1:3500; BD Biosciences Transduction Laboratories), and mouse monoclonal anti-Cdc42 (Santa Cruz Biotechnology, CA). Whole-cell lysates (2% of input) were analyzed in parallel. Autoradiographs, scanned using the Imaging Densitometer GS-700 (Bio-Rad), were quantified by means of MultiAnalist software and normalized as a function of the total proteins loaded in the assay.
The Akt kinase assay was performed by using the nonradioactive kit purchased by Cell Signaling Technology (Danvers, MA) and following the manufacturer's instructions. Briefly, cells were lysed and centrifuged for 10 min at 4°C. Akt was immunoprecipitated by adding 20 μl of anti-Akt antibody bead slurry to 200 μl of cell lysates. After overnight at 4°C, bound Akt was pelleted and incubated with 1 μg of glycogen synthase kinase 3 (GSK-3) fusion protein together with 10 mM ATP for 30 min at 30°C. The reaction was terminated by adding 25 μl of 3× sample buffer, and samples were boiled for 5 min at 95°C. Proteins were resolved on SDS-PAGE and immunoblotted with a specific anti-phospho-GSK-3 α/β (Cell Signaling Technology).
Statistical Analysis
The values reported in graphs are the means ± SD from three separate experiments performed in duplicate. Student's t test was used for analysis of statistical significance. A p value <0.05 was considered significant.
RESULTS
The Ability of CNF1 to Protect HEp-2 Cells from UVB-induced Apoptosis Requires the Inactivation of Rho Protein
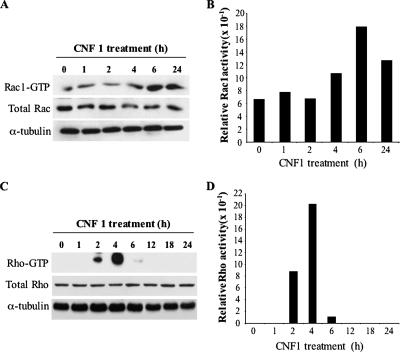
CNF1 activates Rho GTPases in all cell systems where its effects have been studied, but the timing and level of activation of distinct members of Rho family differ depending on the cell type (Doye et al., 2002; Boyer et al., 2006). Hence, to investigate the role played by these proteins in the ability of cells to survive UVB radiation, we monitored the activation state of Rho, Rac, and Cdc42 in HEp-2 cells. For this purpose, cells were treated with CNF1 for different time lengths, and, at each time point, the active forms of GTP-bound Rho, Rac1, and Cdc42 GTPases were detected by pull-down assays (see Materials and Methods). In accordance with the work by Doye et al. (2002) and Boyer et al. (2006), the results obtained clearly evidenced that CNF1 induced a sustained activation of Rac1 (Figure 1, A and B), this GTPase being active at least until 24 h of toxin challenge. By contrast, Rho (Figure 1, C and D) and Cdc42 (data not shown) proteins were activated by CNF1 in a transient manner. Indeed, the immunoblot reported in Figure 1C shows that the active form of Rho, virtually absent in control cells, significantly increased after 2 h of toxin exposure, reached a maximum at 4 h, and then dramatically decreased and disappeared after 12 h of treatment (Figure 1, C and D).
Figure 1.
Activation kinetics of Rac1 and Rho GTPases in HEp-2 cells. Immunoblots showing the amount of activated Rac1 (A) and Rho (C) (top) after different times of CNF1 exposure, obtained by pull-down experiments. The total amount of Rac1 and Rho proteins loaded in the assay was determined by Western blot (middle). Equal protein loading was confirmed by detecting α-tubulin (bottom). (A) CNF1 induces a sustained activation of Rac1 that remains activated up to 24 h of toxin exposure. (C) Rho is activated by CNF1 in a transient manner. The results in A and C were analyzed by densitometry. The histograms represent the Rac1 (B) and Rho (D) activity normalized for the amount of total protein loaded.
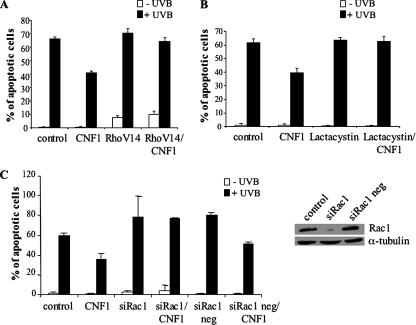
Thus, we wondered whether the disappearance of Rho-GTP that we observed (Figure 1, C and D) could be somehow necessary to achieve an efficient protection from apoptosis by CNF1. To verify this hypothesis, we transfected cells with the constitutively active form of the protein RhoA V14. Twenty-four hours after transfection, cells were treated with CNF1 for further 24 h, exposed to UVB radiation, and then stained with the nuclear dye Hoechst 33258 to evaluate, by fluorescence microscopy, the percentage of apoptotic nuclei. Figure 2A clearly shows that, when cells were transfected with the constitutively active form RhoA V14 before CNF1 exposure, the toxin was not able to exert its protective effect against UVB-induced apoptosis, the number of apoptotic nuclei being similar to that of control cells. Because activated RhoA is degraded via the proteasome (Doye et al., 2006; Boyer et al., 2006), to further confirm the need of Rho deactivation for protection from apoptosis, we pretreated HEp-2 cells for 30 min with 2.5 μM proteasome inhibitor lactacystin, and then we added CNF1 for 24 h before UVB irradiation. As shown in Figure 2B, pretreatment with lactacystin abolished the protective effect of CNF1.
Figure 2.
RhoA and Rac1 involvement in the CNF1-mediated protection against apoptosis. (A and B) Graphs showing the percentages (±SD) of apoptosis in cells transfected with the constitutively active form of RhoA (A) or treated with the proteasome inhibitor lactacystin (B) before CNF1 exposure. In both cases, the CNF1-induced protecting effect against UVB-induced apoptosis is prevented. (C) Graph showing the percentages (±SD) of apoptosis in cells transfected with the siRNA against Rac1 and then exposed to CNF1. Silencing of Rac1, confirmed by Western blot (C, right), counteracts the CNF1-induced protection against UVB-induced apoptosis.
The degradation of RhoA may possibly render Rac1 the predominant form of activated Rho GTPases, suggesting a pivotal role for Rac1 in the protection from apoptosis. To verify this hypothesis, cells were transfected with siRNA to deplete endogenous Rac1, treated with CNF1, and finally exposed to UVB. We found that in HEp-2 cells depleted for Rac1, CNF1 was not able to protect from UVB-induced apoptosis (Figure 2C). We obtained the same results also by using a second siRNA with a different target sequence in the Rac1 gene (data not shown; this siRNA sequence is reported in Materials and Methods), whereas a negative siRNA, lacking the specific sequence that recognizes Rac1 mRNA, failed in counteracting the CNF1-induced protection against apoptosis. Taken together, these results indicate the presence of Rac1-GTP as well as inactivation of RhoA as critical events for the capacity of CNF1 to promote cell survival.
Involvement of the PI3K/Akt/IKK/NF-κB Pathway in the CNF1-induced Protection against UVB-induced Apoptosis
It is worth noting that the ability of lactacystin to impair the CNF1-induced effects on cell death could be explained by two different, but probably overlapping, mechanisms: 1) the impairment of Rho-GTP degradation, which, in turn, prevents the consequent Rac-GTP prevalence, and 2) the inhibition of IκBα degradation and thus of NF-κB activation. Despite the demonstration that CNF1 stimulates the transactivation of NF-κB in a Rac-dependent way (Boyer et al., 2004), it is still unproved whether NF-κB could play a role in the CNF1-promoted protection against apoptotic stimuli. We have thus dissected one of the pathways that lead to the activation of this transcription factor and that could be involved in the CNF1-dependent cell survival, particularly the PI3K/Akt pathway that is known to be activated by Rac (Chan et al., 2002). Hence, we asked whether CNF1 could activate the PI3K/Akt pathway, and, eventually, whether this pathway could be implicated in CNF1-mediated NF-κB activation.
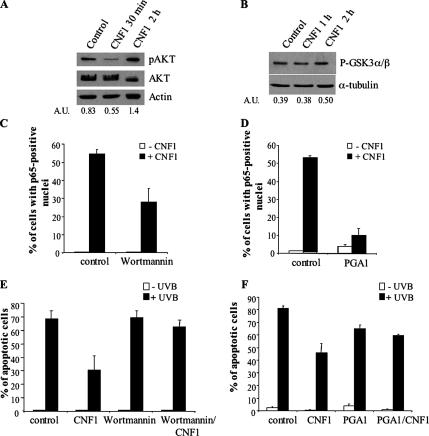
To answer the first question, HEp-2 cells were treated with CNF1, and the amount of phospho-Akt, the activated form of Akt, was evaluated. We found that CNF1 exposure increased the amount of phospho-Akt (Figure 3A). We thus confirmed that the Akt kinase was activated by using a nonradioactive method based on the GSK-3α/β fusion protein as Akt substrate. The immunoblot reported in Figure 3B shows that CNF1 was able to increase the amount of phospho-GSK-3α/β starting from 2 h of toxin treatment, this activation lasting for several hours (data not shown). Together, these data demonstrate that the PI3K/Akt pathway was activated by CNF1.
Figure 3.
CNF1 acts via the PI3K/Akt/IKK/NF-κB pathway. Immunoblots showing the amount of the activated form of Akt (pAkt) (A) or the amount of phosphorylated GSK-3α/β (B), a substrate of activated Akt, after CNF1 exposure (A.U., arbitrary units). (C and D) Graphs showing the percentage (±SD) of p65-positive nuclei in cells pretreated with wortmannin (C) or PGA1 (D) before CNF1 exposure. (E and F) Graphs showing the percentage (±SD) of apoptotic nuclei in cells pretreated with wortmannin (E) or PGA1 (F), before CNF1 exposure and UVB irradiation. Note that inhibition of the PI3K/Akt/IKK/NF-κB pathway counteracts the prosurvival activity of CNF1.
To verify whether Akt activity was involved in the CNF1-mediated NF-κB activation, HEp-2 cells, pretreated with 100 ng/ml PI3K inhibitor wortmannin, were exposed to CNF1 for 4 h, and then they were stained with an antibody directed against the NF-κB subunit p65. The ability of p65 to translocate into the nucleus was evaluated by fluorescence microscopy. As shown in Figure 3C, pretreatment with wortmannin significantly counteracted the CNF1-promoted nuclear translocation of the NF-κB subunit, being the percentage of p65-positive nuclei lower than that of cells treated with CNF1 alone.
Akt represents one of the kinases responsible for the activation of IKK (Romashkova and Makarov, 1999), a kinase that in turn phosphorylates the NF-κB inhibitor IκBα. We thus investigated whether this kinase was involved in the CNF1-dependent NF-κB activation, by pretreating cells with 30 μM IKK inhibitor PGA1. As expected, the inactivation of IKK strongly reduced the ability of CNF1 to stimulate p65 translocation into the nucleus (Figure 3D).
At this point, to verify whether the PI3K/Akt/IKK/NF-κB pathway was involved in the CNF1-mediated protection against apoptotic cell death, we pretreated HEp-2 cells with wortmannin or with PGA1 before exposure to CNF1 for 24 h and subsequent UVB radiation. We then evaluated the percentage of apoptotic cells, and we found that both wortmannin and PGA1 prevented the prosurvival effect of CNF1 (Figure 3, E and F), indicating the involvement of the PI3K/Akt/IKK/NF-κB pathway in the prosurvival activity exerted by CNF1.
Bcl-2, but Not Bcl-XL, Is Necessary for the CNF1-induced Cell Survival
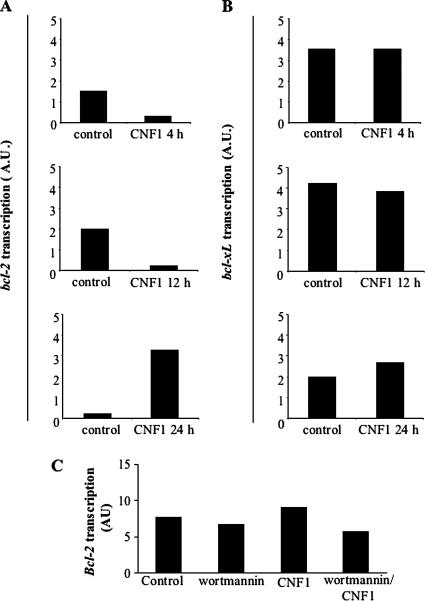
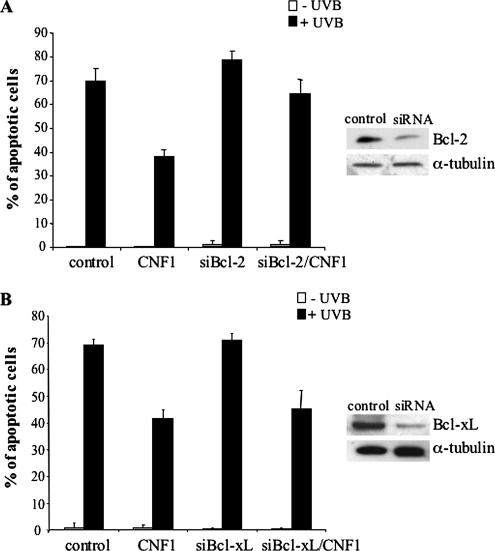
As cited above, NF-κB is involved in the transcription of antiapoptotic genes, such as those belonging to the Bcl-2 family (Ghosh et al., 1998). In this context, we previously reported that CNF1 increases the amount of Bcl-2 and Bcl-XL proteins in HEp-2 cells (Fiorentini et al., 1998). Hence, we analyzed the ability of CNF1 to regulate the expression of these antiapoptotic factors at the transcriptional level. For this purpose, HEp-2 cells were challenged with CNF1, and, at different time points (4, 12, and 24 h), the amount of Bcl-2 and Bcl-XL mRNA levels were analyzed by RT-PCR. Interestingly, we found that CNF1 was able to increase the transcription of the Bcl-2 gene only starting from 24 h of treatment (Figure 4A), but Bcl-XL was not varied whatever the time of CNF1 exposure (Figure 4B). Thus, we wondered whether the observed increase in Bcl-2 mRNA was dependent on the PI3K/Akt pathway. For this purpose, we pretreated HEp-2 cells with wortmannin before CNF1 exposure for 24 h. We found that the increase in Bcl-2 mRNA was inhibited by wortmannin, this finding strongly suggesting the involvement of the PI3K/Akt pathway (Figure 4C). To verify the involvement of Bcl-2 and Bcl-XL in the CNF1-induced protection from apoptosis, we blocked the expression of both antiapoptotic proteins by means of siRNAs before CNF1 exposure and UVB radiation, and then we evaluated the percentage of apoptotic cells. Bcl-2 silencing significantly reduced the ability of CNF1 to protect HEp-2 cells from UVB-induced apoptosis (Figure 5A), whereas silencing Bcl-XL did not appreciably influence the antiapoptotic activity of the toxin (Figure 5B). All in all, these results demonstrate that Bcl-2 protein, but not Bcl-XL, is crucial for CNF1-induced survival against apoptotic cell death.
Figure 4.
CNF1 increases Bcl-2 transcription via the PI3K pathway. Graphs showing Bcl-2 (A) and Bcl-XL (B) mRNA levels after different times of CNF1 exposure in HEp-2 cells, normalized to GAPDH mRNA levels. The transcription of the Bcl-2 gene increases starting from 24 h of CNF1 treatment, whereas Bcl-XL gene transcription is not varied. (C) Graph showing Bcl-2 mRNA level after wortmannin exposure and CNF1 treatment. Note that blocking the PI3K pathway renders cells not responsive to the CNF1-induced Bcl-2 mRNA increase (AU, arbitrary units).
Figure 5.
Bcl-2 is necessary for the CNF1-induced cell-survival. Graphs showing the percentage (±SD) of apoptotic cells after silencing of Bcl-2 (A) or Bcl-XL (B) proteins. (A) Transfection with Bcl-2 siRNA significantly reduces the effect of CNF1 against UVB-induced apoptosis. (B) Transfection with Bcl-XL siRNA does not influence the antiapoptotic activity of CNF1. Silencing of Bcl-2 and Bcl-XL was confirmed by Western blot analysis (right).
CNF1 Alters Mitochondrial Morphology in HEp-2 Cells
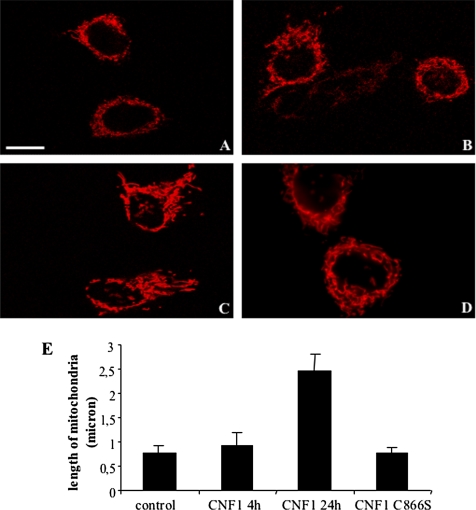
It is well known that Bcl-2, besides blocking the activity of the proapoptotic members of Bcl-2 family, is also involved in the regulation of mitochondrial morphology. In fact, it has been reported that Bcl-2–overexpressing mitochondria show both increased volume and structural complexity (Kowaltowski et al., 2002). Hence, we decided to verify whether CNF1 treatment could provoke a modification in the mitochondrial network morphology. For this purpose, HEp-2 cells were transfected with DsRed-Mitochondria, a plasmid DNA encoding DsRed-tagged protein that specifically recognizes and binds mitochondria, treated with CNF1, and, finally, analyzed by confocal microscopy. The results showed that CNF1 was able to induce changes in the mitochondrial morphology, mitochondria becoming much more elongated and organized in a complex network with respect to those present in control cells (Figure 6A and Supplemental Figure 6A.mov). This effect, although starting after 4 h of toxin challenge (Figure 6B and Supplemental Figure 6B.mov), became particularly evident at 24 h (Figure 6C and Supplemental Figure 6C.mov) of CNF1 exposure and was absent when cells were exposed to a nontoxic mutant of CNF1 (CNF1C866S) that lacks the enzymatic activity (Figure 6D). To quantify changes in mitochondrial length, we used ImageJ software (Research Services Branch, National Institutes of Mental Health, Bethesda, MD), and the results clearly showed that the average length of mitochondria increased after CNF1 exposure (Figure 6E).
Figure 6.
CNF1 alters mitochondrial morphology in HEp-2 cells. (A–D) Confocal microscopy analysis of control (A) or CNF1-treated cells for 4 h (B) and 24 h (C) transfected with DsRed-Mitochondria. In cells challenged with CNF1, mitochondria are more elongated than those present in control cells, and they seem organized in a complex network. This effect becomes particularly evident at 24 h (C) of CNF1 exposure and is not present in HEp-2 cells treated with the mutant CNF1 (CNF1 C866S, D). (E) Graph showing the average length (±SD) of mitochondria in control, CNF1-treated or CNF1 C866S-treated cells. Measurements of the average length of mitochondria were performed by using ImageJ software. Bar, 10 μm.
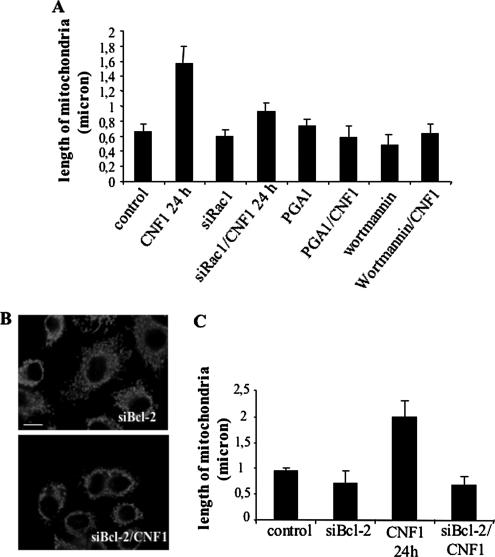
Given the above-mentioned results, we wondered whether the ability of CNF1 to alter mitochondrial morphology was dependent on the activation of the Rac1/PI3K/Akt/IKK pathway. To answer this question, we performed the following pretreatments: 1) suppression of Rac1 with siRNA, 2) pretreatment with wortmannin, and 3) pretreatment with PGA1. Each of these samples was challenged with CNF1, and finally the mitochondrial morphology was analyzed by confocal microscopy and quantified in terms of mitochondrial length by the ImageJ software. All these strategies led to the inhibition of mitochondrial changes caused by CNF1 as quantified in the graph of Figure 7A. We thus wondered whether the mitochondrial rearrangement herein described could be dependent on the expression of Bcl-2. To address this point, we used siRNA to silence Bcl-2, and we found that the ability of the toxin to alter the mitochondrial morphology was significantly counteracted by this strategy. In particular, cells transfected with the siBcl-2 before CNF1 treatment exhibited mitochondria smaller with respect to CNF1-treated cells (Figure 7, B and C). All in all, these results demonstrate that the pathway involved in the prosurvival activity of CNF1 (Rac1/PI3K/Akt/IKK/NF-κB/Bcl-2) is also controlling the CNF1-induced mitochondrial changes.
Figure 7.
CNF1-induced mitochondria alteration depends on the Rac1/PI3K/Akt/IKK pathway and Bcl-2 up-regulation. (A) Graph showing the average length (±SD) of mitochondria after inhibition of the Rac1/PI3K/Akt/IKK pathway before CNF1 exposure. (B) Confocal microscopy analysis of cells transfected with siBcl-2 before CNF1 exposure for 24 h and stained for mitochondria. (C) Graph showing the average length (±SD) of mitochondria after silencing of Bcl-2 before CNF1 exposure. CNF1 induces an increase in mitochondrial length via activation of the Rac1/PI3K/Akt/IKK pathway and increase in Bcl-2 proteins.
DISCUSSION
In this work, we present evidence that the bacterial protein toxin CNF1 protects epithelial cells from apoptosis via activation of the Rac1/PI3K/Akt/IKK/NF-κB pathway. This leads to the up-regulation of the antiapoptotic protein Bcl-2, which is responsible, either directly or indirectly, of a remarkable change in the architecture of the mitochondrial network, mainly consisting in the formation of elongated and interconnected mitochondria.
Many bacterial pathogens have evolved strategies to manipulate the fate of the host cell toward their own benefit. The mechanisms by which the different bacteria and bacterial toxins interfere with apoptosis are various, and they consist of caspase activation, mitochondrial targeting, and regulatory protein modulation as well as transcription factor control (for review, see Gao and Kwaik, 2000). Our results demonstrate that the activation of Rho proteins by CNF1 plays a pivotal role in the protection against apoptosis. As reported previously (Doye et al., 2002), CNF1 causes activation and subsequent degradation of Rho, Rac, and Cdc42 with a dynamic that varies between cell types. In HEp-2 cells, this is represented by activation of all the three proteins and the subsequent proteasomal degradation of Rho and Cdc42 (Doye et al., 2002, 2006; Boyer et al., 2006), which renders Rac1 the prevalent form of Rho GTPase activated. We herein show that in HEp-2 cells, deactivation of Rho and the presence of Rac1-GTP are necessary to achieve cell death protection. In fact, the ability of CNF1 to protect from apoptosis was abrogated by transfecting active dominant RhoA (RhoAV14) into cells as well as by preincubating cells with the proteasomal inhibitor lactacystin that impairs Rho degradation and forces cells to keep high level of activated RhoA. In keeping, transfection with siRNA for Rac1 renders cells unable to be protected by CNF1. It is well known that Rho family members antagonize each other (Sander et al., 1998; Sander et al., 1999), thus it is conceivable that degradation of Rho (and of Cdc42) shifts the dynamic balance of the three GTPases in favor of activated Rac. Rho GTPases have a role in a number of cellular processes such as cytoskeleton remodeling, transcriptional activation, growth control, metastasis, development, and apoptosis. In particular, several groups have indicated Rac as either positive or negative player in the apoptotic program, depending on the cell line investigated and on the stimulus applied (Nishida et al., 1999; Embade et al., 2000; Deshpande et al., 2000; Pervaiz et al., 2001; Harrington et al., 2002; Murga et al., 2002; Zhang et al., 2004). However, the majority of the reports ascribe a protective role to active Rac1. Importantly, CNF1-induced Rac activation protects from apoptosis by stimulating the survival pathway PI3K/Akt/IKK/NF-κB and leads to the production of the antiapoptotic protein Bcl-2. Although Rac proteins are usually reported to act downstream from PI3Ks, it is now well established that Rac itself can enhance the activity of PI3K (Bokoch et al., 1996), in turn activating Akt. Akt signaling inactivates several proapoptotic factors, but it also activates transcription factors that up-regulate antiapoptotic genes. Among the others, Akt activates the IKK to phosphorylate IκBα, leading to its proteasomal degradation and NF-κB nuclear localization (Romashkova and Makarov, 1999; Hennessy et al., 2005). We have already reported the ability of CNF1 to activate NF-κB in a Rac-dependent manner (Boyer et al., 2004). Now, we add the novel finding that CNF1, through the PI3K/Akt pathway, activates the classical pathway of NF-κB, which involves IKK and IκBα (for review, see Hayden and Ghosh, 2004) and that this pathway is responsible for protection against apoptosis. This is in line with a role for IKK as a link between inflammation and cancer, as highlighted recently (Greten et al., 2004), strongly suggesting a possible role of CNF1-producing E. coli in tumor development.
The antiapoptotic protein Bcl-2 is one of the main players in the CNF1-induced protection from apoptosis. We have previously reported that CNF1 increases the amount of both antiapoptotic proteins Bcl-2 and Bcl-XL (Fiorentini et al., 1998), but, in the present work, we found that CNF1-induced protection from apoptosis was dependent on the Bcl-2 protein only. This is in keeping with literature data showing that Bcl-2 and Bcl-XL, although sharing a high degree of homology, can be regulated by different signals and can display distinct anti-cell death functions (Grad et al., 2000; Kim, 2005). For example, the increased expression of Bcl-XL, but not Bcl-2, suppresses tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in some tumor cells (Keogh et al., 2000). More importantly, Bcl-2 (and not Bcl-XL) has been implied in the control of mitochondrial morphology (Kong et al., 2005). In particular, overexpression of Bcl-2 is related to larger mitochondria and larger matrix volumes, which may develop from an increase in crystal folds, resulting in resistance to cytochrome c loss (Kowaltowski et al., 2002). In living cells, mitochondria continuously divide (fission) and fuse (fusion) with one another (Cereghetti and Scorrano, 2006), and Bcl-2 family members are involved in these processes, with the proapoptotic members regulating fission, and the antiapoptotic members regulating fusion of mitochondria (Delivani et al., 2006). We show herein the ability of the bacterial toxin CNF1 to induce a modification of the mitochondrial network mainly consisting in the appearance of elongated and interconnected mitochondria that spread throughout the cell body. This phenomenon is dependent on the presence of Bcl-2 protein and is controlled by the pathway Rac1/PI3K/Akt/IKK, also involved in the CNF1-dependent cell survival. As already stated, cell survival is achieved via the NF-κB activation; however, we cannot rule out the possibility that Akt is inhibiting apoptosis also via other survival pathways, such as, for example, the regulation of hexokinase–mitochondria interaction (Majewski et al., 2004).
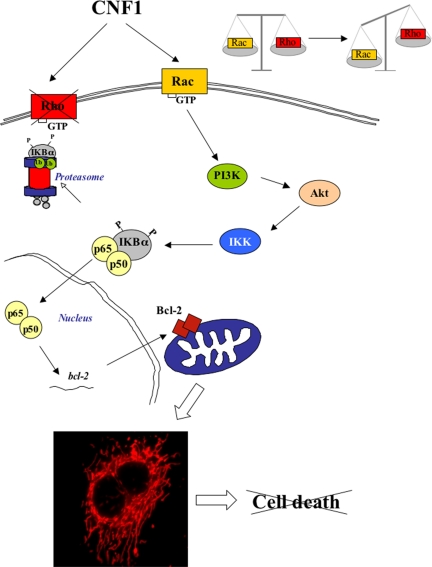
Together, we can propose the model shown in Figure 8, in which CNF1-activated Rac stimulates PI3K/Akt that in turn activates IKK and the subsequent transactivation of NF-κB. This is responsible of the overexpression of the Bcl-2 protein that favors the reshaping of the mitochondrial network and the following protective effect of CNF1 to normally deadly stimuli.
Figure 8.
Hypothetical model on how CNF1 may prevent apoptosis in HEp-2 cells. CNF1 activates Rac and Rho GTPases. The inactivation of Rho protein, which occurs after 12 h of toxin challenge, allows the prevalence of Rac that induces, in turn, the activation of the transcription factor NF-κB via the PI3K/Akt/IKK pathway. Thus, freed NF-κB is translocated into the nucleus where it activates the transcription of the Bcl-2 gene. Finally, Bcl-2 overexpression induces dramatic changes in the mitochondrial morphology, thus preventing UVB-induced cell death.
In the microbial world, a number of bacterial toxins have been reported to manipulate the host cell apoptosis, either inducing or protecting from this type of cell death (Fiorentini et al., 2003; Pirbhai et al., 2006). CNF1-producing E. coli can benefit from prolonged host cell survival because they obtain a protected niche wherein they survive (Rajalingam et al., 2001), and they form a persistent reservoir that can account for recurrent infections. In this context, the ability of CNF1 to block the cell cycle, thus slowing the removal of epithelial cells can be a further bacterial strategy to prolong tissue colonization (Falzano et al., 2006). The pathogenesis of CNF1-producing E. coli can also be reinforced by the toxin-induced release of proinflammatory cytokines from epithelial (Falzano et al., 2003) or endothelial (Munro et al., 2004) cells, thus causing an inflammatory reaction that, once chronically established, can potentially be implicated in transformation of infected tissues. Finally, because the mitochondrial remodeling is of direct relevance for the role of these organelles in cell physiology and the mitochondrial dysfunction can contribute to a number of human disorders, including cancer (Alirol and Martinou, 2006), the role of CNF1 as a factor favoring transformation is further supported by the herein reported novel findings.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Elisa Straface for providing scientific experience in the UVB irradiation experiments and criticisms.
Abbreviations used:
- CNF1
cytotoxic necrotizing factor 1
- FITC
fluorescein isothiocyanate
- IKK
IκB kinase
- NF-κB
nuclear factor-κB
- PGA1
prostaglandin A1
- PI3K
phosphoinositide 3-kinase
- TBS-T
Tris-buffered saline-Tween 20.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-10-0910) on May 16, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Alirol E., Martinou J. C. Mitochondria and cancer: is there a morphological connection? Oncogene. 2006;25:4706–4716. doi: 10.1038/sj.onc.1209600. [DOI] [PubMed] [Google Scholar]
- Baldwin A. S., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Beg A. A., Ruben S. M., Scheinman R. I., Haskill S., Rosen C. A., Baldwin A. S., Jr I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- Bokoch G. M., Vlahos C. J., Wang Y., Knaus U. G., Traynor-Kaplan A. E. Rac GTPase interacts specifically with phosphatidylinositol 3-kinase. Biochem. J. 1996;315:775–779. doi: 10.1042/bj3150775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L., Travaglione S., Falzano L., Gauthier N. C., Popoff M. R., Lemichez E., Fiorentini C., Fabbri A. Rac GTPase instructs nuclear factor-kappaB activation by conveying the SCF complex and IkBalpha to the ruffling membranes. Mol. Biol. Cell. 2004;15:1124–1133. doi: 10.1091/mbc.E03-05-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L., Turchi L., Desnues B., Doye A., Ponzio G., Mege J. L., Yamashita M., Zhang Y. E., Bertoglio J., Flatau G., Boquet P., Lemichez E. CNF1-induced ubiquitylation and proteasome destruction of activated RhoA is impaired in Smurf1−/− cells. Mol. Biol. Cell. 2006;17:2489–2497. doi: 10.1091/mbc.E05-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghetti G. M., Scorrano L. The many shape of mitochondrial death. Oncogene. 2006;25:4717–4724. doi: 10.1038/sj.onc.1209605. [DOI] [PubMed] [Google Scholar]
- Chan T. O., Rodeck U., Chan A. M., Kimmelman A. C., Rittenhouse S. E., Panayotou G., Tsichlis P. N. Small GTPases and tyrosine kinases coregulate a molecular switch in the phosphoinositide 3-kinase regulatory subunit. Cancer Cell. 2002;1:181–191. doi: 10.1016/s1535-6108(02)00033-8. [DOI] [PubMed] [Google Scholar]
- Contamin S., Galmiche A., Doye A., Flatau G., Benmerah A., Boquet P. The p21 Rho-activating toxin cytotoxic necrotizing factor 1 is endocytosed by a clathrin-independent mechanism and enters the cytosol by an acidic-dependent membrane translocation step. Mol. Biol. Cell. 2000;11:1775–1787. doi: 10.1091/mbc.11.5.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delivani P., Adrain C., Taylor R. C., Duriez P. J., Martin S. J. Role for CED-9 and Egl-1 as regulators of mitochondrial fission and fusion dynamics. Mol. Cell. 2006;21:761–773. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Deshpande S. S., Angkeow P., Huang J., Ozaki M., Irani K. Rac1 inhibits TNF-alpha-induced endothelial cell apoptosis: dual regulation by reactive oxygen species. FASEB J. 2000;14:1705–1714. doi: 10.1096/fj.99-0910com. [DOI] [PubMed] [Google Scholar]
- Doye A., Mettouchi A., Bossis G., Clement R., Buisson-Touati C., Flatau G., Gagnoux L., Piechaczyk M., Boquet P., Lemichez E. CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111:553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- Doye A., Boyer L., Mettouchi A., Lemichez E. Ubiquitin-mediated proteasomal degradation of Rho proteins by the CNF1 toxin. Methods Enzymol. 2006;406:447–456. doi: 10.1016/S0076-6879(06)06033-2. [DOI] [PubMed] [Google Scholar]
- Embade N., Valeron P. F., Aznar S., Lopez-Collazo E., Lacal J. C. Apoptosis induced by Rac GTPase correlates with induction of FasL and ceramides production. Mol. Biol. Cell. 2000;11:4347–4358. doi: 10.1091/mbc.11.12.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadeel B., Orrenius S. Apoptosis: a basic biological phenomenon with wide-ranging implications in human disease. J. Intern. Med. 2005;258:479–517. doi: 10.1111/j.1365-2796.2005.01570.x. [DOI] [PubMed] [Google Scholar]
- Falzano L., Filippini P., Travaglione S., Giamboi-Miraglia A., Fabbri A., Fiorentini C. Escherichia coli cytotoxic necrotizing factor 1 blocks cell cycle G2/M transition in uroepithelial cells. Infect. Immun. 2006;74:3765–3772. doi: 10.1128/IAI.01413-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzano L., Fiorentini C., Donelli G., Michel E., Kocks C., Cossart P., Cabanie L. M., Oswald E., Boquet P. Induction of phagocytic behavior in human epithelial cells by Escherichia coli cytotoxic necrotizing factor 1. Mol. Microbiol. 1993;9:1247–1254. doi: 10.1111/j.1365-2958.1993.tb01254.x. [DOI] [PubMed] [Google Scholar]
- Falzano L., Quaranta M. G., Travaglione S., Filippini P., Fabbri A., Viora M., Donelli G., Fiorentini C. Cytotoxic necrotizing factor 1 enhances Reactive Oxygen Species-dependent transcription and secretion of proinflammatory cytokines in human uroepithelial cells. Infect. Immun. 2003;71:4178–4181. doi: 10.1128/IAI.71.7.4178-4181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini C., Arancia G., Caprioli A., Falbo V., Ruggeri F. M., Donelli G. Cytoskeletal changes induced in HEp-2 by the cytotoxic necrotizing factor of Escherichia coli. Toxicon. 1988;26:1047–1056. doi: 10.1016/0041-0101(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Fiorentini C., Falzano L., Fabbri A., Stringaro A., Logozzi M. A., Travaglione S., Contamin S., Arancia G., Malorni W., Fais S. Activation of Rho GTPases by cytotoxic necrotizing factor 1 induces macropinocytosis and scavenging activity in epithelial cells. Mol. Biol. Cell. 2001;12:2061–2073. doi: 10.1091/mbc.12.7.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini C., Falzano L., Travaglione S., Fabbri A. Hijacking Rho GTPases by protein toxins and apoptosis: molecular strategies of pathogenic bacteria. Cell Death Differ. 2003;10:147–152. doi: 10.1038/sj.cdd.4401151. [DOI] [PubMed] [Google Scholar]
- Fiorentini C., Matarrese P., Straface E., Falzano L., Fabbri A., Donelli G., Cossarizza A., Boquet P., Malorni W. Toxin-induced activation of Rho GTP-binding protein increases Bcl-2 expression and influences mitochondrial homeostasis. Exp. Cell Res. 1998;242:341–350. doi: 10.1006/excr.1998.4057. [DOI] [PubMed] [Google Scholar]
- Flatau G., Lemichez E., Gauthier M., Chardin P., Paris S., Fiorentini C., Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- Gao L., Kwaik Y. A. Hijacking of apoptotic pathways by bacterial pathogens. Microb. Infect. 2000;2:1705–1719. doi: 10.1016/s1286-4579(00)01326-5. [DOI] [PubMed] [Google Scholar]
- Ghosh S., May M., Kopp E. B. NF-kB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Grad J. M., Zeng X. R., Boise L. H. Regulation of Bcl-xL: a little bit of this and a little bit of STAT. Curr. Opin. Oncol. 2000;12:543–549. doi: 10.1097/00001622-200011000-00006. [DOI] [PubMed] [Google Scholar]
- Greten F. R., Eckmann L., Greten T. F., Park J. M., Li Z. W., Egan L. J., Kagnoff M. F., Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Hall A. Small GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Harrington A. W., Kim J. Y., Yoon S. O. Activation of Rac GTPase by p75 is necessary for c-jun N-terminal kinase-mediated apoptosis. J. Neurosci. 2002;22:156–166. doi: 10.1523/JNEUROSCI.22-01-00156.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M. S., Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hennessy B. T., Smith D. L., Ram P. T., Lu Y., Mills G. B. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- Keogh S. A., Walczak H., Bouchier-Hayes L., Martin S. J. Failure of Bcl-2 to block cytochrome c redistribution during TRAIL-induced apoptosis. FEBS Lett. 2000;471:93–98. doi: 10.1016/s0014-5793(00)01375-2. [DOI] [PubMed] [Google Scholar]
- Kim R. Unknotting the roles of Bcl-2 and Bcl-xL in cell death. Biochem. Biophys. Res. Commun. 2005;333:336–343. doi: 10.1016/j.bbrc.2005.04.161. [DOI] [PubMed] [Google Scholar]
- Kim K. J., Chung J. W., Kim K. S. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J. Biol. Chem. 2005;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- Kong D., Xu L., Yu Y., Zhu W., Andrews D. W., Yoon Y., Kuo T. H. Regulation of Ca2+-induced permeability transition by Bcl-2 is antagonized by Drpl and hFis1. Mol. Cell. Biochem. 2005;272:187–299. doi: 10.1007/s11010-005-7323-3. [DOI] [PubMed] [Google Scholar]
- Kowaltowski A. J., Cosso R. G., Campos C. B., Fiskum G. Effect of Bcl-2 overexpression on mitochondrial structure and function. J. Biol. Chem. 2002;277:42802–42807. doi: 10.1074/jbc.M207765200. [DOI] [PubMed] [Google Scholar]
- Lacerda H. M., Pullinger G. D., Lax A. J., Rozengurt E. Cytotoxic necrotizing factor 1 from Escherichia coli and dermonecrotic toxin from Bordetella bronchiseptica induce p21(rho)-dependent tyrosine phosphorylation of focal adhesion kinase and paxillin in Swiss 3T3 cells. J. Biol. Chem. 1997;272:9587–9596. doi: 10.1074/jbc.272.14.9587. [DOI] [PubMed] [Google Scholar]
- Landraud L., Gauthier M., Fosse T., Boquet P. Frequency of Escherichia coli strains producing the cytotoxic necrotizing factor (CNF1) in nosocomial urinary tract infections. Lett. Appl. Microbiol. 2000;30:213–216. doi: 10.1046/j.1472-765x.2000.00698.x. [DOI] [PubMed] [Google Scholar]
- Lerm M., Selzer J., Hoffmeyer A., Rapp U. R., Aktories K., Schmidt G. Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor 1, activation of c-jun N-terminal kinase in HeLa cells. Infect. Immun. 1999;67:496–503. doi: 10.1128/iai.67.2.496-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski N., Nogueira V., Bhaskar P., Coy P. E., Skeen J. E., Gottlob K., Chandel N. S., Thompson C. B., Robey R. B., Hay N. Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol. Cell. 2004;16:819–830. doi: 10.1016/j.molcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Malorni W., Rivabene R., Straface E., Rainaldi G., Monti D., Salvioli S., Cossarizza A., Franceschi C. 3-Aminobenzamide protects cells from UV-B-induced apoptosis by acting on cytoskeleton and substrate adhesion. Biochem. Biophys. Res. Commun. 1995;207:715–724. doi: 10.1006/bbrc.1995.1246. [DOI] [PubMed] [Google Scholar]
- Munro P., Flatau G., Doye A., Boyer L., Oregioni O., Mege J. L., Landraud L., Lemichez E. Activation and proteasomal degradation of rho GTPases by cytotoxic necrotizing factor-1 elicit a controlled inflammatory response. J. Biol. Chem. 2004;279:35849–35857. doi: 10.1074/jbc.M401580200. [DOI] [PubMed] [Google Scholar]
- Murga C., Zohar M., Teramoto H., Gutkind J. S. Rac1 and RhoG promote cell survival by the activation of PI3K and Akt, independently of their ability to stimulate JNK and NF-kappaB. Oncogene. 2002;21:207–216. doi: 10.1038/sj.onc.1205036. [DOI] [PubMed] [Google Scholar]
- Nishida K., Kaziro Y., Satoh T. Anti-apoptotic function of Rac in hematopoietic cells. Oncogene. 1999;18:407–415. doi: 10.1038/sj.onc.1202301. [DOI] [PubMed] [Google Scholar]
- Pervaiz S., Cao J., Chao O. S., Chin Y. Y., Clement M. V. Activation of the RacGTPase inhibits apoptosis in human tumor cells. Oncogene. 2001;20:6263–6268. doi: 10.1038/sj.onc.1204840. [DOI] [PubMed] [Google Scholar]
- Pirbhai M., Dong F., Zhong Y., Pan K. Z., Zhong G. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J. Biol. Chem. 2006;281:31495–31501. doi: 10.1074/jbc.M602796200. [DOI] [PubMed] [Google Scholar]
- Rajalingam K., Al-Younes H., Muller A., Meyer T. F., Szczepek A. J., Rudel T. Epithelial cells infected with Chlamydophila pneumoniae (Chlamydia pneumoniae) are resistant to apoptosis. Infect. Immun. 2001;69:7880–7888. doi: 10.1128/IAI.69.12.7880-7888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romashkova J. A., Makarov S. S. NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature. 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- Sander E. E., ten Klooster J. P., van Delft S., van der Kammen R. A., Collard J. G. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander E. E., vanDelft S., ten Clooster J. P., Reid T., van der Cammen R. A., Michiels F., Collard J. G. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J. Cell Biol. 1998;143:1385–1398. doi: 10.1083/jcb.143.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G., Sehr P., Wilm M., Selzer J., Mann M., Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor 1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- Schmidt G., Selzer J., Lerm M., Aktories K. The Rho-deamidating cytotoxic necrotizing factor 1 from Escherichia coli possesses transglutaminase activity: cysteine 866 and histidine 881 are essential for enzyme activity. J. Biol. Chem. 1998;273:13669–13674. doi: 10.1074/jbc.273.22.13669. [DOI] [PubMed] [Google Scholar]
- Shishodia S., Aggarwal B. B. Nuclear factor-κB activation: a question of life or death. J. Biochem. Mol. Biol. 2002;35:28–40. doi: 10.5483/bmbrep.2002.35.1.028. [DOI] [PubMed] [Google Scholar]
- Travaglione S., Messina G., Fabbri A., Falzano L., Giammarioli A. M., Grossi M., Rufini S., Fiorentini C. Cytotoxic necrotizing factor 1 hinders skeletal muscle differentiation in vitro by perturbing the activation/deactivation balance of Rho GTPases. Cell Death Differ. 2005;12:78–86. doi: 10.1038/sj.cdd.4401522. [DOI] [PubMed] [Google Scholar]
- Zhang B., Zhang Y., Shacter E. Rac1 inhibits apoptosis in human lymphoma cells by stimulating Bad phosphorylation on Ser-75. Mol. Cell. Biol. 2004;24:6205–6214. doi: 10.1128/MCB.24.14.6205-6214.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.