Abstract
Cytoplasmic tRNAs have recently been found to accumulate in the nucleus during amino acid starvation in yeast. The mechanism and regulation by which tRNAs return to the nucleus are unclear. Here, we show accumulation of cytoplasmic tRNA in the nucleus also occurs during glucose starvation. Nuclear accumulation of tRNA in response to acute glucose or amino acid starvation is rapid, reversible, requires no new transcription, and is independent of the aminoacylation status of tRNA. Gradual depletion of nutrients also results in the accrual of tRNA in the nucleus. Distinct signal transduction pathways seem to be involved in the accumulation of cytoplasmic tRNA in the nucleus in response to amino acid versus glucose starvation. These findings suggest tRNA nucleocytoplasmic distribution may play a role in gene expression in response to nutritional stress.
INTRODUCTION
Export of newly transcribed tRNA from the nucleus to its site of activity in the cytoplasm is an essential process, long thought to be a unidirectional event. However, recent studies conducted with the Saccharomyces cerevisiae model system demonstrated that mature tRNA from the cytoplasm can accumulate in the nucleus in response to amino acid deprivation (Shaheen and Hopper, 2005) or when nuclear export of tRNA is defective (Shaheen and Hopper, 2005; Takano et al., 2005). The discovery of this phenomenon, referred to as “retrograde tRNA nuclear import” has led to numerous important questions concerning the significance, regulation, and mechanism of the retrograde process.
One important question concerns whether nutrient availability, in general, governs the retrograde process. In addition to affecting tRNA localization, amino acid starvation results in the repression of translation in yeast (Ashe et al., 2000; Holmes et al., 2004). Translational repression also occurs in response to glucose deprivation (Ashe et al., 2000). The similar repression of protein synthesis due to both amino acid and glucose deprivation led to the question, Does cytoplasmic tRNA accumulate in nuclei in response to glucose deprivation as well as to amino acid starvation? Alternatively, because tRNAs are coupled to amino acids, amino acid availability may uniquely affect tRNA cellular localization. As such, uncharged tRNA, in particular, may return to the nucleus under these conditions.
Another important question concerns the kinetics of cytoplasmic tRNA accumulation in the nucleus. If mature tRNA remains in the cytoplasm until the intracellular pools of nutrients are exhausted, then there may be a delay between the withdrawal of extracellular nutrients and nuclear accumulation of tRNA. Alternatively, the ability of the cell to sense and signal the loss of extracellular nutrients may lead to a rapid nuclear accrual of cytoplasmic tRNA, which in turn could define a newly discovered means by which cells control gene expression in response to nutrient availability.
The fate of cytoplasmic tRNA imported into the nucleus is another unresolved issue. Translational repression in response to glucose deprivation is coupled with the formation and increase of cytoplasmic foci containing nontranslating mRNAs called processing bodies (P-bodies) (Teixeira et al., 2005). P-bodies contain mRNA decay factors, yet they also seem to be sites of mRNA storage, because mRNAs can exit P-bodies and act in translation if conditions improve (Brengues et al., 2005). Similarly, tRNA may be recruited from the cytoplasm, stored in the nucleus under unfavorable nutrient conditions, and subsequently returned to the cytoplasm when conditions are favorable. Conversely, because two distinct pathways have been identified that degrade hypomodified tRNA species (Kadaba et al., 2004; Alexandrov et al., 2006), one of which functions in the nucleus (Kadaba et al., 2004), tRNA imported into the nucleus from the cytoplasm may be subject to degradation.
The mechanism(s) and regulation of nuclear import of cytoplasmic tRNA remains undefined. tRNA aminoacylation and the Ran pathway have been implicated in the retrograde process. Because defects in tRNA 3′ CCA formation prerequisite for aminoacylation or defects in aminoacylation per se cause nuclear accumulation of cytoplasmic tRNAs (Azad et al., 2001; Feng and Hopper, 2002; Shaheen and Hopper, 2005) and because tRNAs can be aminoacylated in the nucleus (Lund and Dahlberg, 1998; Sakar et al., 1999; Grosshans et al., 2000, Ko et al., 2000), the aminoacylation status of tRNA might provide a mechanism to regulate the distribution of tRNA between the nucleus and the cytoplasm. The role of the Ran pathway is somewhat controversial. Both Yoshihisa's group and our laboratory demonstrated that nuclear import of tRNA in yeast is an energy-dependent process; however, we differ as to whether this process is Ran dependent (Shaheen and Hopper, 2005; Takano et al., 2005). Furthermore, it is not known whether the import of cytoplasmic tRNA into the nucleus is constitutive, regulated, or both. Finally, it is unknown whether previously described signaling pathways that regulate protein synthesis in response to nutrient deprivation also function in the regulation to the tRNA retrograde process.
In this study, we begin to address several of the key questions. In addition to amino acid starvation, we found that cytoplasmic tRNA accumulates in the nucleus during glucose deprivation. Furthermore, we demonstrate that nuclear accumulation of tRNA occurs during acute or gradual starvation and that it is specific for loss of glucose as a carbon source. Amassing of mature tRNA in the nucleus is rapid, reversible, and independent of transcription. Contrary to predictions, we show that tRNA aminoacylation in the nucleus is not sufficient for tRNA reexport to the cytoplasm. Finally, investigations of the involvement of possible signal transduction pathways lead us to propose that amino acid and glucose deprivation may signal the retrograde process through separate pathways.
MATERIALS AND METHODS
Strains and Growth Conditions
The following yeast strains were used: BY4742 (MATα his3Δ1 leu2Δ lys2Δ ura3Δ), BY4741 (MATa his3Δ1 leu2Δ met15Δ ura3Δ), and reg1Δ (MATα his3Δ1 leu2Δ lys2Δ ura3Δ reg1Δ) (Open Biosystems, Huntsville, AL) (Winzeler et al., 1999); MS739 (MATα ade2-101 leu2-3, 112 ura3-52 kar1-1; provided by M. Rose, Princeton University); P2504 (MATa his3Δ1 leu2Δ met15Δ ura3Δ gcn2Δ; provided by Ralph L. Keil, Pennsylvania State College of Medicine, Hershey, PA); and SP1 (MATa his3 leu2 ura3 trp1 ade8 can1) and RS13–58A-1 (MATa his3 leu2 ura3 trp1 ade8 tpk1w1 tpk2::HIS3 tpk3::TRP1 bcy1::LEU2; both provided by David Engelberg, Hebrew University of Jerusalem, Jerusalem, Israel) (Cameron et al., 1988). Strains were grown on either standard yeast extract/peptone (YEP) or synthetic complete (SC) medium supplemented with 2% carbon source as indicated previously (Guthrie and Fink, 1991). Amino acid starvation experiments were performed with synthetic medium containing yeast nitrogenous base/ammonium sulfate/2% glucose. Carbon starvation experiments were performed with synthetic medium containing yeast nitrogenous base/ammonium sulfate/amino acids. Thiolutin was used at a final concentration of 5 μl/ml from a 1 mg/ml stock in dimethyl sulfoxide with 1 h incubation. Rapamycin was used at a final concentration of 200 ng/ml from a 1 mg/ml stock in 90% ethanol/10% Tween 20 with 1-h incubation.
Fluorescence In Situ Hybridization (FISH)
FISH was performed as described previously (Sarkar and Hopper, 1998) with the following modifications. Cells were grown in defined medium at 23°C, with prehybridization at 37°C, hybridization at 43.5°C, and 2× SSC washes performed at 50°C. Published probes for monitoring tRNATyr and tRNAGlu-D were used (Shaheen and Hopper, 2005). Probes were generated to monitor tRNAiMet (5′ TCGGTTTCGATCCGAGGACATCAGGGTTATGAGCCCTGCGCGCTTCCACTGCGCCA) and tRNACAALeu (CTTGCATCTTACGATACCTGAGCTTG). Images were acquired using a Nikon Microphot-FX microscope in conjunction with a SenSys charge-coupled device camera (Photometrics, Tucson, AZ) and QED imaging software (QED Imaging, Pittsburgh, PA). For growth curve FISH analysis, a Nikon Eclipse E1000 microscope was used in conjunction with an Orca Extended Range charge-coupled device camera (Hamamatsu, Bridgewater, NJ) for image capture and Image-Pro Plus software, version 4.1 (Media Cybernetics, Silver Spring, MD). Experiments to assess the subcellular distribution of tRNAiMet and tRNACAALeu and the role of the glucose derepression pathway used a Nikon 90i equipped with a CoolSNAP HQ2 digital camera, and MetaMorph software (Molecular Devices, Sunnyvale, CA). Images were assembled using Photoshop 6.0 (Adobe Systems, San Jose CA).
Heterokaryon Assay
Heterokaryon analysis of tRNA retrograde movement was performed as described previously (Shaheen and Hopper, 2005) by mating BY4741 with MS739 (kar1-1) with modifications to the medium to monitor effects of glucose starvation for 90 min.
RNA Isolation and Northern Blot Analysis
Nonaminoacylated tRNAs were extracted from log phase yeast cells by phenol extraction (Hopper et al., 1980). RNAs were separated by electrophoresis on a 10% polyacrylamide, pH 8.0, 8 M urea gel. Aminoacylated tRNAs were prepared from log phase yeast cells under acidic conditions (0.3 M NaOAc, pH 4.5, and 10 mM EDTA) via glass bead lysis (Sarkar et al., 1999). RNAs were separated by electrophoresis on a 10% polyacrylamide, pH 4.5, 8 M urea gel. RNAs were transferred onto Hybond N+ membrane (GE Healthcare, Little Chalfont, Buckinghamshuire, United Kingdom) and hybridized with appropriate antisense γ-32P terminally labeled probes as described previously (Wang and Hopper, 1988). The probes for tRNAIle, tRNAeMet, and tRNATyr were as described previously (Sarkar and Hopper, 1998); the probe for tRNACAALeu was the same as used for FISH analyses (see above).
RESULTS
Nuclear Import of tRNAs upon Glucose Starvation
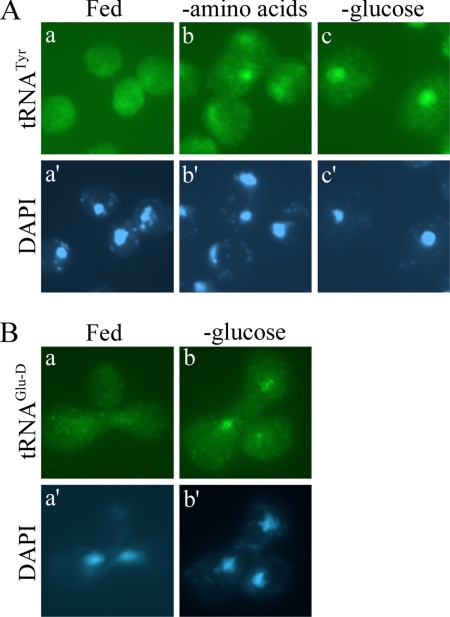
Previously, we demonstrated that nuclear accumulation of cytoplasmic mature tRNAs occurs during acute amino acid deprivation (Shaheen and Hopper, 2005). To determine whether this phenomenon is specific for amino acid deprivation or a general response to nutrient starvation, we assessed the subcellular location of tRNA during glucose starvation. Yeast cells were grown in rich medium and shifted to medium lacking glucose for 1 h, and tRNA localization was determined by FISH analysis. Similar to the response to amino acid deprivation, tRNATyr accumulated in the nucleus of cells deprived of glucose, whereas cells that remained satiated had an even distribution of tRNATyr throughout the entire cell (Figure 1A).
Figure 1.
Nuclear import of tRNA upon glucose starvation. (A) BY4742 cells were grown in SC medium, collected, and resuspended in SC (a), SC without amino acids (b), or SC without glucose (c) for 1 h. FISH analysis was performed by probing for tRNATyr. (B) Heterokaryon zygotes were generated by mating BY4741 + tRNAGlu-D and MS739 (kar1-1) in SC medium. Zygotes were incubated in SC (a) or SC −glucose (b) for 90 min followed by FISH using probes against tRNAGlu-D. 4,6-Diamidino-2-phenylindole (DAPI) staining of DNA shows the location of the nucleus for the respective cells (A, a′–c′; B, a′ and b′).
Nuclear accumulation of tRNA upon glucose deprivation could be the result of defective export of newly synthesized tRNA or the result of accumulation of tRNA that previously resided in the cytoplasm. To distinguish between these two possibilities, we used a modified heterokaryon assay in conjunction with FISH described previously (Shaheen and Hopper, 2005). Briefly, an exogenous tRNAGlu-D gene from Dictostelium discodeum was expressed from a centromere-containing plasmid in a strain harboring the kar1-1 mutation that prevents nuclear fusion in zygotes. This strain was then fused by mating to a wild-type yeast strain and shifted to medium lacking glucose for 90 min. The location of tRNAGlu-D was subsequently monitored via FISH. If glucose starvation results in movement of cytoplasmic tRNA back into the nucleus, we expected to detect tRNAGlu-D in both nuclei of the heterokaryon. However, if the nuclear accumulation of tRNA upon glucose removal is a result of an export blockage of newly synthesized tRNA, we expected to observe tRNAGlu-D in only one of the nuclei (the nucleus expressing the tRNAGlu-D gene). Our experiments show the former to be the case, because tRNAGlu-D was observed in two nuclei in these zygotes (Figure 1B). Therefore, cytoplasmic tRNA contributes to the nuclear pool of tRNA that accumulates during glucose starvation.
Rapid and Reversible Response to Glucose Removal
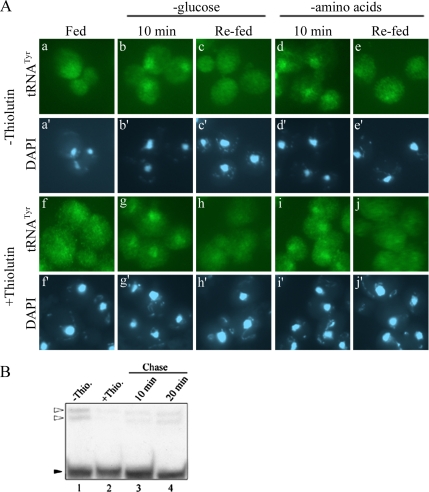
Nuclear accumulation of cytoplasmic tRNA upon glucose deprivation could be the end result of nutrient starvation or it could function in preparing cells for nutrient deprivation. For the former possibility, nuclear tRNA accumulation would likely follow other responses to deprivation such as inhibition of protein synthesis (Ashe et al., 2000) and formation of P-bodies (Teixeira et al., 2005). In the latter possibility, tRNA nuclear accumulation would be expected to precede or be concurrent with inhibition of protein synthesis and P-body formation. To distinguish between these two possibilities, we evaluated the rapidity of tRNA movement in response to glucose availability. We found tRNAs to accumulate in the nucleus within 10 min of glucose removal (Figure 2Ab), rather concurrently with the reported responses for inhibition of protein synthesis and P-body formation. Nuclear accumulation of tRNA in response to amino acid starvation was also prompt (Figure 2Ad).
Figure 2.
Rapid and reversible tRNA nuclear accumulation in response to nutrient availability. (A) FISH analysis for tRNATyr of BY4742 cells grown in SC with or without the addition of thiolutin for 1 h as indicated. Cells were resuspended in SC (a and f), SC −glucose (b, c, g, and h), or SC −amino acids (d, e, i, and j) for 10 min (no thiolutin). One half of the cultures incubated in SC −glucose (c and h) or SC −amino acids (e and j) were returned to SC for 10 min. Location of the nucleus as assessed by DAPI staining (a′–j′). (B) Northern blot analysis for tRNAIle of BY4742 cells treated with thiolutin for 1 h (lane 2) then chased with SC (lanes 3 and 4). White triangles, precursor tRNAs; black triangle, mature tRNA.
If the retrograde process is indeed a response to nutrient availability, it might be reversible. To assess this, we reintroduced nutrients to cells that had been deprived of glucose or amino acids for 10 min. We found that within 10 min of refeeding the nuclear pools of tRNA were no longer detectable (Figure 2A, c and e), perhaps indicating that imported cytoplasmic tRNA redistributed to the cytoplasm after readdition of glucose or amino acids.
To confirm that the absence of a nuclear pool of tRNATyr upon readdition of nutrients is caused by tRNA “reexport” to the cytoplasm, we tracked the localization of only “old” tRNA during nutrient starvation and replenishment by inhibiting new transcription of RNAs with thiolutin, an RNA polymerase inhibitor (Herrick et al., 1990; Takano et al., 2005). To assess whether thiolutin indeed inhibited RNA synthesis, RNAs were extracted for Northern blot analysis under conditions that mimic the 1-h incubation in the presence of thiolutin and 10- or 20-min incubation without thiolutin used for FISH. tRNAIle was monitored by Northern blot analysis, because precursor forms of this tRNA were more readily detectable than tRNATyr. Treatment with thiolutin did indeed halt synthesis of new tRNA as shown by the absence of pre-tRNA species via Northern blot analysis (Figure 2B, lane 2). Yet, this treatment is not toxic, and it is reversible as evidenced by the reappearance of newly synthesized pre-tRNA within 20 min upon removal of thiolutin (Figure 2B, lane 4).
We then located old tRNATyr in cells treated with thiolutin. Cells grown in complete medium were treated with thiolutin for 1 h followed by 10 min of starvation for either amino acids or glucose (−thiolutin). Cells were then returned to complete medium for 10 min (−thiolutin), and tRNA localization was determined by FISH. FISH analysis of cells treated with thiolutin exhibited the same tRNA nucleocytosolic distributions as cells that did not receive treatment (Figure 2A, third row). The data show that transcription of tRNA, or RNA in general, is not required for the intracellular movement of tRNA in response to nutrient levels, supporting the conclusion that the retrograde tRNA process is rapid and reversible.
Aminoacylated and Deaminoacylated tRNAs Accumulate in the Nucleus upon Nutrient Deprivation
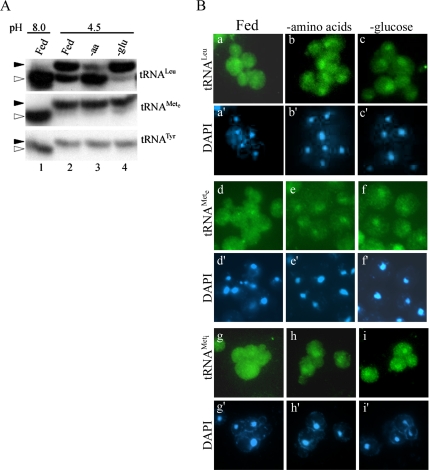
We previously proposed that uncharged tRNA generated during nutrient deprivation would move to the nucleus and remain there until conditions permit nuclear tRNA aminoacylation (Shaheen and Hopper, 2005). This hypothesis is consistent with a recent report that in permeabilized vertebrate cells only tRNA with defective 3′ termini are imported into nuclei (Zaitseva et al., 2006). To examine the in vivo aminoacylation status of tRNA in response to amino acid or glucose starvation, we extracted RNAs from acutely starved cells and performed Northern blot analysis. RNAs were isolated and analyzed at 4°C and pH 4.5 to stabilize the tRNA-aminoacyl bond. The aminoacylation status differed for various tRNA families. Our yeast strains are prototrophic for methionine and tyrosine, and we observed that nearly all of the cognate elongator tRNAMet (tRNAeMet) and tRNATyr remained charged even after 1 h of amino acid starvation (Figure 3A, lane 3). However, our strain is auxotrophic for leucine and we observe that the cognate tRNALeu became mostly uncharged after starvation for amino acids (Figure 3A, lane 3). Glucose deprivation did not result in the accumulation of uncharged tRNA for any species of tRNA examined (Figure 3A, lane 4). Regardless of the aminoacylation status, FISH revealed that both uncharged (tRNALeu) and charged (tRNAeMet) tRNA species accumulate in the nucleus during amino acid and glucose starvation (Figure 3B, a–f). Moreover, both initiator (tRNAiMet) and elongator species of tRNA (tRNAeMet and tRNALeu) accumulate in the nucleus during amino acid or glucose starvation (Figure 3B, g and h). Although we are unable to decipher whether tRNAs are aminoacylated at the time of nuclear import, these results indicate that aminoacylation of tRNA is not sufficient for tRNA nuclear reexport. Also, we do not observe tRNA degradation products by Northern blot analysis (Figure 3A), because the levels of mature tRNAs are similar comparing starved versus fed cells. This suggests that tRNAs are not rapidly degraded after accumulating in the nucleus during acute starvation.
Figure 3.
Nuclear accumulation of mature tRNA during starvation is independent of tRNA charging. (A) tRNA was isolated at pH 8 from SC grown BY4742 cells (lane 1). Cells grown in SC were also shifted to fresh SC, SC lacking amino acids, or glucose for 1 h, and tRNAs were isolated at pH 4.5 (lanes 2–4, respectively). tRNAs were subjected to acid gel Northern blot analysis, and they were probed for various tRNAs as indicated. Black triangles indicate aminoacylated tRNAs, and white triangles indicate nonaminoacylated tRNAs. (B) FISH analysis for tRNALeu, tRNAeMet, and tRNAiMet of BY4741 cells shifted to fresh SC, SC lacking amino acids, or glucose for 1 h.
Nuclear Accumulation of tRNA Occurs during Gradual Nutrient Depletion
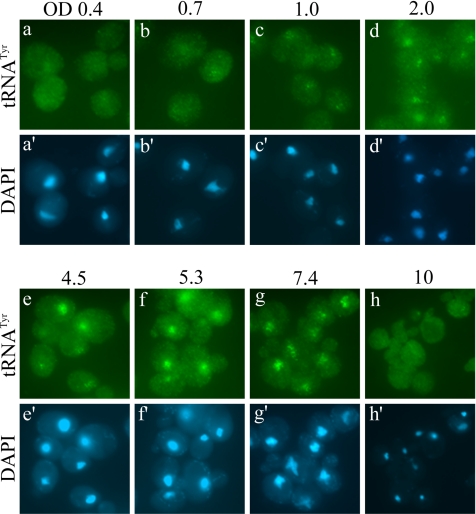
The previous experiments examined the localization of tRNA upon an acute loss of nutrients. We were also interested in whether the phenomenon of tRNA nuclear accumulation occurred during gradual nutrient deprivation. To test this, we examined the localization of tRNA in cells at various stages of a culture growth curve. We observed that cells in early- to mid-log phase of growth (OD600 0.4 and 0.7) have an even distribution of tRNA throughout the cell (Figure 4, a and b). However, as cell density increased there was a gradual increase in the nuclear accumulation of tRNA beginning around OD 1.0 (Figure 4c) that became more prominent as the culture entered late-log phase and near-stationary phase (Figure 4, d–g). Surprisingly, cells in extended stationary phase (OD600 10) no longer exhibited nuclear accumulation of tRNA, but rather had a cellular distribution of tRNA that resembles early-log phase cells (Figure 4h). Thus, tRNA nuclear accumulation occurs in response to both acute and gradual nutrient deprivation. However, cells under chronic starvation no longer accumulate tRNA in the nucleus. Possible explanations for this observation will be addressed (see Discussion).
Figure 4.
FISH analysis showing tRNATyr localization at various stages of cell growth. BY4742 cells were grown in YEPD to various densities as indicated by OD600 (OD) units followed by FISH probing against tRNATyr (a–h).
tRNA Nuclear Accumulation upon Carbon Starvation Is Specific for Glucose
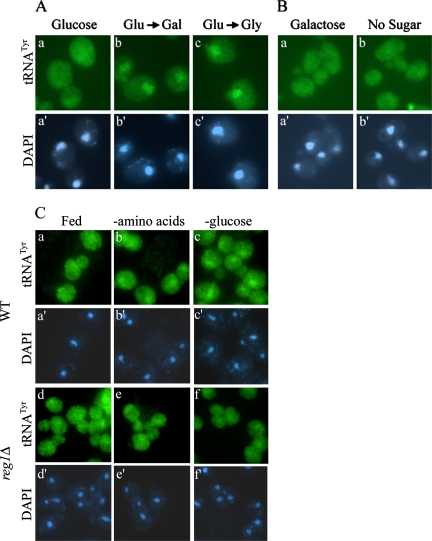
To determine whether nuclear accumulation of mature tRNA was the result of decreasing rates of metabolism or sensing the loss of particular nutrients, we examined what effect sudden changes in carbon source may have on tRNA localization. Cells grown in medium with glucose as the sole carbon source were shifted to medium containing either galactose or glycerol as the carbon source for 30 min. This sudden switch of carbon source led to nuclear accumulation of tRNA comparable to that exhibited with acute removal of all sugar (Figure 5A). However, when cells grown in galactose-containing medium were shifted to glucose-containing medium or medium lacking any carbon source for 30 min, there was no nuclear accumulation of tRNA (Figure 5B). Similarly, cells grown on glycerol did not accumulate tRNA in the nucleus in response to carbon withdrawal (Supplemental Figure S1). To rule out the possibility that cells grown in media with galactose as the carbon source are delayed in responding to carbon starvation, we shifted these cells to medium lacking sugar for 2 or 4 h. Once again, no nuclear accumulation of tRNA was observed (data not shown). Together, these results highlight the uniqueness of glucose as a carbon source, and they suggest that sensing the absence of glucose, not a general loss of extracellular carbon, specifically triggers tRNA nuclear accumulation. The data also suggest that glucose-derepressed cells fail to alter tRNA cellular distribution in response to carbon starvation.
Figure 5.
Nuclear accumulation of tRNATyr is specific for loss of glucose. (A) FISH analysis of BY4742 cells grown in SC (glucose) (a) and then shifted to SC containing galactose (b) or glycerol (c) as a carbon source for 30 min. (B) FISH analysis of BY4742 cells grown in SC galactose (a) shifted to SC without sugar (b) for 30 min. (C) FISH analysis of BY4742 (WT) and reg1Δ cells. Cells were grown in SC and shifted to SC, SC −amino acids, or SC −glucose for 1 h.
The main glucose repression/derepression pathway controls the expression of a number of genes involved in the metabolism of alternative carbon sources when glucose is lacking. The Snf1 protein kinase complex becomes activated in the absence of glucose and facilitates glucose derepression through inhibition of transcriptional repressors and modulation of a transcriptional activator (Lesage et al., 1996). Reg1 maintains the Snf1 protein kinase in its inactive form; therefore, reg1Δ results in constitutive glucose derepression (Tu and Carlson, 1995). To further examine whether tRNA localization remains unchanged during starvation in derepressed cells, we subjected a reg1Δ mutant strain to amino acid and glucose starvation. FISH analysis revealed that in contrast to wild-type cells, tRNATyr did not accumulate in the nucleus in response to amino acid or glucose starvation in reg1Δ cells (Figure 5C). This result is consistent with the data described above that glucose-derepressed cells are unaffected by loss of carbon.
Nutrient Signaling Pathways and tRNA Nuclear Accumulation
Due to the rapidity and reversibility of tRNA nuclear accumulation in response to changes in nutrient availability, we hypothesized that the regulation of the tRNA retrograde pathway was likely controlled via a signal transduction pathway(s). We focused our search on known signal transduction pathways that respond to either glucose or amino acid availability.
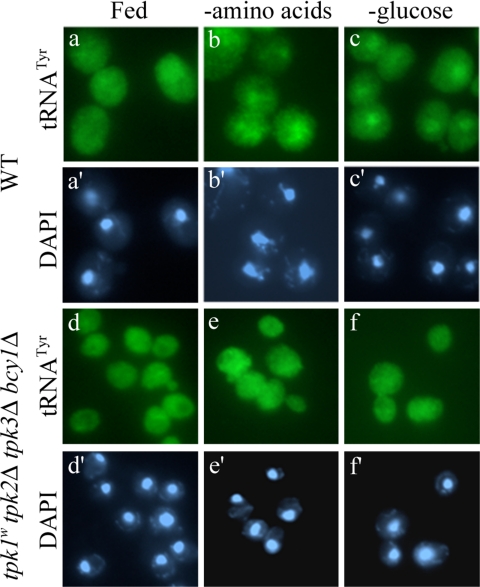
The cyclic AMP-dependent protein kinase or protein kinase A (PKA) pathway is particularly sensitive to glucose or sucrose, and it plays major roles in a variety of cellular processes, including regulating metabolism, proliferation, and stress response (Thevelein and de Winde, 1999). Nitrogen or amino acids have also been implicated in signaling through the PKA pathway (Klein and Struhl, 1994; Griffioen and Thevelein, 2002). In yeast, three genes (TPK1, TPK2, and TPK3) encode the PKA catalytic subunit with BCY1 encoding the regulatory subunit (Toda et al., 1987a,b). Although similar, these three catalytic subunits have largely distinct specificities (Robertson et al., 2000; Ptacek et al., 2005). Using the tpk1w1 (tpk1w1 tpk2Δ tpk3Δ bcy1Δ) strain (Cameron et al., 1988), which has constitutive low PKA activity, we analyzed tRNA cellular localization during amino acid or glucose starvation. Not only did we observe even distribution of tRNA in satiated tpk1w1 cells but also tRNA did not accumulate in the nucleus after amino acid or glucose starvation as it does in the parent strain (Figure 6, e and f). One possible interpretation for this result is that the PKA pathway is required for signaling tRNA nuclear accumulation when amino acids or glucose become unavailable. Alternatively, it is possible that tpk1w1 cells are resistant to acute nutrient starvation because they exist as if in a chronic state of starvation similar to stationary phase and therefore, are impervious to further nutritional stress (see Discussion).
Figure 6.
The PKA pathway is required for nuclear accumulation of tRNA in response to amino acid or glucose starvation. FISH analysis for tRNATyr of a wild-type strain (SP1 [WT]) and RS13-58A-1 [tpk1w1 tpk2Δ tpk3Δ bcy1Δ]. Cells were grown in SC and shifted to SC, SC −amino acids, or SC −glucose for 30 min.
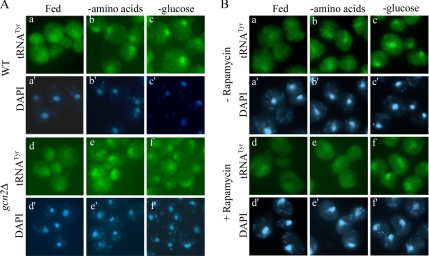
Starvation for amino acids has been shown to inhibit translation initiation through the activation of the kinase Gcn2 (Tzamarias et al., 1989; Dever et al., 1992; Hinnebusch, 1993). Activation of Gcn2 requires the binding of uncharged tRNA, and it results in the induction of the general amino acid control pathway (Wek et al., 1995; Dong et al., 2000; Qiu et al., 2001). However, because FISH analysis with a gcn2Δ mutant revealed that these cells respond to loss of nutrients (amino acid or glucose) identically to wild-type cells (Figure 7A, e and f), Gcn2 is not required for nuclear import of tRNA in response to amino acid or glucose removal.
Figure 7.
TOR kinase activity is necessary for nuclear accumulation of tRNA during amino acid starvation specifically, whereas activation of the general amino acid control is not. (A) FISH analysis for tRNATyr of BY4741 (WT) and gcn2Δ (P2504) cells shifted to fresh SC, SC −amino acids, or SC −glucose for 1 h. (B) FISH analysis for tRNATyr of BY4742 cells grown in SC with or without the addition of rapamycin for 1 h as indicated. Cells were then transferred to SC, SC −amino acids, or SC −glucose, in the presence of rapamycin where indicated (d–f), for 30 min.
The target of rapamycin (TOR) kinase responds to nutrient availability by inhibiting starvation-specific gene expression. TOR has been shown to be activated by nitrogen or carbon sources in yeast, and it is responsive to amino acids in mammalian cells (Crespo and Hall, 2002; Inoki et al., 2005). The mechanism by which TOR senses changes in nutrient availability remains poorly understood. Treatment with rapamycin inactivates TOR, inhibits protein synthesis, and is thought to mimic starvation (Barbet et al., 1996). We treated yeast cells with rapamycin for 1 h, and then we assessed cellular localization of tRNA by FISH. Surprisingly, treatment of fully fed cells with rapamycin did not mimic amino acid starvation with regard to tRNA nuclear accumulation, because tRNA remained evenly distributed throughout the cell (Figure 7Bd). However, treatment with rapamycin rendered cells unresponsive to amino acid starvation by our assay, given that tRNA did not accumulate in the nucleus (Figure 7Be). In contrast, treatment with rapamycin did not prevent nuclear accumulation of tRNA upon glucose starvation (Figure 7Bf). The results suggest that although inhibition of TOR with rapamycin does not cause nuclear accumulation of tRNA, TOR is required for signaling nuclear accretion of tRNA during acute amino acid starvation. Furthermore, the data indicate that there are distinct signal transduction pathways for prompting tRNA accumulation in the nucleus in response to amino acid versus glucose deprivation.
DISCUSSION
Nuclear accumulation of mature cytoplasmic tRNAs is not restricted to amino acid deprivation (Shaheen and Hopper, 2005), because we showed that accumulation of cytoplasmic tRNA also occurs after removal of glucose. We demonstrated that accumulation of tRNA in the nucleus upon sudden removal of amino acids or glucose occurs rapidly and that it is reversible when nutrients are restored. Neither tRNA nuclear accumulation nor reexport of tRNA to the cytoplasm requires transcription. Furthermore, tRNA that enters the nucleus is not immediately degraded. The data support a model whereby cytoplasmic tRNA is recruited into the nucleus during starvation for either storage or sequestration from the cellular translational machinery, but it may return to the cytoplasm once conditions improve.
Surprisingly, nuclear accumulation of cytoplasmic tRNA during starvation seems to be independent of the aminoacylation status of tRNA. No tRNA species we tested became deacylated upon glucose deprivation. For amino acid deprivation of the tRNAs tested, only tRNALeu became largely deaminoacylated, likely because our yeast strain is auxotrophic for leucine. We cannot rule out the possibility that only uncharged tRNAs are able to reenter the nucleus, because tRNA may become charged while in the nucleus (Sarkar et al., 1999; Grosshans et al., 2000; Ko et al., 2000; Azad et al., 2001). At the very least, we are able to conclude that both charged and uncharged tRNAs accumulate in the nucleus during amino acid or glucose starvation. Previous studies have found that aminoacylation of tRNA is important for nuclear export (Lund and Dahlberg, 1998; Sarkar et al., 1999; Azad et al., 2001). However, the data presented here indicate that the tRNA aminoacylation alone is not sufficient for nuclear reexport of imported tRNAs.
Nuclear import of tRNA during starvation may play a role in regulation of translation. Previous studies showed that acute starvation for amino acids or glucose led to rapid inhibition of translation initiation (Tzamarias et al., 1989; Ashe et al., 2000). In glucose withdrawal, the effect on translation is also rapidly reversible with the readdition of glucose (Ashe et al., 2000). The mechanism by which glucose availability affects translation is not yet known. It is possible that reducing tRNA pools from the cytoplasm may be part of this translational regulation. Our studies show that both initiator tRNAMet and elongator tRNAs amass in the nucleus during nutrient deprivation. However, inhibition of translation initiation is not always accompanied by nuclear accumulation of tRNA. Previous studies revealed that cells grown on galactose shifted to medium lacking a carbon source exhibit inhibition of translation initiation (Ashe et al., 2000). In contrast, when we shifted cells grown on galactose to medium lacking carbon, tRNA did not accumulate in the nucleus even after 4 h of starvation. Likewise, our studies of the pathways that may signal the tRNA retrograde process reinforce the conclusion that there is not a strict correlation between efficiency of protein translation and tRNA subcellular distribution. Although tpk1w1 cells maintained tRNA in the cytoplasm during starvation, the same correlation was not obtained for gcn2Δ cells. gcn2Δ cells are resistant to inhibition of translation initiation caused by severe amino acid starvation (Holmes et al., 2004), but they still accumulate tRNA in the nucleus under this condition. Therefore, it seems that sequestering significant amounts of tRNA in the nucleus does not necessarily cause global inhibition of translation. Nevertheless, nuclear accumulation of tRNA in response to starvation may play an as yet unidentified role for specific gene regulation.
Intriguingly, tRNAs are not the only RNA species that relocate during nutritional stress. Nontranslating mRNAs are rapidly recruited into cytoplasmic foci called P-bodies during acute glucose removal (Teixeira et al., 2005). Although P-bodies possess mRNA decay factors, mRNAs that enter P-bodies may be quickly released and translated when nutrients are restored (Brengues et al., 2005). Striking similarities also exist between the appearance of P-bodies and nuclear accumulation of tRNA during gradual nutrient depletion. P-bodies increase in size and number as cell densities increase (Teixeira et al., 2005) at approximately the same stages of growth as tRNA becomes increasingly nuclear. Thus, it seems as though there may be a correlation between the regulation of P-body formation and nuclear accumulation of tRNA.
We attempted to identify the particular signal transduction pathway required for nuclear accumulation of tRNA in response to amino acid or glucose deprivation by examining known nutrient-sensitive signaling pathways. Alterations of two glucose-sensitive signal transduction pathways interfered with nuclear accumulation of tRNA in response to amino acid or glucose starvation. Diminished PKA activity or constitutive activation of Snf1 rendered cells unresponsive to starvation as the cellular localization of tRNA remained unchanged. Alterations of these two pathways were previously reported to result in resistance to the inhibition of translation initiation normally observed upon glucose removal (Ashe et al., 2000). Whether either pathway is directly involved in the nuclear import of tRNA requires further study.
Alternatively, mutations in the PKA or Snf1 pathways may indirectly affect the nuclear accumulation of tRNA during starvation. The tpk1w1 mutant used in our studies exhibits a “low PKA” phenotype. This is characterized by several typical manifestations of stationary phase cells including accumulation of trehalose and glycogen, expression of stress responsive element-controlled genes, and high resistance to stress (Thevelein and de Winde, 1999). We propose that because the tpk1w1 mutant exists as if in chronic stationary phase, that it has already activated stress responses and is therefore impervious to actual nutritional stress. Consistent with this hypothesis is our finding that cells in stationary phase do not accumulation tRNA in nuclei. Stationary phase cells are generally glucose-derepressed because the last carbon sources used are ethanol and acetate after the diauxic shift. Additionally, we showed that glucose-derepressed cells grown on galactose or glycerol are impervious to the withdrawal of carbon from the medium by our tRNA localization assay. Constitutively derepressed reg1Δ cells behave similarly.
Although mutations in the PKA or Snf1 pathways affect the nuclear accumulation of tRNA in response to both amino acid and glucose starvation, inhibition of the TOR kinase pathway specifically inhibits amino acid starvation from signaling this response. Inactivation of the TOR kinase with rapamycin did not result in the nuclear accumulation of tRNA as expected, but it rendered cells impervious to the withdrawal of amino acids from the medium. Rapamycin-treated cells were still responsive to the loss of glucose by our assay. These results suggest that inhibiting TOR with rapamycin does not genuinely mimic the effect of amino acid starvation on tRNA localization and that TOR kinase activity is required for the signaling of tRNA to the nucleus in the absence amino acids. Furthermore, these results suggest that there are distinct pathways for triggering nuclear accumulation of tRNA depending on which nutrient is lacking. However, we speculate that ultimately these pathways operate via a shared mechanism for tRNA retrograde accumulation, because a possible tRNA nuclear importer, importin-β family member Mtr10, has been found to be required for nuclear accumulation of tRNA in response to amino acid (Shaheen and Hopper, 2005) and glucose deprivation (our unpublished data).
That nuclear accumulation of tRNA from the cytoplasm to the nucleus is rapid, reversible, and independent of new transcription in response to nutrient levels points to the exciting possibility that tRNA nucleocytoplasmic distribution plays a role in gene expression in response to nutritional stress. Further research is needed to understand the signaling processes that control the localization of tRNA. Additionally, our results open questions regarding the mechanism by which tRNA enters and exits the nucleus according to nutrient availability as well as whether other environmental stresses might also cause tRNA nuclear accumulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ralph Keil for yeast strains and scientific contributions, Scot Kimball for insightful discussion, and Athula Murthi and Ziqian Zhou for discussions and comments on the manuscript. We are grateful to Sergei Grigoryev for use and instruction of the Nikon Eclipse E1000 microscope and imaging system. This work was supported by a National Institutes of Health grant GM-27930 (to A.K.H.) and by an American Heart Association predoctoral fellowship (to R.L.H.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0006) on May 2, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Alexandrov A., Chernyakov I., Gu W., Hiley S. L., Hughes T. R., Grayhack E. J., Phizicky E. M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Ashe M. P., De Long S. K., Sachs A. B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad A. K., Stanford D. R., Sarkar S., Hopper A. K. Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol. Biol. Cell. 2001;12:1381–1392. doi: 10.1091/mbc.12.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N. C., Schneider U., Helliwell S. B., Stansfield I., Tuite M. F., Hall M. N. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S., Levin L., Zoller M., Wigler M. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell. 1988;53:555–566. doi: 10.1016/0092-8674(88)90572-7. [DOI] [PubMed] [Google Scholar]
- Crespo J. L., Hall M. N. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2002;66:579–591. doi: 10.1128/MMBR.66.4.579-591.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A. G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- Feng W., Hopper A. K. A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:5412–5417. doi: 10.1073/pnas.082682699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffioen G., Thevelein J. M. Molecular mechanisms controlling the localisation of protein kinase A. Curr. Genet. 2002;41:199–207. doi: 10.1007/s00294-002-0308-9. [DOI] [PubMed] [Google Scholar]
- Grosshans H., Hurt E., Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 2000;14:830–840. [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol. Microbiol. 1993;10:215–223. doi: 10.1111/j.1365-2958.1993.tb01947.x. [DOI] [PubMed] [Google Scholar]
- Holmes L. E., Campbell S. G., De Long S. K., Sachs A. B., Ashe M. P. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol. Cell Biol. 2004;24:2998–3010. doi: 10.1128/MCB.24.7.2998-3010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Schultz L. D., Shapiro R. A. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980;19:741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- Inoki K., Ouyang H., Li Y., Guan K. L. Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 2005;69:79–100. doi: 10.1128/MMBR.69.1.79-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G., Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Struhl K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell Biol. 1994;14:1920–1928. doi: 10.1128/mcb.14.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko Y. G., Kang Y. S., Kim E. K., Park S. G., Kim S. Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J. Cell Biol. 2000;149:567–574. doi: 10.1083/jcb.149.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage P., Yang X., Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol. Cell Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- Ptacek J., et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Qiu H., Dong J., Hu C., Francklyn C. S., Hinnebusch A. G. The tRNA-binding moiety in GCN2 contains a dimerization domain that interacts with the kinase domain and is required for tRNA binding and kinase activation. EMBO J. 2001;20:1425–1438. doi: 10.1093/emboj/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L. S., Causton H. C., Young R. A., Fink G. R. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA. 2000;97:5984–5988. doi: 10.1073/pnas.100113397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Azad A. K., Hopper A. K. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1999;96:14366–14371. doi: 10.1073/pnas.96.25.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Hopper A. K. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol. Biol. Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen H. H., Hopper A. K. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2005;102:11290–11295. doi: 10.1073/pnas.0503836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A., Endo T., Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005;309:140–142. doi: 10.1126/science.1113346. [DOI] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M., de Winde J. H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., Hurwitz M., Krebs E. G., Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell Biol. 1987a;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987b;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- Tu J., Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D., Roussou I., Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989;57:947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Wang S. S., Hopper A. K. Isolation of a yeast gene involved in species-specific pre-tRNA processing. Mol. Cell Biol. 1988;8:5140–5149. doi: 10.1128/mcb.8.12.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek S. A., Zhu S., Wek R. C. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zaitseva L., Myers R., Fassati A. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 2006;4:e332. doi: 10.1371/journal.pbio.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.