Abstract
Recent experiments have shown that mRNAs can move between polysomes and P-bodies, which are aggregates of nontranslating mRNAs associated with translational repressors and the mRNA decapping machinery. The transitions between polysomes and P-bodies and how the poly(A) tail and the associated poly(A) binding protein 1 (Pab1p) may affect this process are unknown. Herein, we provide evidence that poly(A)+ mRNAs can enter P-bodies in yeast. First, we show that both poly(A)− and poly(A)+ mRNA become translationally repressed during glucose deprivation, where mRNAs accumulate in P-bodies. In addition, both poly(A)+ transcripts and/or Pab1p can be detected in P-bodies during glucose deprivation and in stationary phase. Cells lacking Pab1p have enlarged P-bodies, suggesting that Pab1p plays a direct or indirect role in shifting the equilibrium of mRNAs away from P-bodies and into translation, perhaps by aiding in the assembly of a type of mRNP within P-bodies that is poised to reenter translation. Consistent with this latter possibility, we observed the translation initiation factors (eIF)4E and eIF4G in P-bodies at a low level during glucose deprivation and at high levels in stationary phase. Moreover, Pab1p exited P-bodies much faster than Dcp2p when stationary phase cells were given fresh nutrients. Together, these results suggest that polyadenylated mRNAs can enter P-bodies, and an mRNP complex including poly(A)+ mRNA, Pab1p, eIF4E, and eIF4G2 may represent a transition state during the process of mRNAs exchanging between P-bodies and translation.
INTRODUCTION
The regulation of mRNA translation and degradation is an important aspect of the control of eukaryotic gene expression. In yeast, the major pathway of mRNA degradation is initiated by shortening of the 3′ poly(A) tail (deadenylation), followed by decapping by the Dcp1/Dcp2 decapping complex, thereby exposing the transcript to a 5′-to-3′ degradation by the exonuclease Xrn1p (for review, see Coller and Parker, 2004). Decapping is a key step of this process, because it precedes and permits the degradation of the body of the mRNA and represents the site of multiple control inputs.
The processes of mRNA decapping and translation are mechanistically intertwined and seem to compete with each other, at least in yeast (for review, see Coller and Parker, 2004). For example, decreasing translation initiation by a variety of means increases the rate of mRNA decapping (LaGrandeur and Parker, 1999; Muhlrad and Parker, 1999; Schwartz and Parker, 1999). Conversely, an inhibition of translation elongation leads to a significant decrease in the rate of decapping (Beelman and Parker, 1994). Moreover, coimmunoprecipitation experiments suggested that before decapping, an mRNA exits translation and then assembles into a translationally repressed messenger ribonucleoprotein (mRNP) complex (Tharun and Parker, 2001).
Additional evidence for a discrete population of nontranslating mRNPs has been that nontranslating mRNAs, and the decapping machinery, accumulate in discrete cytoplasmic foci, called P-bodies (also referred as GW182 or Dcp bodies) (Ingelfinger et al., 2002; Lykke-Andersen, 2002; Sheth and Parker, 2003; Cougot et al., 2004). P-bodies have now been observed in yeast, insect cells, nematodes, and mammalian cells, and they contain various proteins involved in mRNA decay, including the decapping enzyme (Dcp1p/Dcp2p); activators of decapping Dhh1p, Pat1p, Lsm1–7p, and Edc3p; and the exonuclease Xrn1p (for review, see Anderson and Kedersha, 2006; Eulalio et al., 2007; Parker and Sheth, 2007). Moreover, P-bodies have been suggested to be functionally involved in mRNA decapping (Sheth and Parker, 2003; Cougot et al., 2004), nonsense-mediated decay (Unterholzner and Izaurralde, 2004; Sheth and Parker, 2006), mRNA storage (Brengues et al., 2005; Bhattacharyya et al., 2006), general translation repression (Holmes et al., 2004; Coller and Parker, 2005), microRNA-mediated repression (Jakymiw et al., 2005; Liu et al., 2005; Pillai et al., 2005), and possibly viral packaging (Beliakova-Bethell et al., 2006). The existence of P-bodies as a discrete cytoplasmic compartment containing untranslating mRNAs suggests that understanding the movement of eukaryotic mRNAs between different cytoplasmic compartments will be important in understanding the control of mRNA translation and degradation.
Nontranslating mRNAs can also accumulate in another cytoplasmic structure termed a stress granule (SG). SGs are formed in response to stress that leads to the phosphorylation of eukaryotic initiation factor (eIF)2α (Kedersha et al., 1999), but they also can form in response to defects in eIF4A or eIF4G function (Dang et al., 2006; Mazroui et al., 2006). Stress granules have not yet been described in Saccharomyces cerevisiae, although they have been described in Schizosaccharomyces pombe (Dunand-Sauthier et al., 2002). Stress granules contain most of the 48S preinitiation complex [e.g., eIF3, eIF4E, eIF4G, and poly(A) binding protein], the RNA binding proteins TIA-1 and TIAR, and poly(A)+ RNA (for review, see Anderson and Kedersha, 2006). Stress granules and P-bodies seem to interact with each other (Kedersha et al., 2005; Wilczynska et al., 2005), although the diversity of different mRNP types and the mechanisms that mediate transitions between stress granules, P-bodies, and polysome-bound mRNAs remain to be established.
The poly(A) tail of mRNA has an important role in mRNA degradation and translation. Removal of the poly(A) tail generally precedes degradation of the mRNA and when deadenylation is inhibited, mRNA are stabilized (Decker and Parker, 1993; Tucker et al., 2001). The poly(A) binding protein 1 (Pab1p) is highly conserved in eukaryotes and its function is essential for viability (Sachs et al., 1987). Pab1p interacts with the translation initiation factors eIF4E and eIF4G, and thereby it is thought to stimulate translation (Jacobson and Peltz, 1996; Tarun and Sachs, 1996; Wells et al., 1998; Gray et al., 2000). Pab1p also plays a role in translation termination via its interaction with eukaryotic release factor 3 (eRF3), suggesting that Pab1p participates in translation in multiple ways (Cosson et al., 2002). Pab1p also participates in the control of mRNA degradation by impeding mRNA decapping until deadenylation is completed (Caponigro and Parker, 1995). An important and unresolved issue is whether the poly(A) tail and the associated poly(A) binding protein Pab1p affect the distribution of mRNAs between P-bodies and polysomes.
In this work, we examine the contribution of the poly(A) tail in controlling the movement of mRNA entering or exiting P-bodies. Our results indicate that poly(A)+ mRNAs can be present in P-bodies under certain conditions. Moreover, the poly(A) binding protein Pab1p is also found in P-bodies in the same conditions. Strains lacking Pab1p show increased P-bodies, suggesting that Pab1p functions directly or indirectly to promote the exit of mRNAs from P-bodies. In addition to Pab1p, the translation initiation factors eIF4E and eIF4G2 can be observed in P-bodies. This suggests the existence of an mRNP complex in P-bodies containing poly(A)+ mRNAs, Pab1p, eIF4E, and eIF4G2, yet lacking other translation initiation components. These results argue that polyadenylated mRNAs can enter P-bodies, and an mRNP complex including poly(A)+ mRNA, Pab1p, eIF4E, and eIF4G2 may represent a transition state during the process of mRNAs exchanging between P-bodies and translation.
MATERIALS AND METHODS
Yeast Strain and Growth Conditions
The genotypes of all strains used in this study are listed in Table 1. Strains expressing proteins C-terminally tagged with green fluorescent protein (GFP) were obtained either from genomic libraries or constructed (Table 1) following the polymerase chain reaction (PCR)-based gene modification method described previously (Longtine et al., 1998). All the constructions were confirmed to be full length and functional (Sheth and Parker, 2003; data not shown). Strains were grown on either standard yeast extract/peptone medium (YP) or synthetic complete medium (SC) supplemented with appropriate amino acids and 2% dextrose (Glu) as carbon source. Strains were grown at 30°C except for temperature-sensitive mutants, which were grown at 23°C and shifted for 1 h at 37 or 39°C to inactivate the mutant allele. For stationary phase (SP) experiments, cells were grown four days in YPGlu or first grown for one day in SCGlu and then three more days in YPGlu to reach stationary phase. For the strain expressing MFA2pG reporter mRNA under the control of the Tet-Off promoter (pRP1192), transcription was blocked by using doxycycline (Sigma-Aldrich, St. Louis, MO) at a final concentration of 2 μg/ml. Yeast strains were transformed by standard techniques, and plasmids were maintained by growth in selective media.
Table 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| yRP840 | MATa leu2–3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG | Hatfield et al. (1996) |
| yRP923 | MATa leu2-3,112 trp1 ura3-52 his4-539 lys2-201 sbp2::URA3 pab1::URA3 | Caponigro and Parker (1995) |
| yRP924 | MATa leu2-3,112 trp1 ura3-52 his4-539 lys2-201 sbp2::URA3 | Caponigro and Parker (1995) |
| yRP1069 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA | Beelman et al. (1996) |
| yRP1131 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG pat1-2 pab1::URA3 | Hatfield et al. (1996) |
| yRP1134 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG pat1-2 | Hatfield et al. (1996) |
| yRP1199 | MATa leu2-3,112 trp1-D1 ura3-52 his4-539 xrn1::URA3 | Anderson and Parker (1998) |
| yRP1321 | MATa leu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [cdc33-42/TRP1] | Schwartz and Parker (1999) |
| yRP1326 | MATa leu2 prt1-63 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG | Schwartz and Parker (1999) |
| yRP1327 | MATa leu2 trp1 ura3 cdc33::LEU2 prt1-63 cup1::LEU2/PGK1pG/MFA2pG [cdc33-42/TRP1] | Schwartz and Parker (1999) |
| yRP1346 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG lys2-201 dcp2::TRP1 | Dunckley and Parker (1999) |
| yRP1365 | MATa trp1 ura3-52 leu2-3,112 lys2-201 cup1::LEU2/PGK1pG/MFA2pG lsm1::TRP1 | Tharun et al. (2000) |
| yRP1372 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG lys2-201 pat1::LEU2 | Tharun et al. (2000) |
| yRP1529 | MATa leu2 trp1 ade2 ura3 his3 lys2 cup1::LEU2/PGK1pG/MFA2pG sbp1::NEO | This study |
| yRP1561 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG lys2-201 dhh1::URA3 | Coller et al. (2001) |
| yRP1616 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG ccr4::NEO | Tucker et al. (2001) |
| yRP1617 | MATa leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG caf1::URA3 | Tucker et al. (2001) |
| yRP1727 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG DCP2-GFP (NEO) | Sheth and Parker (2003) |
| yRP1745 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG edc3::NEO | Kshirsagar and Parker (2004) |
| yRP1943 | MATa leu2 ura3 his3 met15 EIF1A-GFP (HIS) | Huh et al. (2003) |
| yRP1944 | MATa leu2 ura3 his3 met15 TIF35-GFP (HIS) | Huh et al. (2003) |
| yRP1945 | MATa leu2 ura3 his3 met15 EIF4B-GFP (HIS) | Huh et al. (2003) |
| yRP1946 | MATa leu2 ura3 his3 met15 EIF4E-GFP (HIS) | Huh et al. (2003) |
| yRP1947 | MATa leu2 ura3 his3 met15 EIF4G1-GFP (HIS) | Huh et al. (2003) |
| yRP1949 | MATa leu2 ura3 his3 met15 EIF5-GFP (HIS) | Huh et al. (2003) |
| yRP1950 | MATa leu2 ura3 his3 met15 EIF5B-GFP (HIS) | Huh et al. (2003) |
| yRP1951 | MATa leu2 ura3 his3 met15 EIF6-GFP (HIS) | Huh et al. (2003) |
| yRP1952 | MATa leu2 ura3 his3 met15 RPG1-GFP (HIS) | Huh et al. (2003) |
| yRP1953 | MATa leu2 ura3 his3 met15 NIP-GFP (HIS) | Huh et al. (2003) |
| yRP2190 | MATa leu2 ura3 his3 met15 PRT1-GFP (HIS) | Huh et al. (2003) |
| yRP2191 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG PAB1-GFP (NEO) | This study |
| yRP2384 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG EIF4G2-GFP (NEO) | This study |
Preparation of Cells for Confocal Microscopy
Cells were grown to an OD600 of 0.3–0.35 in the appropriate medium and prepared for microscopy. For observation, cells were washed twice with SC plus amino acids supplemented with glucose and resuspended in the same medium. For glucose depletion conditions, cells were washed in YP or SC supplemented with appropriate amino acids without Glu and resuspended in the same medium for 10 min before being collected. Cells were then washed with SC without Glu and resuspended in the same medium. For temperature shift experiments, cultures were shifted 1 h at the indicated temperature in SC medium plus glucose or 50 min in SC medium plus a 10-min shift at the same temperature in SC medium without glucose. Cells were washed in the same medium and analyzed. For stationary phase experiments, the culture was washed and resuspended in SC with no Glu before observation. Observations were made using a Nikon PCM 2000 confocal microscope by using 100× objective with 3× zoom using Compix imaging software (Compix, Sewickley, PA). All images are a Z-series compilation of 6–10 images, except for the colocalization experiments, which are made from a single Z-plane image.
Fluorescence In Situ Hybridization (FISH) of Poly(A)+ RNA
The subcellular distribution of poly(A)+ RNA was examined by in situ hybridization by using a digoxigenin-conjugated oligo(dT) probe and indirect immunofluorescence microscopy as described previously (Marfatia et al., 2003) with the following modifications. Cells were fixed with 4% Formalin solution for 15 min at room temperature. Oligo(dT)50 (MWG Biotech, High Point, NC) was 3′ end labeled with digoxigenin (Roche Applied Science), and rhodamine-conjugated anti-digoxigenin antibody (1:200 dilution; Roche Applied Science, Indianapolis, IN) was used to visualize poly(A)+ RNA in cells grown to mid-log phase.
Plasmids
The URA3 and TRP1 PAB1-GFP plasmids pRP1362 and pRP1363 were created by PCR amplification (oRP1319: 5′-CTGTATGAAGCCACAAAGCATCTAGATCAATCATG-3′; oRP1320: 5′-CTAGCGGATCTGCCTCTAGAGGTGTGGT-3′) from yRP2191 (PAB1-GFP) and ligation (XbaI) into pRP1304 (TRP1) or pRP1305 (URA3).
Polysome Analysis
Polysome analyses were performed as a modification of the protocol described previously (Kuhn et al., 2001). Cell lysates were prepared from 200 ml of exponential growth culture (0.3 OD600). Cycloheximide (CYH) was added at the time of harvest to a final concentration of 100 μg/ml in the presence of ice. Cells were centrifuged at 4000 rpm for 5 min at 4°C, washed in 20 ml of lysis buffer (20 mM Tris-HCl, pH,8, 140 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 1% Triton X-100, 0.1 mg/ml CYH, and 1 mg/ml heparin), and frozen in liquid nitrogen. The pellet was resuspended in 400 μl of lysis buffer. A 400-μl volume of glass beads was added, and the cell suspension was vortexed at full speed for 2 min followed by incubation on ice for 2 min, for a total of three times. Excess cells debris and glass beads were removed by centrifugation for 2 min at 4000 rpm at 4°C. Approximately 20 A254 units were loaded on a 15–50% sucrose gradient suspended in lysis buffer lacking Triton X-100. Samples were separated using a Beckman SW41 rotor at 4°C for 2.5 h at 39,000 rpm and collected, while using A254 value to monitor the fractionation.
RNA Analysis
RNA was extracted from polysome fractions (900 μl) by vortexing for 2 min at maximum speed in the presence of an equal volume of phenol/CHCl3/LET (LET: 100 mM LiCl, 25 mM Tris-HCl, pH 8, and 20 mM EDTA). The aqueous phase was extracted with an equal volume of CHCl3, and RNA was recovered by precipitation with 2 volumes of 100% ethanol (EtOH) and 1/10 volume of 3 M sodium acetate, pH 5.2. Pellets were washed in 75% EtOH and resuspended in diethyl pyrocarbonate-treated H2O. RNA was separated by agarose gel electrophoresis and transferred to nylon membrane (Whatman Schleicher and Schuell, Keene, NH). RNA was detected using radiolabeled oligonucleotide probes directed against the MFA2pG mRNA reporter (oRP140) and RPL41A mRNA (oRP1249: 5′-TTAGAGTTATTTACTCATAATCCGC-3′). mRNAs were quantified by PhosphorImager analysis (Typhoon 9410; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
RESULTS
Both Poly(A)+ and Poly(A)− mRNAs Can Be Translationally Repressed during Glucose Deprivation
To determine any contribution of the poly(A) tail and Pab1p with regard to mRNAs being targeted to P-bodies, we analyzed the control of translation during glucose deprivation. During glucose deprivation, translation is repressed and untranslated mRNAs accumulate in P-bodies (Teixeira et al., 2005; Ashe et al., 2000). This raises two possibilities with regard to the function of the poly(A) during glucose deprivation. First, polyadenylated mRNAs might be resistant to translation repression under these conditions and only deadenylated mRNAs are translationally repressed and targeted to P-bodies. Alternatively, translation repression could function on both adenylated and deadenylated mRNAs. To distinguish between these two possibilities, we used sucrose gradients to separate the translating and untranslating populations of mRNAs, and then we examined the poly(A) lengths of reporter mRNAs before and after glucose deprivation.
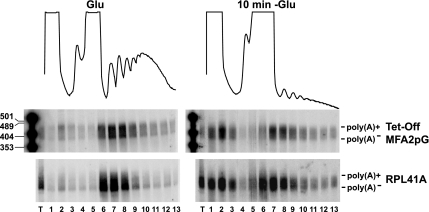
As shown previously (Ashe et al., 2000; Brengues et al., 2005), glucose deprivation leads to a rapid loss of polysomes, and movement of the MFA2P-U1A reporter mRNA, as well as the endogenous RPL41A mRNA, from the polysome region of the gradient to the mRNP fractions (where P-body components run under these conditions). An important result was that the distribution of the poly(A) tail lengths on the MFA2P-U1A and RPL41A mRNAs were identical in the mRNP fraction and in the fractions associated with polysomes (Figure 1). This result indicates that both polyadenylated and oligoadenylated mRNAs are translationally repressed to the same extent. Moreover, because translation repression under these conditions requires components of P-bodies (Holmes et al., 2004; Coller and Parker, 2005), this result implies that poly(A)+ mRNAs as well as poly(A)− mRNAs are entering P-bodies.
Figure 1.
poly(A)+ and poly(A)− mRNAs can be translationally repressed. WT cells expressing the plasmid MFA2pG under the control of a tetracycline promoter (Tet-Off MFA2pG) were grown in SC medium plus Glu, shifted for 10 min to SC with no glucose containing doxycycline (10 min −Glu). Polysomes profiles of the collected sucrose gradients are shown. Fraction 1 corresponds to the top of the gradient. Northern blots of polyacrylamide gels for the indicated mRNA are shown.
Polyadenylated mRNAs Colocalize with P-Bodies during Glucose Deprivation
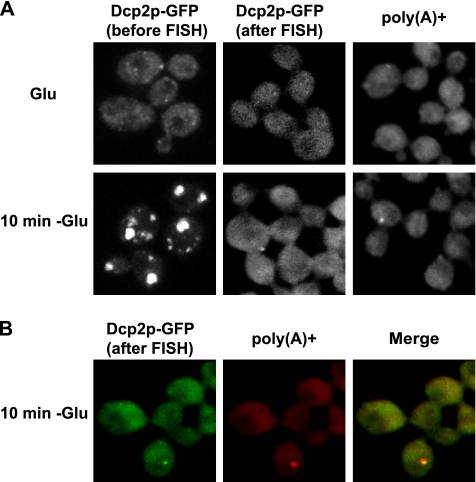
To directly examine whether adenylated mRNAs accumulate in P-bodies during glucose deprivation, we used FISH with an oligo(dT) probe to determine whether poly(A)+ mRNA could be detected in P-bodies. When starting this experiment, we noted that no report had been made of poly(A)+ granules in yeast cytoplasm, despite numerous analyses of poly(A)+ mRNA in yeast nuclei. This suggested that yeast P-bodies might be sensitive to the conditions used for in situ hybridization. To test this possibility, we first examined how the integrity of P-bodies survived in situ hybridization by using a GFP-tagged Dcp2p as a P-body marker. We observed that under standard in situ conditions, no P-bodies survived to the end of the in situ procedure, although P-bodies could be observed immediately after cell fixation (data not shown). However, by modification of the protocol we were able to develop a method where at least some of the P-bodies survived the in situ process, although there was still a 10–20× reduction in the number of cells with P-bodies (Figure 2A, after FISH), in comparison with fixed cells (Figure 2A, before FISH). That Dcp2p foci decline dramatically indicates that yeast P-bodies are sensitive to the FISH procedure. Nevertheless, because some P-bodies could survive the process, we were able to determine whether they contained poly(A)+ mRNAs.
Figure 2.
poly(A)+ mRNAs colocalize with P-bodies. WT cells expressing a GFP-tagged version of Dcp2p were grown in glucose containing medium (Glu) (A), shifted for 10 min without glucose (10 min −Glu) (A and B). Poly(A)+ RNA was visualized by oligo(dT) hybridization and indirect immunofluorescence. (B) Colocalization of poly(A)+ mRNAs with Dcp2-GFP. Left, Dcp2-GFP. Middle, poly(A)+. Right, merge generated by Adobe Photoshop (Adobe Systems, Mountain View, CA).
Using our modified procedure, an important result was that during glucose deprivation, poly(A)+ RNA was concentrated in small foci in ∼5% of the cells with an average of one to two foci per cell (Figure 2A), which is similar to the number of P-bodies that survive the in situ procedure as assessed by monitoring Dcp2-GFP. Moreover, these poly(A)+ foci colocalized with the Dcp2p-GFP foci that remained after the in situ procedure (Figure 2B). This observation indicates that polyadenylated mRNA are present in P-bodies during glucose deprivation.
Pab1p Can Be Present in P-Bodies
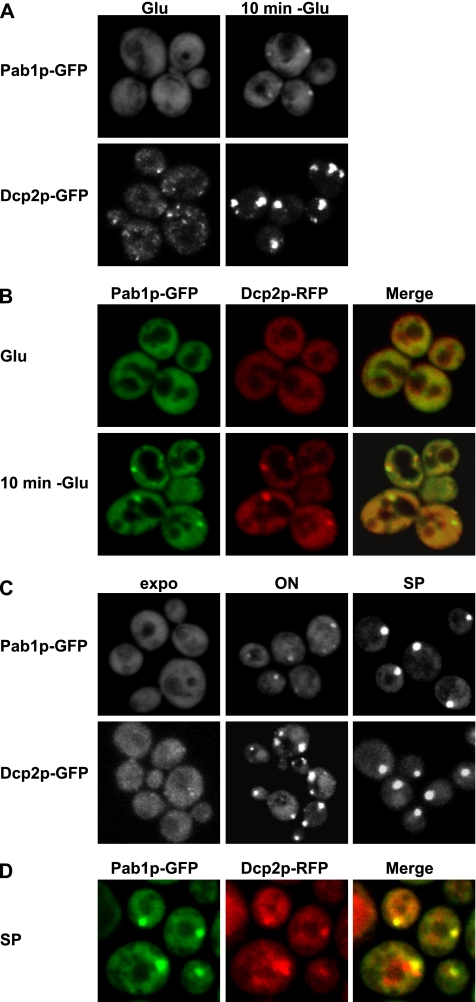
The presence of poly(A)+ mRNA in P-bodies led us to determine whether the poly(A) binding protein Pab1p could also be observed in P-bodies. To do this, we followed the distribution of Pab1p-GFP before and after glucose depletion as well as in stationary phase, where P-bodies are large and contain mRNAs that can reenter translation once growth resumes (Brengues et al., 2005; Teixeira et al., 2005). We observed that after glucose depletion, a small amount of Pab1p localizes in small foci (Figure 3A), which colocalized with Dcp2-red fluorescent protein (RFP) (Figure 3B). It should be noted that although Dcp2p is robustly in P-bodies under these conditions, only a small percentage of the Pab1p was detected in P-bodies with the majority being diffuse in the cytoplasm. This suggests that Pab1p is associated with a smaller subset of mRNAs localized to P-bodies compared with Dcp2p.
Figure 3.
The poly(A) binding protein Pab1p is present in P-bodies under stress. WT cells expressing a GFP-tagged version of Dcp2p or Pab1p (A) or coexpressing a GFP-tagged version of Pab1p and a RFP-tagged version of Dcp2p (B) were grown in glucose-containing medium (Glu), shifted for 10 min without glucose (10 min −Glu). (B) Colocalization of Pab1p-GFP with Dcp2-RFP. Left, Pab1p-GFP. Middle, Dcp2p-RFP. Right, merge generated by Adobe Photoshop. (C) Cells expressing a GFP-tagged version of Dcp2p or Pab1p were observed at different stages of cellular growth, as described. (D) Colocalization of Pab1p-GFP with Dcp2-RFP during stationary phase.
We also observed that in stationary phase (4 d in culture), Pab1p-GFP was highly concentrated in foci, in a manner similar to Dcp2p (Figure 3C, right). However, the accumulation of Pab1p in foci at higher cell densities was delayed compared with Dcp2p. Specifically, Dcp2p is highly concentrated in large P-bodies after overnight growth of a culture (Figure 3D, middle). In contrast, at a similar time Pab1p is still somewhat diffuse and is only present in smaller foci after overnight growth (Figure 3C, middle), but it becomes highly concentrated in large foci as the cells reach stationary phase.
To determine whether these Pab1p foci in stationary phase also colocalize with P-bodies, we examined their subcellular distribution relative to Dcp2p-RFP. We observed that the majority of Pab1p foci in stationary phase colocalized with P-bodies (Figure 3D), although in some cases the overlap was incomplete or the Pab1p-GFP foci was adjacent to the Dcp2p-RFP foci (Figure 3D). These results indicate that Pab1p foci in stationary phase colocalize with P-bodies similar to what we observed during glucose deprivation. However, the presence in stationary phase of some Pab1p foci that only partially overlap with Dcp2p suggests that these Pab1p foci may, at least in some cases, represent accumulations of mRNPs slightly different from those in P-bodies.
Pab1p Can Affect the Distribution of P-Bodies
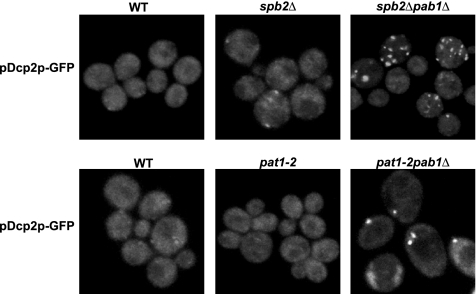
Because Pab1p functions to enhance mRNA translation, the presence of Pab1p in P-bodies suggested that Pab1p might play a role in associating with mRNAs and in promoting their exit from P-bodies to allow entry into translation. To examine this possibility, we determined whether loss of Pab1p from cells altered P-bodies. Because the PAB1 gene is essential for cell viability, we examined pab1Δ strains that harbor a bypass suppressor of the pab1 deletion. First, we used the spb2Δ strain. The SPB2 gene encodes the Rpl46 subunit of the 60S ribosome, and its depletion suppresses the pab1Δ lethality by an unknown mechanism (Sachs and Davis, 1990). Using Dcp2p-GFP as a marker for P-bodies, comparison of the spb2Δ and the spb2Δpab1Δ strains revealed that the pab1Δ increased the number and size of P-bodies in cells during mid-log growth. Specifically, in the spb2Δ strain, Dcp2p-GFP is present in small foci in 10% of the cells (Figure 4). In contrast, in the double mutant spb2Δpab1Δ, 50% of the cells contain either one big foci or multiple small foci of Dcp2p-GFP (Figure 4). These results argue that Pab1p normally functions to reduce the number of mRNAs found in P-bodies. However, this experiment has the caveat that the spb2Δ strain alone had an increased size of P-bodies compared with wild-type cells in mid-log growth, raising the possibility that the effect of the pab1Δ was specific to the spb2Δ strain.
Figure 4.
Absence of Pab1p affects P-bodies distribution. WT, sbp2Δ, sbp2Δpab1Δ, pat1-2, and pat1-2pab1Δ cells expressing a GFP-tagged version of Dcp2p were grown in YPGlu.
To address the caveats with the spb2Δ strain, we also examined how the pab1Δ affected P-bodies using the pat1-2 allele (also referred to as mrt1-2, Hatfield et al., 1996), which is a nonsense mutation in the PAT1 gene that suppresses the lethality of the pab1Δ by an unknown mechanism (Hatfield et al., 1996; Bonnerot et al., 2000; Tharun et al., 2000). We observed that Dcp2p-GFP was not concentrated in foci in the pat1-2 strain (Figure 4). However, in the pat1-2pab1Δ strain, Dcp2-GFP concentrated in foci like in the spb2Δpab1Δ strain (Figure 4). Thus, the pab1Δ leads to an increase in P-bodies in both the spb2Δ and pat1-2 bypass suppressor strains. This suggests that Pab1p plays a direct or indirect role in shifting the equilibrium of mRNAs away from P-bodies and into translation (see Discussion).
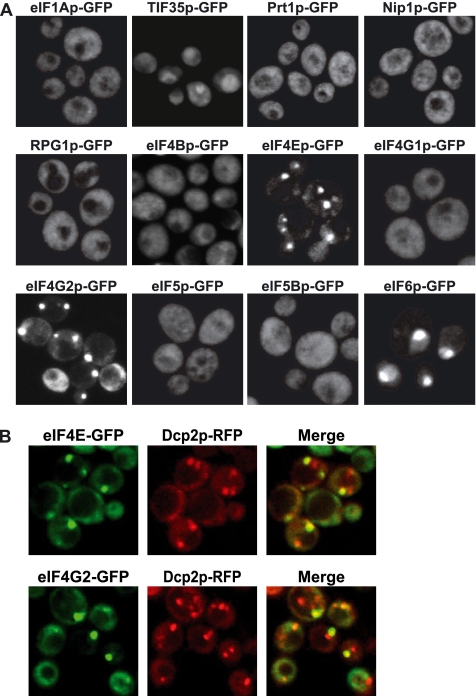
Translation Initiation Factors eIF4E, eIF4G1, and eI4G2 Can Be Seen in P-Bodies during Glucose Deprivation and Stationary Phase
The observation that Pab1p clearly accumulated in P-bodies during stationary phase, and to a small extent during glucose deprivation, suggested there might be other translation initiation factors present in P-bodies under these conditions. We have reported previously that many translation initiation factors do not accumulate in P-bodies during glucose deprivation (Brengues et al., 2005; Teixeira et al., 2005). However, we reexamined these experiments with greater awareness, because the Pab1p accumulation under glucose deprivation is not strong, and other factors with a similar localization pattern might have been missed. In addition, the robust accumulation of Pab1p in stationary phase provided an ideal way to increase any possible signal from initiation factors that might be associated with P-bodies.
We first followed the distribution of several translation factors during stationary phase, because it represents the condition where Pab1p is most easily visualized in P-bodies. Similar to what we had previously observed during glucose deprivation of mid-log cultures (Brengues et al., 2005), we observed a relatively homogenous cytoplasmic distribution of the GFP constructs for eIF1A, Tif35 (which did show some clumping), Prt1 (eIF3 subunit), Nip1 (eIF3 subunit), Rpg1 (eIF3 subunit), eIF4B, eIF5, eIF5B, and eIF4G1, and the expected nuclear localization for eIF6 (Figure 5A). However, we did observe that the cap binding protein eIF4E and its binding partner eIF4G2 were clearly in foci. The accumulation of eIF4G2 but not eIF4G1 may simply be due to the increased expression of eIF4G2 at high cell density compared with eIF4G1 expression (http://genome-www.stanford.edu/). Similar to our results with Pab1p in stationary phase, we observed that the eIF4E and eIF4G2 foci in stationary phase colocalize with the P-body marker Dcp2p, although in some cases the overlap is again not complete (Figure 5B). These results indicate that eIF4E and eIF4G2 foci in stationary phase generally colocalize with P-bodies.
Figure 5.
eIF4E and eIF4G2 are present in P-bodies during stationary phase. (A) Cells were grown in YPGlu (Glu) for 4 d. Cells expressing GFP-tagged versions of eIF1Ap, TIF35p, Prt1p, Nip1p, RPG1p, eIF4Bp, eIF4Ep, eIF4G1p, eIF4G2p, eIF5p eIF5Bp, and eIF6p are shown. (B) Colocalization of eIF4E-GFP and eIF4G2-GFP with Dcp2-RFP.
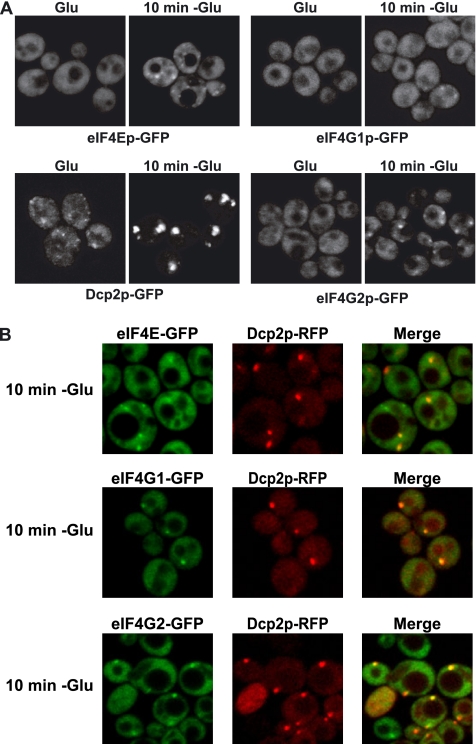
The presence of eIF4E and eIF4G2 in P-bodies in stationary phase led us to reexamine the localization of all the translation factors in glucose deprivation during exponential growth. Again most factors were not seen in P-bodies (data not shown), although low levels of eIF4E, eIF4G2, and some eIF4G1 could be seen in foci (Figure 6A). Moreover, these eIF4E, eIF4G2, and eIF4G1 foci essentially all colocalized with Dcp2p-RFP (Figure 5B), which is similar to the results seen with Pab1p (Figure 3D). These results indicate that low levels of Pab1p, eIF4E, and eIF4G can be present in P-bodies during glucose deprivation. Because these three proteins are known to interact and enhance translation rates, the simplest interpretation is that an mRNP bound by these factors can accumulate in or near P-bodies and that it might be an intermediate in the exchange between polysomes and P-bodies (see Discussion). However, we cannot rule out the possibility that eIF4E, eIF4G, and Pab1p are each individually associating with a different subset of mRNAs in/near P-bodies.
Figure 6.
eIF4E, eIF4G1, and eIF4G2 are present in P-bodies under stress. (A) Cells were grown in YPGlu (Glu) and then shifted in YP without Glu for 10 min (10 min −Glu). Cells expressing GFP-tagged versions of eIF4Ep, eIF4G1p, eIF4G2p, and Dcp2p are shown. (B) Colocalization of eIF4E-GFP, eIF4G1-GFP, and eIF4G2-GFP with Dcp2-RFP under stress.
Differential Effects of Factors on Pab1p Accumulation in P-Bodies
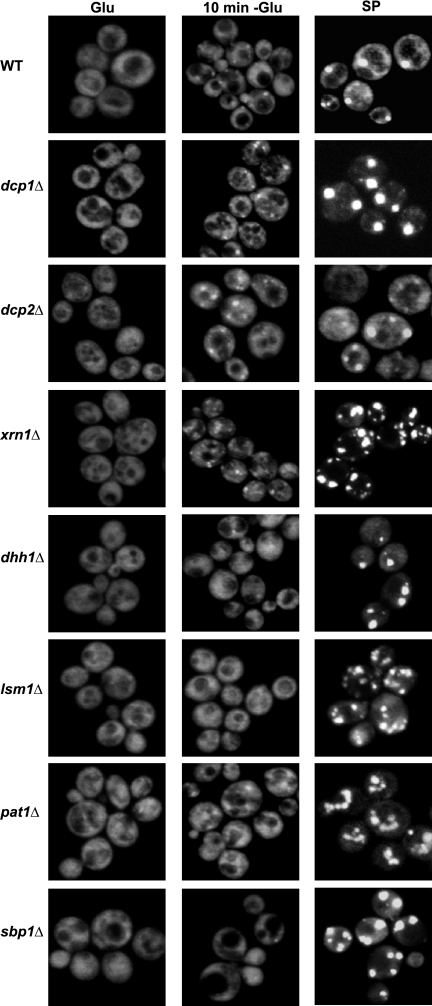
The presence of Pab1, eIF4E, and eIF4G proteins in P-bodies suggested that an mRNP bound by these factors might be an intermediate in the exchange between P-bodies and polysomes. In principle, such an intermediate could be explained by several possible models. Using Pab1p as a marker for this mRNP, we have explored possible explanations for the accumulation of these proteins in P-bodies. In one model, Pab1p might remain associated with mRNPs targeted to P-bodies and then be removed by degradation of the mRNA. Note that in this model, Pab1p might remain associated with deadenylated mRNAs by protein–protein interactions and thus be part of the mRNP targeted for degradation. A prediction of this model is that Pab1p would accumulate in P-bodies in strains defective in the catalytic steps of mRNA degradation. To test this possibility, we examined the accumulation of Pab1p during mid-log growth in P-bodies in dcp1Δ and dcp2Δ strains, which are lacking the decapping enzyme, or an xrn1Δ strain, which is deficient in 5′-to-3′ degradation of mRNAs after decapping. We observed that Pab1p was not present in P-bodies during mid-log growth in dcp1Δ, dcp2Δ and xrn1Δ strains (Figure 7), despite these strains having large P-bodies as judged by other protein markers (Sheth and Parker, 2003; Teixeira and Parker, 2007). Moreover, Pab1p still accumulated in dcp1Δ, dcp2Δ, and xrn1Δ strains during glucose deprivation or stationary phase (Figure 7), indicating that these proteins are also not required for the accumulation of Pab1p in P-bodies. These results argue that Pab1p is not removed from P-bodies by degradation of its bound mRNA.
Figure 7.
Effect of different factors on Pab1p accumulation in P-bodies. WT or mutant cells (as described) expressing a GFP-tagged version of Pab1p were grown in YPGlu (Glu) and then shifted in YP without Glu for 10 min (10 min −Glu) or were grown in SC medium plus Glu for 24 h and then shifted in YPGlu for three more days (SP).
Another possible model for Pab1p accumulating in P-bodies is that Pab1p is removed from translationally repressed mRNPs after their accumulation in P-bodies by one of the P-body components. A prediction of this model is that the accumulation of Pab1p should be increased in strains deficient at Pab1p removal from the mRNP. To test this possibility, we examined the accumulation of Pab1p in lsm1Δ, pat1Δ, sbp1Δ, and dhh1Δ strains, all of which are deleted for proteins involved in the formation of P-bodies. We observed that Pab1p was not increased in P-bodies in lsm1Δ, pat1Δ, sbp1Δ, and dhh1Δ strains during glucose deprivation in comparison with the wild-type (WT) strain (Figure 7). Indeed, the amount of Pab1p in these mutants strains in P-bodies was somewhat reduced compared with the wild-type strain, which could be explained by the partial reduction in translation repression seen in these strains in response to glucose deprivation (Holmes et al., 2004; Coller and Parker, 2005, Segal et al., 2006). In addition, the lsm1Δ, pat1Δ, dhh1Δ, sbp1Δ, strains all accumulated significant amounts of Pab1p in P-bodies in stationary phase (Figure 7), which indicates that the Lsm1p, Pat1p, Dhh1p, or Sbp1p are not required for Pab1p accumulation in P-bodies. These results are inconsistent with a model wherein Pab1p is removed by the action of any single specific protein during mRNAs being targeted to P-bodies.
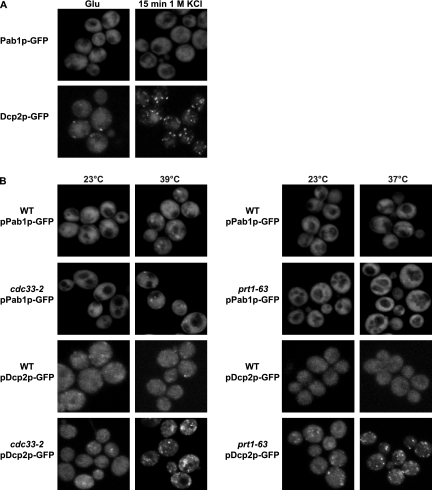
Another possible explanation for Pab1p accumulation in P-bodies is that Pab1p could be part of an mRNP that poised to exit the P-body and enter translation. This would be consistent with Pab1p accumulating in P-bodies during glucose deprivation or stationary phase where translation is inhibited (Ashe et al., 2000; Brengues et al., 2005). Such an accumulation could be due to any defect in translation initiation or to a specific alteration induced by glucose deprivation or stationary phase. To examine this possibility further, we examined whether Pab1p accumulated in P-bodies when translation initiation was blocked in other manners. We observed that Pab1p was not detectable in P-bodies in response to osmotic stress (Figure 8A), despite the robust inhibition of translation initiation and increase in P-bodies seen under these conditions (Uesono and Toh-e, 2002; Teixeira et al., 2005). We also observed that inhibition of translation initiation by using temperature-sensitive alleles of eIF4E or Prt1p was not sufficient to lead to Pab1p accumulation in P-bodies, despite the clear accumulation of Dcp2p-GFP in P-bodies (Figure 8B). These results indicate that any defect in translation initiation is not sufficient to trigger Pab1p accumulation in P-bodies, and they suggest that the accumulation of Pab1p in P-bodies is due to an alteration in translational control triggered by glucose deprivation or stationary phase.
Figure 8.
Effect of translation initiation on Pab1p accumulation in P-bodies. (A) WT strain expressing a GFP-tagged version of Pab1p or Dcp2p grown in YPGlu (Glu) were exposed to 1 M KCl for 15 min. Cells were washed and resuspended in SC plus Glu supplemented or not with the same concentration of KCl and observed. (B) WT, cdc33–2 and prt1-63 strains expressing a GFP-tagged version of Pab1p or Dcp2p grown in SC medium plus glucose at 23°C were shifted to 39 or 37°C, respectively, for 1 h.
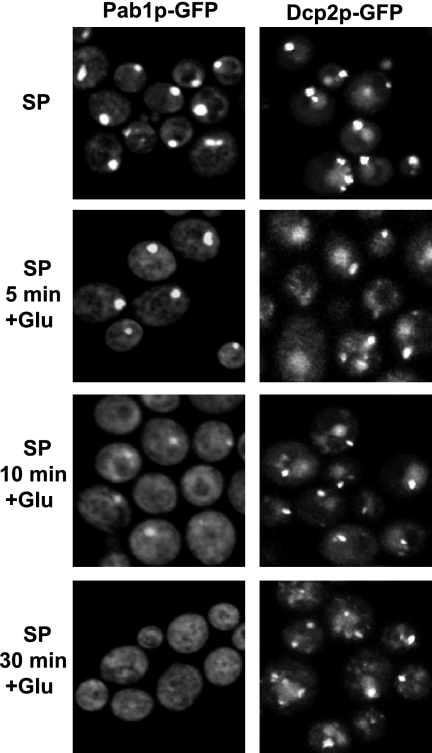
The accumulation of an mRNP containing eIF4E, eIF4G, and Pab1p in P-bodies during stationary phase suggests that these mRNPs might represent a subset of translationally repressed mRNAs that would be poised to reenter translation when growth resumes. This would be consistent with recent work showing that mRNAs stored within P-bodies can reenter translation once growth resumes (Brengues et al., 2005). Moreover, the different kinetics and specificity of Dcp2p and Pab1p accumulation in P-bodies suggests that the P-body mRNAs bound by Pab1p represent a subset of the transcripts in these structures that would rapidly reenter translation once growth was resumed. To test this possibility, we examined the rates at which Pab1p and Dcp2p exited P-bodies in stationary phase cells when fresh nutrients were provided. Strikingly, we observed that Pab1p was much more rapidly lost from P-bodies than Dcp2p after nutrient addition (Figure 9). This argues that Pab1p is associated with a specific subset of stored mRNAs within or near P-bodies in stationary phase cells and that these mRNAs preferentially exit the repressed state, and presumably reenter translation, after resumption of growth.
Figure 9.
Pab1p exits P-bodies faster than Dcp2p. WT strains expressing a GFP-tagged version of Pab1p or Dcp2p were grown in YPGlu for 4 d (SP). Then, glucose was added to the culture, and the cells were observed at different time points as indicated.
DISCUSSION
poly(A)+ mRNAs Can Be Detected in P-Bodies
Several lines of evidence argue that polyadenylated mRNAs can be in P-bodies during stress. One key observation is that FISH experiments with an oligo(dT) probe showed that poly(A)+ mRNAs colocalize with P-body components during glucose depletion (Figure 2). It should be noted that in these in situ experiments, we also observed a significant amount of oligo(dT) staining in the cytoplasm after glucose deprivation (Figure 2A). However, because we also observed that the in situ procedure led to a spreading of the Dcp2p-GFP signal in a similar manner (Figure 2B), the simplest interpretation is that at least some of this diffuse cytoplasmic staining reflects poly(A)+ mRNAs that were localized in P-bodies but that have delocalized as a result of the in situ procedure. A second observation supporting the presence of polyadenylated mRNAs in P-bodies was that polysome analysis showed that polyadenylated mRNAs as well as oligoadenylated mRNAs can be translationally repressed during glucose deprivation, suggesting that poly(A)+ mRNAs are present in P-bodies with the pool of untranslated mRNA (Figure 1). Consistent with poly(A)+ mRNAs being able to enter P-bodies, we also observe Pab1p in P-bodies at low levels during glucose deprivation and at high levels in stationary phase cells (Figure 3). Based on these observations, we conclude that poly(A)+ mRNAs are present in P-bodies at least during glucose deprivation and stationary phase. An implication of these observations is that polyadenylated mRNAs can enter P-bodies even during mid-log growth but that the presence of the poly(A) tail promotes their exit from P-bodies and return to translation. Such a role for poly(A) tails in limiting the accumulation of mRNAs in P-bodies is supported by the observation that loss of the Pab1p from yeast cells led to the accumulation of P-bodies in mid-log cultures in two different genetic backgrounds (Figure 4).
The presence of poly(A)+ mRNAs in P-bodies suggests that the poly(A) tail will be a feature of mRNAs that affects whether mRNAs are decapped, stored, or return to translation once they enter a P-body. Specifically, because removal of the poly(A) tail is required before mRNAs can be decapped (Decker and Parker, 1993; Muhlrad et al., 1994, 1995), it is likely that any polyadenylated mRNA that enters a P-body will be resistant to decapping and could be recycled back into translation. Interestingly, the use of poly(A) as a marker for exiting P-bodies and returning to translation is analogous to the readenylation of stored maternal or neuronal mRNAs for their entry into translation (Richter, 1999). This similarity, and the common composition of P-bodies in yeast or somatic cells with maternal or neuronal RNA storage granules (Barbee et al., 2006), suggest that a common mechanism will be used for the exit of mRNAs from these granules to reenter translation.
An unresolved issue is how the poly(A) tail inhibits decapping and may promote mRNAs exiting from P-bodies and reentering translation. One possibility is that the interaction of Pab1p with the poly(A) tail might facilitate the assembly of other translation initiation factors and thereby promote the disassembly of the repression/decapping complex that accumulates in P-bodies. This possibility is supported by the fact that eIF4E and eIF4G2, which can interact with Pab1p, also can be observed within P-bodies. Alternatively, or in addition, the poly(A) tail might nucleate interactions that inhibit the decapping enzyme. Interestingly, the available evidence suggests that there might be two manners by which the poly(A) tail can inhibit decapping. One mechanism seems to require the poly(A) binding protein, because pab1Δ strains can decap some transcripts before deadenylation (Caponigro and Parker, 1995). However, several observations suggest there is likely to be a Pab1p-independent mechanism by which the poly(A) tail can inhibit decapping. First, in pab1Δ strains, the majority of the transcripts persist and undergo decay after deadenylation (Caponigro and Parker, 1995). Second, biochemical analyses suggest that poly(A) tails can inhibit decapping independently of Pab1p (Wilusz et al., 2001). Finally, the low abundance of Pab1p seen in P-bodies during glucose deprivation would imply an additional mechanism by which the poly(A) tail can inhibit decapping. Although currently unknown, additional proteins that bind poly(A) tail and that are present in P-bodies would be a good candidate for being part of this process.
Significance of eIF4E, eIF4G, and Pab1p in P-Bodies
Our results demonstrate that eIF4E, eIF4G1, eIF4G2, and Pab1p can all be detected to accumulate to some degree in P-bodies under glucose deprivation, and more strikingly in stationary phase for eIF4E, eIF4G2, and Pab1p. In principle, this accumulation could have been due to the formation of a stress granule particle, which then colocalizes with a P-body. However, this is unlikely because the classic stress granules described in more complex eukaryotes also contain other factors such as eIF3 (Anderson and Kedersha, 2006), which we do not observe in foci under glucose deprivation or stationary phase (Figure 5). Another formal possibility is that eIF4E, eIF4G2, and Pab1p are components of the basic translationally repressed mRNP that accumulates in P-bodies. However, this possibility is also unlikely because Pab1p is absent from P-bodies in dcp1Δ cells (Figure 7), or during osmotic stress (Figure 8A), and are only present at low levels in P-bodies during glucose deprivation (Figure 6). These latter observations argue that eIF4E, eIF4G2, and Pab1p are present on a subpopulation of mRNAs within P-bodies. Such a subpopulation could either be a specific class of transcripts, or it could be a certain percentage of all mRNAs within P-bodies that are trapped in an intermediate state of transition between polysomes and P-bodies.
The accumulation of Pab1p in P-bodies seems to be due to a specific alteration in the process of mRNAs exiting P-bodies and entering translation that is inhibited during glucose deprivation or stationary phase, leading to the accumulation of an mRNP intermediate between P-bodies and translation. This is based on the observations that Pab1p accumulates in P-bodies during glucose deprivation and stationary phase (Figure 3), but not when translation initiation is inhibited due to osmotic stress or defects in translation initiation factors (Figure 8). Moreover, because Pab1p rapidly dissociates from stationary phase P-bodies when growth resumes (Figure 9), we suggest that this process allows cells to accumulate a pool of mRNAs within P-bodies that are poised to reenter translation. Such a mechanism would provide a possible mechanism for rapidly restarting growth, and thus it may be an important aspect of stationary phase translational control.
A clear implication is that the same eIF4E, eIF4G, Pab1p-containing intermediate mRNP seen associated with P-bodies during glucose deprivation or in stationary phase is formed during the normal cycling of mRNAs between P-bodies and polysomes, but it is too transient to be detected. This model is consistent with the known functions of eIF4E, eIF4G2, and Pab1p in promoting translation initiation. Moreover, this could explain why cells lacking Pab1p show an increase in the number and size of P-bodies even during mid-log growth (Figure 4). Interestingly, earlier results suggest that there is an mRNP transition that occurs during mRNA decapping that includes loss of Pab1p, eIF4E, and eIF4G from the mRNA and an association of the decapping activator complex (Tharun and Parker, 2001). Given this, one anticipates that the distribution of mRNAs between polysomes and P-bodies will reflect a continual competition between different types of mRNP complexes. An important goal of future work will be to define the various mRNP complexes that occur in this process and the nature of transitions between them.
ACKNOWLEDGMENTS
We thank the members of the Parker laboratory for helpful discussions, especially Carolyn Decker and Jeff Dahlseid for critical reading of the manuscript. We also thank Denise Muhlrad for help in the laboratory and the Department of Molecular and Cellular Biology for the use of the confocal facility. This work was supported by Howard Hughes Medical Institute and National Institutes of Health grant GM-45443.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1149) on May 2, 2007.
REFERENCES
- Ashe M. P., De Long S. K., Sachs A. B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. S., Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee S. A., et al. Staufen and FMRP containing neuronal RNPs are structurally and functionally related to somatic P-bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C. A., Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- Beelman C. A., Stevens A., Caponigro G., LaGrandeur T. E., Hatfield L., Fortner D. M., Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Beliakova-Bethell N., Beckham C., Giddings T. H., Jr, Winey M., Parker R., Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bonnerot C., Boeck R., Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G., Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- Coller J. M., Tucker M., Sheth U., Valencia-Sanchez M. A., Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B., Couturier A., Chabelskaya S., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell. Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W. K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J. O. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–33288. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- Decker C. J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Dunand-Sauthier I., Walker C., Wilkinson C., Gordon C., Crane R., Norbury C., Humphrey T. Sum1, a component of the fission yeast eIF3 translation initiation complex, is rapidly relocalized during environmental stress and interacts with components of the 26S proteasome. Mol. Biol. Cell. 2002;13:1626–1640. doi: 10.1091/mbc.01-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T., Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Gray N. K., Coller J. M., Dickson K. S., Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L., Beelman C. A., Stevens A., Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes L. E., Campbell S. G., De Long S. K., Sachs A. B., Ashe M. P. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol. Cell. Biol. 2004;24:2998–3010. doi: 10.1128/MCB.24.7.2998-3010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Russell W., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D., Arndt-Jovin D. J., Luhrmann R., Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Jacobson A., Peltz S. W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J. C., Fritzler M. J., Chan E. K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M., Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn K. N., DeRisi J. L., Brown P. O., Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 2001;21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur T., Parker P. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. RNA. 1999;5:420–433. doi: 10.1017/s1355838299981748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F. V., Wohlschlegel J., Yates J. R., 3rd, Parker R., Hannon G. J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Mckenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle P. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfatia K. A., Crafton E. B., Green D. M., Corbett A. H. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J. Biol. Chem. 2003;278:6731–6740. doi: 10.1074/jbc.M207571200. [DOI] [PubMed] [Google Scholar]
- Mazroui R., Sukarieh R., Bordeleau M. E., Kaufman R. J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Parker R. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol. Biol. Cell. 1999;10:3971–3978. doi: 10.1091/mbc.10.11.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. P-bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Richter J. D. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W., Kornberg R. D. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. D., Davis R. W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal S. P., Dunckley T., Parker R. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:5120–5130. doi: 10.1128/MCB.01913-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S. Z., Jr, Sachs A. B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., He W., Mayes A. E., Lennertz P., Jean D., Beggs J. D., Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun S., Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M. A., Staples R. R., Chen J., Denis C. L., Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Uesono Y., Toh-e A. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 2002;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Wilusz C. J., Gao M., Jones C. L., Wilusz J., Peltz S. W. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]