Abstract
Translationally controlled Tumor Protein (TCTP) is an evolutionally highly conserved protein which has been implicated in many cellular functions that are related to cell growth, death, and even the allergic response of the host. To address the physiological roles of TCTP, we generated TCTP knockout mice by targeted gene disruption. Heterozygous mutants appeared to be developmentally normal. However, homozygous mutants (TCTP−/−) were embryonic lethal. TCTP−/− embryos were smaller in size than the control littermates at all postimplantation stages examined. Although TCTP is widely expressed in both extraembryonic and embryonic tissues, the most prominent defect of the TCTP−/− embryo at embryonic stage day 5.5 (E5.5) was in its epiblast, which had a reduced number of cells compared with wild-type controls. The knockout embryos also suffered a higher incidence of apoptosis in epiblast starting about E6.5 and subsequently died around E9.5–10.5 with a severely disorganized structure. Last, we demonstrated that TCTP−/− and control mouse embryonic fibroblasts manifested similar proliferation activities and apoptotic sensitivities to various death stimuli. Taken together, our results suggest that despite that TCTP is widely expressed in many tissues or cell types, it appears to regulate cell proliferation and survival in a tissue- or cell type–specific manner.
INTRODUCTION
Translationally controlled Tumor Protein (TCTP) has been identified in many eukaryotes, including yeast, fungus, insects, plants, and mammals (Yenofsky et al., 1983; Bohm et al., 1989; MacDonald et al., 1995; Thaw et al., 2001). The extremely high degree of sequence conservation of TCTP during evolution suggests that it plays an essential role in the development or normal functioning of various organisms (Thaw et al., 2001). TCTP is widely expressed in many tissues and cell types, and its protein levels are highly regulated in response to a wide range of extracellular signals and cellular conditions (Bommer and Thiele, 2004). Overexpression or down-regulation of TCTP perturbs cell growth (Gachet et al., 1999; Kamath et al., 2003; Tuynder et al., 2004), suggesting that TCTP has a growth-related function. On the other hand, the human homolog of TCTP (termed histamine-releasing factor [HRF]) was found to be the component present in patients' biological fluids that could stimulate histamine release from basophils. The latter study suggests that TCTP plays a role in the acute allergic response (MacDonald et al., 1995).
Many cellular proteins have been reported to interact with TCTP. For example, tubulin (Gachet et al., 1999), the mammalian Plk (Yarm, 2002), translation elongation factors eEF1A and eEF1Bbeta (Cans et al., 2003; Langdon et al., 2004), Mcl-1 (Li et al., 2001; Liu et al., 2005), TSAP6 (Amzallag et al., 2004), Na,K-ATPase (Jung et al., 2004), and Bcl-XL (Yang et al., 2005). Studies on TCTP's interaction with tubulin, Plk and translation elongation factors suggest that TCTP plays a role in cell cycle progression (Gachet et al., 1999; Yarm, 2002) and protein synthesis (Cans et al., 2003; Langdon et al., 2004). On the other hand, an anti-apoptotic activity has been reported for TCTP, which may be related to its interaction with Mcl-1 and/or Bcl-XL (Yang et al., 1996; Li et al., 2001; Liu et al., 2005).
The widely expressed patterns and diverse cellular functions described for TCTP prompted us to generate a gene-disrupted mouse model to address its physiological roles. In this study, we provide genetic evidence that TCTP plays a critical role in the normal proliferation and survival of early mouse embryos. However, to our surprise, we found that the regulatory functions of TCTP in cellular proliferation and survival controls are dispensable in some cell types like mouse embryonic fibroblasts.
MATERIALS AND METHODS
Antibodies
Anti-TCTP antibody was generated as previously described (Liu et al., 2005). Other antibodies used in this study include those specific to cyclin B1 and cyclin E1 (both from eBioscience, San Diego, CA); cyclin D2 (Abcam, Cambridge, MA); cyclin E2 and cyclin D3 (both from Cell Signaling, Beverly, MA); β-actin (Sigma, St. Louis, MO); and total or active form of S6K1 (P-T389), Akt (P-S473), and ERK (all from Cell Signaling).
Generation of TCTP Gene Disruption Allele
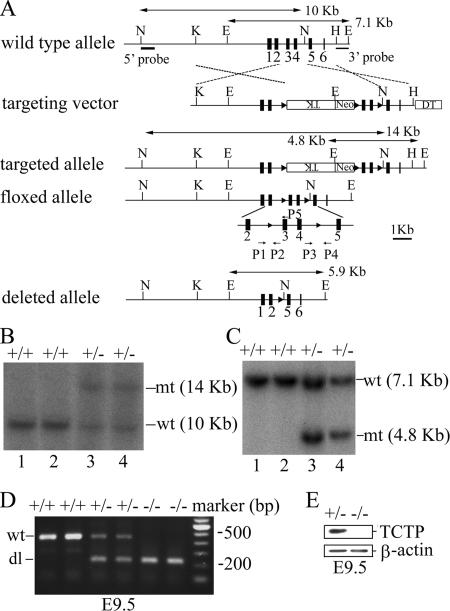
To generate TCTP gene disruption allele, two overlapping genomic fragments harboring the TCTP locus were isolated from a 129/Svj mouse genomic library and used to construct the targeting vector as depicted in Figure 1A. This targeting vector was constructed by PCR assisted cloning in such a way that a floxed cassette containing both Neo and TK selection markers was introduced into intron 2 and the third loxP site into intron 4 of the TCTP gene. Lying outside of the selection cassette and the third loxP site were two homology arms (4.8 and 1.5 kb) and the gene encoding the diphtheria toxin (DT) for negative selection. This targeting vector was then electroporated into R1 embryonic stem (ES) cells, and Southern blotting using 5′ and 3′ probes as indicated in Figure 1A was carried out to select clones that had undergone homologous recombination at the TCTP locus. Two positive clones, 248 and 280, harboring the “targeted allele” were subsequently transiently transfected with a cytomegalovirus promoter–driven Cre expression vector to generate either the “floxed allele” or the “deleted allele” of the TCTP gene as indicated in Figure 1A. Two ES cell clones harboring the deleted allele “−” (248.2 and 280.4), and two with the floxed allele “f” (248.41 and 248.101) were further microinjected into C57BL/6 blastocysts to generate chimeric mice. The male chimeric mice were backcrossed with C57BL/6 females to generate TCTP+/− or TCTPf/+ mice. Genotyping was performed by PCR using primers P1 (5′-TCTAGAAAAGTGGAGGCGGAGC-3′) and P5 (5′-GGTGACTACTGTGCTTTCGGTA-3′) for the wild-type (450 base pairs) and floxed alleles (520 base pairs), P1 and P4 (5′-AAAGCAGATCCAGAATAACCCC-3′) for the deleted allele (250 base pairs). All animal experiments were performed in accordance with the guidelines set by Academia Sinica Institutional Animal Care and Utilization Committee.
Figure 1.
Targeted disruption of the TCTP gene. (A) The structures of the wild-type, targeting vector, and recombinant alleles are shown together with some relevant restriction sites (E, EcoRI; H, HindIII; K, KpnI; N, NdeI). The 5′ and 3′ probes and the predicted length of EcoRI or NdeI restriction fragments in Southern blot analysis are as indicated. (B) Southern blot analysis of the recombinant ES cell clones harboring the “targeted allele.” Genomic DNA extracted from ES cell clones (lanes 1 and 2, nonrecombinants; lanes 3 and 4, clones 248 and 280) was digested with NdeI and probed with the 5′probe. (C) Same as in B except that the genomic DNA was digested with EcoRI and probed with the 3′ probe. The predicted signals for the wild-type (wt) and targeted allele (mt) are as indicated. (D) Representative genotypic analysis of E9.5 embryos harboring the wt (+) or deleted allele (dl or “−”) of the TCTP gene from a TCTP+/− intercross. Genotyping was performed by PCR using primers P1 and P5 for the wild-type (wt, 450 base pairs) and P1 and P4 for the deleted allele (dl, 250 base pairs). (E) Immunoblotting analysis of representative E9.5 embryos with the indicated genotypes using antibodies specific to TCTP or β-actin.
Embryo Dissection, Histological Analysis, and Immunofluorescence Microscopy
Timed mating was performed with TCTP+/− mice on a mixed genetic background (C57BL/J × 129/Svj). Females with copulation plugs were considered to be at day 0.5 of gestation, and embryos present in this pregnant female were designated at embryonic stage day 0.5 (E0.5). Pregnant females were killed at various times of gestation, and the dissected embryos were photographed and genotyped by PCR. For histological analysis, embryos within decidua were fixed with 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. To genotype embryos for histological analysis, embryo sections were boiled in 50 mM Tris, pH 8.0, for 30 min before they were stained first with guinea pig anti-mouse TCTP (Liu et al., 2005) and then with Alexa Fluor 555–conjugated goat anti-guinea pig immunoglobulin G (Molecular Probes, Eugene, OR). Stained sections were analyzed with confocal fluorescence microscopy (Zeiss type LSM-meta 510, Thornwood, NY).
Apoptosis Measurement in Mouse Embryos
Cells in embryos undergoing apoptosis were analyzed by the terminal deoxynucleotidyl transferase–mediated dUTP nick end-labeling (TUNEL) assay essentially as previously described (Gavrieli et al., 1992). After TUNEL staining, embryo sections were counterstained with hematoxylin. The percentage of TUNEL-positive cells was determined by counting at least six sections of the same embryo excluding cells at the ectoplacental cone region.
Whole Mount In Situ Hybridization
Whole mount in situ hybridization of the mouse embryos was carried out essentially as described by Shen et al. (1997). The riboprobes for Brachyury and Shh corresponding to the entire open reading frame of the cDNA were synthesized with a DIG RNA labeling kit (Roche, Indianapolis, IN). For each marker, at least three embryos from each genotype were examined.
Real-Time Quantitative PCR
To analyze the expression of cyclins in control and TCTP knockout embryos, total RNA was isolated using the Trizol reagent (Invitrogen, Carlsbad, CA) from morphologically normal or abnormal E9.5 embryos from the intercrosses of TCTP heterozygous mutants. RNA from four or eight embryos with normal or abnormal phenotypes, respectively, was pooled for the generation of cDNA using random hexamers and the Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen), and the expression levels of cyclin D1, D3, E1, TCTP, and hprt mRNA were analyzed by real-time quantitative PCR on the LightCycler Real-Time PCR System according to the manufacturer's protocol (Roche Applied Science, Indianapolis, IN). The sequences of the primers used in this assay are as follows: cyclin D1 sense, 5′-CAGAAGTGCGAAGAGGAGGTC-3′, and anti-sense, 5′-TCATCTTAGAGGCCACGAACAT-3′; cyclin D3 sense, 5′-AAAGGAGATCAAGCCGCACAT-3′, and antisense, 5′-GTTCATAGCCAGAGGGAAGACATC-3′; cyclin E1 sense, 5′-TCTCCTCACTGGAGTTGATGCA-3′, and antisense, 5′-AACGGAACCATCCATTTGACA; TCTP sense, 5′-ATGATCATCTACCGGGACCTCA-3′, and antisense, 5′-GGTGACTACTGTGCTTTCGGTA; Hprt sense, 5′-GTTGGATACAGGCCAGACTTTGTTG-3′, and antisense, 5′-GAGGGTAGGCTGGCCTATAGGCT-3′. The specificity of the PCR was verified by gel electrophoresis of the PCR products on 1.8% agarose gel. The LightCycler software version 3.0 was used for data analysis. The transcript expression level of the gene of interest was normalized to that of Hprt (hypoxanthine guanine phosphoribosyl transferase) by subtracting the crossover point (CT) value of Hprt from that of the target gene (ΔCT = CT target gene − CT Hprt). The relative transcript level of genes of interest in abnormal versus normal embryos was presented in the results.
Generation of TCTP−/− Mouse Embryonic Fibroblasts by Retroviral Infection
Retrovirus producing Cre recombinase was generated by transient transfection of Phoenix-Eco cells (gift from Dr. Garry Nolan, Stanford University School of Medicine) with a mouse stem-cell virus-based bicistronic retroviral vector (Shah et al., 2002) coexpressing GFP and Cre. Two days after transfection, culture supernatants were used to infect TCTPf/f mouse embryonic fibroblasts (MEFs). Infected cells (GFP-positive) were sorted out by flow cytometry and processed for subsequent analysis, including cell proliferation and survival assays as indicated in the individual figure.
RESULTS
Targeted Disruption of the Mouse TCTP Gene
To generate a disrupted or conditional allele of the mouse TCTP gene, a targeting vector as depicted in Figure 1A was constructed and electroporated into the R1 ES cells (Nagy et al., 1993). Two positive clones (248 and 280) harboring the “targeted allele” (Figure 1, B and C, lanes 3 and 4) were further transiently transfected with a Cre expression vector to generate ES subclones containing either the “floxed allele” or the “deleted allele” of the TCTP gene (Figure 1A; see Materials and Methods). The “deleted allele” is a null allele, because embryos homozygous with this allele (TCTP−/−) did not produce any TCTP proteins (Figure 1, D and E), whereas the “floxed allele” behaves like a wild-type allele (see below).
Loss of TCTP Results in Early Embryonic Lethality
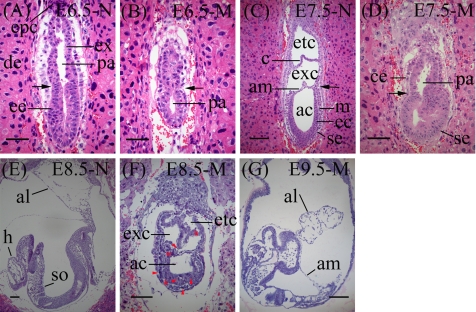
Mice heterozygous for the deleted allele of TCTP (TCTP+/−) were viable, fertile and morphologically no different from the wild type littermates. Genotype analysis of progeny from heterozygote intercrosses revealed that 32.6% were wild type, 67.4% heterozygous and none homozygous (Table 1), indicating that the TCTP−/− mutation manifests a recessive embryonic lethal trait. We next carried out experiments to determine the gestation stage where the TCTP−/− embryos might have become lethal. In this experiment, embryos from heterozygote intercrosses were collected at various developmental stages and genotyped. As illustrated in Table 1, no homozygous mutants could be identified from embryos collected between E11.5 and E14.5, and during this period of time a significantly high percentage (∼23%) of embryo resorption was noted. On the other hand, for embryos collected between E 6.5 and E9.5, the three genotypes (wild type, heterozygous, and homozygous) were identified in an expected Mendelian ratio (1:2:1), albeit the morphology of the homozygote always appeared to be abnormal, i.e., they were severely growth-retarded and became increasingly disorganized with age (Figure 2). Of note, homozygous mutants with an abnormal phenotype like that observed at the E9.5 stage (Figure 2D) could sometimes be identified at the E10.5 stage, but with a frequency that was much lower than that predicted by the Mendelian law (Table 1). This abnormal phenotype was identical in mutant embryos derived from two independent ES clones, i.e., 248.2 and 280.4.
Table 1.
Genotype analysis of offspring from TCTP+/− intercrosses
| Stage | Genotype |
Resorption | Total | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| Newborn | 112 | 231 | 0 | 0 | 354 |
| E11.5–E14.5 | 12 | 21 | 0 | 10 | 43 |
| E10.5 | 7 | 18 | 4 | 4 | 33 |
| E9.5 | 10 | 24 | 11 | 1 | 46 |
| E8.5 | 19 | 35 | 18 | 0 | 72 |
| E7.5 | 15 | 29 | 13 | 0 | 57 |
| E6.5 | 5 | 14 | 7 | 0 | 26 |
Figure 2.
Morphological analysis of normal and TCTP null embryos. Photomicrographs of littermates dissected at various stages (E6.5–E9.5) as indicated. al, allantois; epc, ectoplacental cone; h, head folds; j, junction between the embryonic and extraembryonic regions. Scale bar in all panels, 100 μm.
TCTP−/− Mutants Undergo Gastrulation But Develop Abnormally and Die before Turning
To further characterize the developmental defects of the TCTP−/− mutants, whole-mount in situ hybridization using probes specific for the mesoderm and endoderm structures, Brachyury (T) and Sonic hedgehog (Shh), was carried out. At the early head fold stage (E7.5), the Brachyury expression in the normal embryos (wild-type or heterozygote) was detectable in the primitive streak (ps) throughout its length, as well as in the node (n) and the axial mesendoderm (am; Wilkinson et al., 1990; Figure 3A), whereas in the homozygous mutants, its expression was only observed in the primitive streak with a pattern as if the embryo had only progressed to the early streak stage (E 6.0 or 6.5; Figure 3A). When normal embryos developed to the E8.5 stage, Brachyury marked the developing notochord and was abundantly expressed in the posterior end. However, at the same stage the expression pattern of Brachyury in the homozygote looked more like the wild-type embryos that had developed to the early or late head fold stage with abnormal sizes (Figure 3B). To further examine the germ layer formation patterns in TCTP−/− mutants, we compared the expression of Shh in normal embryos at E8.5 and mutants at E9.5, because their gross appearance was similar (Figure 2, C and D). As shown in Figure 3C, the Shh expression of the E9.5 mutant was markedly reduced in the anterior end, and its expression in the posterior end was also partially defective. These results suggest that targeted disruption of TCTP in mouse results in defects in mesoderm and endoderm patterning.
Figure 3.
Brachyury and Shh expression in normal and TCTP-null embryos. Whole-mount in situ hybridization of wild-type and TCTP-null embryos at various stages as indicated was carried out using riboprobes specific to Brachyury (A and B) or Shh (C). (C) Left, E8.5 TCTP+/− embryo; right, E9.5 TCTP−/− embryo. am, axial mesendoderm; nt, notochord; n, node; ps, primitive streak; endo, endoderm. Scale bar in all panels, 100 μm.
To further analyze the TCTP−/− mutant phenotype, we examined histological sections of embryos collected in utero from E5.5 to E9.5 stages. All these embryos were “genotyped” by immunofluorescence staining using TCTP-specific antibodies (Figure 4, B and D, and data not shown for embryos from E6.5 to E9.5). At the E5.5 stage, mutant embryos “M” without TCTP expression appeared to have a reduced number of cells in the epiblast compared with control embryos “N” with TCTP expression (∼20 vs. ∼28 cells in the most central 4-μm section of the M or N embryos, respectively; Figure 4, A and C). Furthermore, cells in the visceral endoderm (ve) of the mutant embryos were significantly more vacuolated than that in the control embryos. At the E6.5 stage, the TCTP null embryos were markedly smaller than the control littermates with positive TCTP staining, even though a characteristic, two-layer structure of ectodermal and endodermal cells enclosing the proamniotic cavity was readily identified in these mutants (compare Figure 5, A and B). When normal embryos develop to the E7.5 stage, other than the emergence of the mesodermal layer, three characteristic cavities, i.e., amniotic (ac), exocolomic (exc), and ectoplacental (etc) cavities, have formed (Figure 5C). However, in the mutant embryos, the proamniotic cavity remained largely undivided (Figure 5D). At the E8.5 stage, although the mutant embryos had developed further to form a structure that slightly resembled the normal E7.5 embryos, e.g., formation of the three distinct cavities, a significant number of pyknotic cells were noted (Figure 5F, arrowheads). At the E9.5 stage, all mutant embryos were severely growth-retarded and disorganized. The chorioallantoic fusion failed to occur, and they all died before turning (Figure 5G).
Figure 4.
Histological analysis of E5.5 control and TCTP-null embryos “genotyped” by immunostaining. Sagittal sections of E5.5 embryos derived from heterozygous intercrosses were first stained with TCTP-specific antibody and visualized by confocal microscopy as described in Materials and Methods. After immunostaining (right panels of B and D), the same section was stained with hematoxylin and eosin (HE; left panels of B and D). (A and C) HE staining of the most central 4-μm sections of the same embryos as that shown in B and D, respectively. TCTP-expressing N (A and B) and nonexpressing M littermates (C and D) are as indicated. ve, visceral endoderm; epi, epiblast.
Figure 5.
Histological analysis of E6.5–E9.5 embryos derived from heterozygous intercrosses. Sagittal sections of TCTP-expressing (N) or nonexpressing (M) littermates at various stages as indicated. Arrows in A–D show the boundary between embryonic and extraembryonic regions. Embryos were “genotyped” by immunostaining using TCTP-specific antibodies as that described in the legend to Figure 4. ac, amniotic cavity; al, allantois; am, amnion; c, chorion; ce, cuboidal visceral endoderm; de, decidua; ec, ectoderm; ee, embryonic ectoderm; epc, ectoplacental cone; etc, ectoplacental cavity; ex, extraembryonic ectoderm; exc, exocoelomic cavity; h, head folds; m, mesoderm; pa, proamniotic cavity; SE, squamous visceral endoderm; so, somite. Arrowheads point to pyknotic cells. Scale bar, 50 μm (A, B, and F) and 100 μm (C and E–G).
Increased Apoptosis of Cells in TCTP−/− Embryos
We next examined whether increased cell death could have also contributed to the abnormally small phenotype of the TCTP−/− mutant embryos. To address this issue, we used the TUNEL assay to compare the percentages of cell death in embryos with (+) or without (−) TCTP expression (again determined by immunostaining with TCTP specific antibody as described in the legend to Figure 4). As shown in Figure 6, at the E5.5 stage, the percentage of TUNEL-positive cells was comparable between the TCTP(+) (5.8 ± 0.9%) and TCTP(−) embryos (5.7 ± 0.8%; Figure 6, A, B, and E, p = 0.8, n = 3). However, at the E6.5 stage, markedly increased cell death was observed in the TCTP(−) mutants (11.7 ± 1.5% vs. 3.1 ± 1.5% in TCTP(+) cells; Figure 6, C, D, and F, p = 0.007, n = 3), especially in the epiblast region. Taken together, this result suggests that the reduced cell number in the epiblast of the E5.5 TCTP-null embryos compared with wild type is mainly due to reduced cellular proliferation beginning at E5.5 or earlier stages, and that increased cell death starting around E6.5 contributes to later progressively smaller and disorganized structure of the TCTP-null embryos.
Figure 6.
Increased apoptosis of TCTP-null mutants at the E6.5 but not E5.5 stage. Sections of TCTP(+) and TCTP(−) embryos at E5.5 (A and B) and E6.5 (C and D) stages were subjected to TUNEL analysis. Embryos were “genotyped” by immunostaining with TCTP-specific antibody. Apoptotic cells were stained brown. The graphs in E and F show mean percentages ± SEs of TUNEL-positive cells from three to four independent experiments. Only the difference between TCTP(+) and TCTP(−) embryos at the E6.5 stage was statistically significant; *p = 0.007; n = 3 (F). (D) An arrow points to one example of apoptotic cells. Scale bar, 25 μm (A and B) and 50 μm (C and D).
Decreased Expression of G1- But Not G2/M-specific Cyclins in E9.5 TCTP−/− Embryos
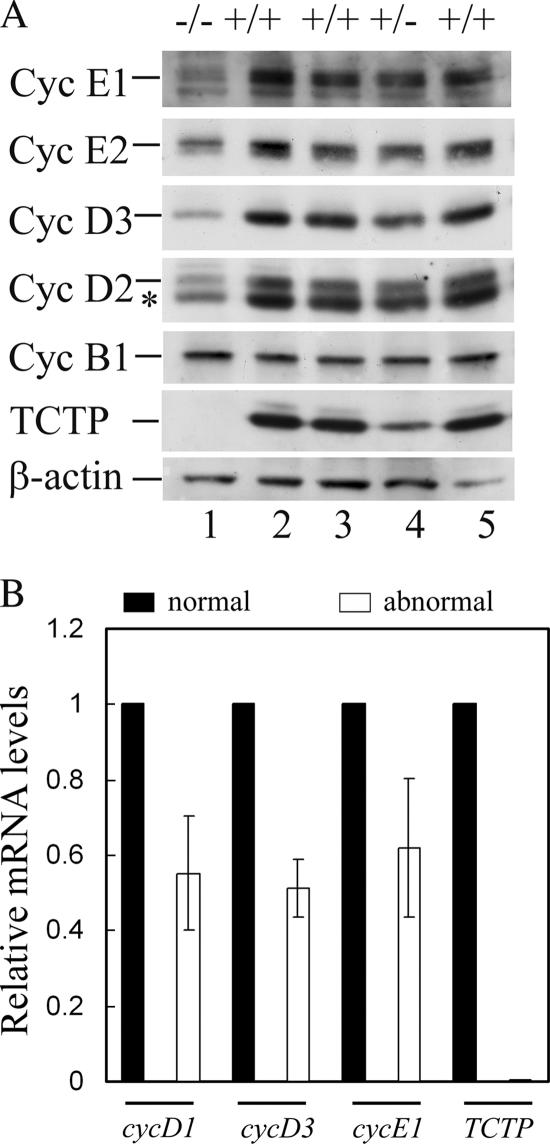
We next examined whether the E9.5 stage TCTP−/− embryos might have encountered a problem in cell cycle progression, given that overexpression or down-regulation of TCTP was shown to perturb cell growth (Gachet et al., 1999; Kamath et al., 2003; Tuynder et al., 2004). To address this issue, immunoblotting analysis (Figure 7A) (or in some cases quantitative RT-PCR, Figure 7B) was carried out to compare the expression levels of various cyclins between the E9.5 normal and TCTP−/− embryos. As shown in Figure 7, all G1 cyclins tested (both D and E types) were markedly reduced in the TCTP−/− embryos, compared with those in either wild-type or heterozygous embryos. However, there was no significant difference in the levels of mitotic cyclin (cyclin B1) among embryos of all three genotypes. These results suggest that TCTP regulates cell proliferation in embryos via a mechanism that is directly or indirectly linked to the synthesis of proteins required for S phase entry.
Figure 7.
Reduced expression of D- and E- but not B-type cyclins in TCTP−/− embryos. (A) Protein extracts from E9.5 embryos with the indicated genotypes were analyzed by immunoblotting using antibodies specific to various proteins as indicated. Lane 1 was the extracts pooled from three embryos with mutant phenotype as shown in Figure 2D. Asterisk (*) indicates cyclin D3 cross-reacted with cyclin D2 antibody. (B) Quantitative RT-PCR was carried out using RNA isolated from E9.5 embryos with normal or abnormal morphology as described in Materials and Methods. Shown here is the relative mRNA level of various genes as indicated in abnormal versus normal embryos. The results (mean ± SD) were plotted from two independent experiments done in triplicate.
TCTP Is Dispensable for Normal Proliferation and Survival of MEFs
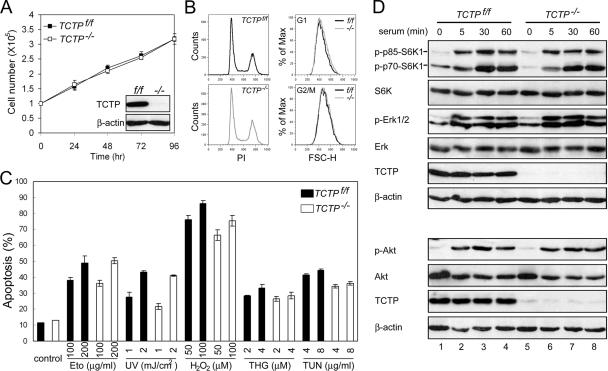
Given that the TCTP−/− embryos were severely growth retarded, we next examined whether TCTP was essential for normal proliferation of MEFs. To address this issue, we initially attempted to isolate MEFs from the E9.5 TCTP−/− embryos. However, it was not successful. We therefore turned to the mutant mice harboring the floxed allele of the TCTP gene (Figure 1A). The TCTPf/f mice were born with a frequency that was very close to the Medelian ratio and appeared to be healthy and fertile like the wild-type littermates. This result indicates that insertion of a loxP site into introns 2 and 4 of the TCTP genomic locus did not disrupt the expression of this gene. We then followed the standard procedure to isolate MEFs from the E15.5 TCTPf/f embryos and infected these cells with retroviruses expressing Cre to generate TCTP−/− MEFs (see Materials and Methods). To our surprise, TCTP−/− MEF proliferated in a way that was very similar to the control TCTPf/f cells (Figure 8A). Also, there was no significant difference in the size of the control and TCTP−/− MEFs, as revealed by the flow cytometric analysis (Figure 8B).
Figure 8.
Control and TCTP−/− MEFs manifest similar proliferation and survival activities. (A) TCTPf/f and TCTP−/− MEFs grow similarly in culture. Control or TCTP−/− MEFs were seeded in complete growth medium, and at various time points after seeding cells were counted by trypan blue exclusion assays. Inset in A shows the TCTP protein levels in cells used in this study. (B) Control and TCTP−/− MEFs display similar cell size. Exponentially growing control or TCTP−/− MEFs were stained by propidium iodide and analyzed by flow cytometry. The DNA content (PI) and the forward scatter height (FSC-H) distributions of cells at the G1 or G2/M phase of the cell cycle are as indicated. (C) Control and TCTP−/− MEFs manifest similar apoptotic sensitivities to death inducers. Control or TCTP−/− cells were treated with two different doses of various apoptotic stimuli as indicated. One day after treatment, cells that had undergone apoptosis were quantified by annexin V staining assay. Data shown here are mean ± SD of triplicates from experiments that were repeated three times with very similar results. Eto, etoposide; THG, thapsigargin; TUN, tunicamycin. (D) Similar activation of S6K1, Akt, and ERK after serum stimulation of control or TCTP−/− MEFs. Cells to be analyzed were starved and restimulated with serum for various times as indicated. The same cell lysates were analyzed by two immunoblots (top and bottom) using antibodies specific to TCTP, β-actin, total, or active form of S6K1 (P-T389), Akt (P-S473), and ERK as indicated.
Overexpression of TCTP was shown to be able to protect apoptosis triggered by growth factor deprivation or treatment of cells with the DNA-damaging agent (Li et al., 2001; Liu et al., 2005). The increased apoptotic cells observed in the TCTP−/− embryos further prompted us to examine whether TCTP−/− MEF would be more sensitive to apoptotic stimuli than the wild-type controls. To address this issue, control or TCTP−/− MEF was subjected to various apoptotic treatments, including those that would damage DNA (etoposide, UV, hydrogen peroxide) and those that would cause ER stress (thapsigargin and tunicamycin). As shown in Figure 8C, in no treatment did we observe any significant increase in the apoptotic percentage of TCTP−/− MEF than that of the control cells. Consistent with the results shown above (Figure 8, A–C), we also failed to observe any significant difference between control and TCTP−/− MEF in terms of serum-induced activation of signaling molecules commonly involved in cell growth and death controls, e.g., phosphorylation of S6K (T389), Akt (S473), and ERK (Figure 8D). Taken together, our results suggest that TCTP is not essential for normal proliferation or survival of cells like MEF, albeit such functions of TCTP are indispensable during early embryo development.
DISCUSSION
The protein sequence of TCTP is highly conserved during evolution of eukaryotes, suggesting that this protein carries an essential function for the normal development of these organisms. It is therefore not surprising to observe that mice deficient in TCTP are embryonic lethal. But, what exactly TCTP does to ensure the normal development of mouse embryos remains to be determined. On the other hand, our observation that TCTP deficiency results in a selective reduction of G1 (both D and E types) but not mitotic cyclins (cyclin B1) in the developing embryos suggests that TCTP plays a role in embryonic cell proliferation via a mechanism that is linked to the regulation of the cell cycle machinery. More experiments will be required to address the molecular mechanisms involved in this intriguing finding.
The apoptosis-prone nature of the TCTP−/− embryos was first noted at the E6.5 stage. We previously reported that TCTP interacts with Mcl-1 and enhances Mcl-1's stability and anti-apoptotic activity (Liu et al., 2005). Mice deficient of Mcl-1 die at peri-implantation stage of embryo development (Rinkenberger et al., 2000). However, TCTP-null embryos characterized in this study can still be detected around the E9.5 stage, albeit with abnormal morphology. The different phenotype of these two knockout models does not support a cofactor role of TCTP in the Mcl-1 functional complex, at least during early embryo development. Given that TCTP is structurally similar to the Mss4/Dss4 family of guanine nucleotide-free chaperones (Thaw et al., 2001) and can interact with many cellular proteins involved in cell growth or survival controls, TCTP is more likely to function as a protein chaperone for its interacting proteins. Under such conditions, in the knockout embryos, those proteins whose functions are normally positively modulated by TCTP may still function partially during early time points. As a result of this partial defect, the growth and survival of the mutant embryos may thus be progressively affected, and later on degeneration of the abnormally developed embryos occur. More experiments are certainly required to test this possibility.
TCTP expression is detected both in the extraembryonic and embryonic regions of E5.5 embryos. However, reduced cell numbers and increased apoptosis of TCTP−/− embryos were much more prominent in epiblast than other parts of the mutant embryos. These results together with the similar biochemical properties of control and TCTP−/− MEFs suggest that the critical role of TCTP in cell proliferation and survival actually manifests in a tissue or cell-type specific manner. One possibility for this observation is that the TCTP function in cells like MEFs may be replaced by another functionally redundant protein that is present (or activated) in MEFs but not in certain cells during early embryonic development. Alternatively, TCTP may be differentially required for the cellular machinery that controls proliferation and/or survival of certain cells in early embryos and in MEFs. Of note, all cell types examined so far (e.g., U937, HeLa, MCF7, U2OS, and Ba/F3), in which TCTP has been shown to influence cell growth or survival (Li et al., 2001; Tuynder et al., 2002; Liu et al., 2005) are cultured cancerous or immortalized cell lines that can rapidly proliferate like certain cells in early embryos. In addition, TCTP has been shown to interact with translation elongation factor, eEF1A, and its guanine nucleotide exchange factor, eEF1Bβ, and such interaction may regulate the function of eEF1A during the elongation cycle of protein translation (Cans et al., 2003). Interestingly, the expression levels of the eEF1A in many tissues like brain, heart, and skeletal muscle are high in embryonic life but gradually decline with postnatal age (Khalyfa et al., 2001); and constitutive expression of eEF1A causes mouse or hamster fibroblasts to become highly susceptible to transformation induced by 3-methylcholanthrene and UV light (Tatsuka et al., 1992). Taken together, these earlier studies together with our findings presented in this report suggest that in rapid growing cells like those in some region of the early embryos or those derived from tumors a distinct set of protein synthesis machinery involving eEF1A is used, whereas in cells that are more toward the end of their developmental stage like MEFs, another set of protein synthesis machinery without the involvement of eEF1A is used. Under such conditions, it would not be surprising to observe a differential requirement for the eEF1A-interacting protein TCTP for the normal growth of early embryos and MEFs. More experiments are certainly required to test this possibility.
While this manuscript was ready for submission, we learned that Drosophila with reduced expression of dTCTP manifested a phenotype with reduced cell size, cell number, and organ size (Hsu et al., 2007). Hsu et al. further demonstrated that dTCTP functions upstream of dS6K and regulates fly cell growth by positively regulating dRheb activity. Their results also suggest that dTCTP likely activates dRheb by functioning as a guanine nucleotide exchange factor (GEF) for this protein. Given that human TCTP can rescue the dTCTP knockdown phenotype in the Drosophila system (Hsu et al., 2007) and that severe growth retardation was observed for the TCTP−/− mouse embryos (this study), the possibility exists that TCTP regulates mouse embryonic development via a mechanism that is similar to that found in the fly system, i.e., involving activation of both Rheb and S6K. However, to our surprise, our preliminary data did not seem to favor this possibility, because unlike that reported for the fly system TCTP-deficient mouse embryos, at least at the E9.5 stage, did not manifest any reduced amounts of activated p70-S6K1 (phosphorylation at T389) compared with normal controls (in fact, the mutant embryos seemed to have slightly more activated form; see Supplementary Figure S1). Furthermore, although we have not directly compared the activation state of Rheb between control and TCTP−/− mouse embryos, the fact that comparable (or slightly more) amount of activated form of p70-S6K1 was observed in the TCTP−/− embryos (Supplementary Figure S1) again does not favor the possibility that TCTP functions as a GEF (or an essential GEF) for Rheb during mouse development, at least not around the E9.5 stage, because p70-S6K1 is known to be activated by mTOR and activation of the mTOR kinase requires the binding of the GTP-charged Rheb to the TOR complex 1 (Long et al., 2005; Avruch et al., 2006). On the other hand, our negative results observed in the E9.5 mouse embryos cannot really rule out the possibility that TCTP may still function as a positive regulator for Rheb in the mouse system, because TCTP may carry out such a function in a cell-type, tissue or even developmental stage-specific manner, a feature that is likely to be very similar to that observed for its roles in cell proliferation and survival. More experiments including examining mouse embryos at stages earlier than E9.5 and tissue-specific TCTP knockout mice will be required to address this issue.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Transgenic Core Facility of Institute of Molecular Biology, Academia Sinica for generation of TCTP knockout mice. This study was supported in part by a major theme project from Academia Sinica (AS23–57) and by grants NHR1-EX91-9119BN and NSC 92-3112-B-001-016 from National Health Research Institutes and the National Science Council of Taiwan to H.-F.Y.-Y.
Abbreviations used:
- TCTP
translationally controlled tumor protein
- Shh
Sonic hedgehog
- MEF
mouse embryonic fibroblasts.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0188) on May 2, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Amzallag N., Passer B. J., Allanic D., Segura E., Thery C., Goud B., Amson R., Telerman A. TSAP6 facilitates the secretion of translationally controlled tumor protein/histamine-releasing factor via a nonclassical pathway. J. Biol. Chem. 2004;279:46104–46112. doi: 10.1074/jbc.M404850200. [DOI] [PubMed] [Google Scholar]
- Avruch J., Hara K., Lin Y., Liu M., Long X., Ortiz-Vega S., Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- Bohm H., Benndorf R., Gaestel M., Gross B., Nurnberg P., Kraft R., Otto A., Bielka H. The growth-related protein P23 of the Ehrlich ascites tumor: translational control, cloning and primary structure. Biochem. Int. 1989;19:277–286. [PubMed] [Google Scholar]
- Bommer U. A., Thiele B. J. The translationally controlled tumour protein (TCTP) Int. J. Biochem. Cell Biol. 2004;36:379–385. doi: 10.1016/s1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- Cans C., et al. Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc. Natl. Acad. Sci. USA. 2003;100:13892–13897. doi: 10.1073/pnas.2335950100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y., Tournier S., Lee M., Lazaris-Karatzas A., Poulton T., Bommer U. A. The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J. Cell Sci. 1999;112(Pt 8):1257–1271. doi: 10.1242/jcs.112.8.1257. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y., Sherman Y., Ben-Sasson S. A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y. C., Chern J. J., Cai Y., Liu M., Choi K. W. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature. 2007;445:785–788. doi: 10.1038/nature05528. [DOI] [PubMed] [Google Scholar]
- Jung J., Kim M., Kim M. J., Kim J., Moon J., Lim J. S., Kim M., Lee K. Translationally controlled tumor protein interacts with the third cytoplasmic domain of Na,K-ATPase alpha subunit and inhibits the pump activity in HeLa cells. J. Biol. Chem. 2004;279:49868–49875. doi: 10.1074/jbc.M400895200. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Khalyfa A., Bourbeau D., Chen E., Petroulakis E., Pan J., Xu S., Wang E. Characterization of elongation factor-1A (eEF1A-1) and eEF1A-2/S1 protein expression in normal and wasted mice. J. Biol. Chem. 2001;276:22915–22922. doi: 10.1074/jbc.M101011200. [DOI] [PubMed] [Google Scholar]
- Langdon J. M., Vonakis B. M., MacDonald S. M. Identification of the interaction between the human recombinant histamine releasing factor/translationally controlled tumor protein and elongation factor-1 delta (also known as eElongation factor-1B beta) Biochim. Biophys. Acta. 2004;1688:232–236. doi: 10.1016/j.bbadis.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Li F., Zhang D., Fujise K. Characterization of fortilin, a novel antiapoptotic protein. J. Biol. Chem. 2001;276:47542–47549. doi: 10.1074/jbc.M108954200. [DOI] [PubMed] [Google Scholar]
- Liu H., Peng H. W., Cheng Y. S., Yuan H. S., Yang-Yen H. F. Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol. Cell. Biol. 2005;25:3117–3126. doi: 10.1128/MCB.25.8.3117-3126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- MacDonald S. M., Rafnar T., Langdon J., Lichtenstein L. M. Molecular identification of an IgE-dependent histamine-releasing factor. Science. 1995;269:688–690. doi: 10.1126/science.7542803. [DOI] [PubMed] [Google Scholar]
- Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkenberger J. L., Horning S., Klocke B., Roth K., Korsmeyer S. J. Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes Dev. 2000;14:23–27. [PMC free article] [PubMed] [Google Scholar]
- Shah A. H., Tabayoyong W. B., Kimm S. Y., Kim S. J., Van Parijs L., Lee C. Reconstitution of lethally irradiated adult mice with dominant negative TGF-beta type II receptor-transduced bone marrow leads to myeloid expansion and inflammatory disease. J. Immunol. 2002;169:3485–3491. doi: 10.4049/jimmunol.169.7.3485. [DOI] [PubMed] [Google Scholar]
- Shen M. M., Wang H., Leder P. A differential display strategy identifies Cryptic, a novel EGF-related gene expressed in the axial and lateral mesoderm during mouse gastrulation. Development. 1997;124:429–442. doi: 10.1242/dev.124.2.429. [DOI] [PubMed] [Google Scholar]
- Tatsuka M., Mitsui H., Wada M., Nagata A., Nojima H., Okayama H. Elongation factor-1 alpha gene determines susceptibility to transformation. Nature. 1992;359:333–336. doi: 10.1038/359333a0. [DOI] [PubMed] [Google Scholar]
- Thaw P., Baxter N. J., Hounslow A. M., Price C., Waltho J. P., Craven C. J. Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones. Nat. Struct. Biol. 2001;8:701–704. doi: 10.1038/90415. [DOI] [PubMed] [Google Scholar]
- Tuynder M., et al. Translationally controlled tumor protein is a target of tumor reversion. Proc. Natl. Acad. Sci. USA. 2004;101:15364–15369. doi: 10.1073/pnas.0406776101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuynder M., Susini L., Prieur S., Besse S., Fiucci G., Amson R., Telerman A. Biological models and genes of tumor reversion: cellular reprogramming through tpt1/TCTP and SIAH-1. Proc. Natl. Acad. Sci. USA. 2002;99:14976–14981. doi: 10.1073/pnas.222470799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D. G., Bhatt S., Herrmann B. G. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Yang T., Buchan H. L., Townsend K. J., Craig R. W. MCL-1, a member of the BLC-2 family, is induced rapidly in response to signals for cell differentiation or death, but not to signals for cell proliferation. J. Cell. Physiol. 1996;166:523–536. doi: 10.1002/(SICI)1097-4652(199603)166:3<523::AID-JCP7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Yang Y., Yang F., Xiong Z., Yan Y., Wang X., Nishino M., Mirkovic D., Nguyen J., Wang H., Yang X. F. An N-terminal region of translationally controlled tumor protein is required for its antiapoptotic activity. Oncogene. 2005;24:4778–4788. doi: 10.1038/sj.onc.1208666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarm F. R. Plk phosphorylation regulates the microtubule-stabilizing protein TCTP. Mol. Cell. Biol. 2002;22:6209–6221. doi: 10.1128/MCB.22.17.6209-6221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenofsky R., Cereghini S., Krowczynska A., Brawerman G. Regulation of mRNA utilization in mouse erythroleukemia cells induced to differentiate by exposure to dimethyl sulfoxide. Mol. Cell. Biol. 1983;3:1197–1203. doi: 10.1128/mcb.3.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.