Abstract
The ubiquitin-associated (UBA) domain of the mRNA nuclear export receptor Mex67 helps in coordinating transcription elongation and nuclear export by interacting both with ubiquitin conjugates and specific targets, such as Hpr1, a component of the THO complex. Here, we analyzed substrate specificity and ubiquitin selectivity of the Mex67 UBA domain. UBA-Mex67 is formed by three helices arranged in a classical UBA fold plus a fourth helix, H4. Deletion or mutation of helix H4 strengthens the interaction between UBA-Mex67 and ubiquitin, but it decreases its affinity for Hpr1. Interaction with Hpr1 is required for Mex67 UBA domain to bind polyubiquitin, possibly by inducing an H4-dependent conformational change. In vivo, deletion of helix H4 reduces cotranscriptional recruitment of Mex67 on activated genes, and it also shows an mRNA export defect. Based on these results, we propose that H4 functions as a molecular switch that coordinates the interaction of Mex67 with ubiquitin bound to specific substrates, defines the selectivity of the Mex67 UBA domain for polyubiquitin, and prevents its binding to nonspecific substrates.
INTRODUCTION
Ubiquitylation has emerged as a major regulatory mechanism for highly diverse cellular functions. Ubiquitin can be conjugated to its target as a monomer or a polyubiquitin chain that can be linked through several different lysine residues. Polyubiquitin chains linked by Lys48 promote proteasome-dependent degradation of target proteins, whereas monoubiquitination or Lys63-linked polyubiquitin is involved in the nonproteolytic regulation of various functions such as vesicular transport or DNA repair (Pickart and Eddins, 2004).
The diverse functions generated by different ubiquitin modifications are mediated through effector proteins that contain ubiquitin binding motifs that can be classified into six major groups according to their precise structural fold (for reviews, see Hicke et al., 2005; Harper and Schulman, 2006). Crystallography and nuclear magnetic resonance (NMR) have shown that, although they show relatively low sequence conservation, three of these domains, “ubiquitin associated” (UBA), “coupling of ubiquitin conjugation to endoplasmic reticulum” (CUE), and “GGA and Tom1” (GAT), adopt a compact helical fold consisting in three α-helices connected by two short loops that generates a large hydrophobic surface, mainly constituted by the C terminus of the helix 1, loop 1, and helix 3, which forms the principal interface with ubiquitin (for review, see Harper and Schulman, 2006). Lys63-linked polyubiquitins interact with UBA domains in a 1:1 stoichiometry, whereas with Lys48-linked diubiquitin, a single UBA domain binds both ubiquitins (Varadan et al., 2004; Trempe et al., 2005; Varadan et al., 2005). A recent analysis of the ubiquitin binding properties of >25 UBA domains defined four classes of UBA domains ranging from no interaction, recognition of linkage-specific chains, to binding of both mono- and polyubiquitin (Raasi et al., 2005), leading to the proposal that non-UBA sequences present in full-length proteins could modulate the interactions of a given UBA domain.
In Saccharomyces cerevisiae, two ubiquitin ligases, Tom1 and Rsp5, are involved in the regulation of poly(A)+ RNA nuclear export (Duncan et al., 2000; Neumann et al., 2003; Rodriguez et al., 2003). The stability of Hpr1 (a component of the THO/TREX complex that couples transcription elongation to mRNA nuclear export) is modulated by the ubiquitin/proteasome pathway in an Rsp5-dependent manner (Gwizdek et al., 2005). Mature yeast messenger ribonucleoproteins (mRNPs) are exported through nuclear pores by the Mex67:Mtr2 heterodimer (TAP:p15 in metazoans). Mex67:Mtr2 is recruited to mRNAs through RNA binding adaptors, including components of the THO/TREX complex (Rodriguez et al., 2004). TAP has a modular structure that includes a 70-residue C-terminal domain with three helices arranged in a classical UBA fold plus an additional fourth helix (Liker et al., 2000; Grant et al., 2002; Grant et al., 2003). We recently showed that Mex67 can interact with ubiquitin and polyubiquitylated proteins both in vitro and in vivo through its UBA domain (Gwizdek et al., 2006). Surprisingly, this UBA domain also interacts with specific partners, including Hpr1. This interaction gives ubiquitylated Hpr1 transient protection from proteosomal degradation. In addition to its mRNP nuclear export function, the Mex67 UBA domain also contributes to the recruitment of Mex67 to transcribing genes, possibly through its interaction with Hpr1. Altering ubiquitylation of Hpr1 affects these UBA-dependent functions of Mex67 (Gwizdek et al., 2006).
Here, we examine the basis for the interaction of the Mex67 UBA domain with both ubiquitin and Hpr1. Our in vitro and in vivo data show that helix H4 influences these interactions and suggest a model in which the affinity of the Mex67 UBA domain for ubiquitin is enhanced through a conformational change induced by its binding to Hpr1.
MATERIALS AND METHODS
Plasmids and Cloning
Plasmid constructs encoding the 57-amino acid (543–599) UBA domain of Mex67 and the 57 amino acid (342–398) UBA domain of Rad23 fused to glutathione S-transferase (GST) or LexA were described previously (Gwizdek et al., 2006). The DNA fragment encoding amino acids 543–589 of the UBA domain of Mex67 (UBAΔH4-Mex67) was amplified by polymerase chain reaction (PCR) by using pGEX-4T-1-UBA-Mex67 as a template, and it was cloned as a BamHI–SalI fragment into pGEX4T-1 and pLex10 vector (Invitrogen, Carlsbad, CA) to generate plasmids encoding GST-UBAΔH4-Mex67 and lexA-UBAΔH4-Mex67, respectively. A NcoI–NotI DNA fragment encoding amino acids 527–589 of Mex67 was amplified by PCR and cloned into the same restriction sites of pGex4T-1-Mex67 to obtain the pGex4T-1-Mex67ΔH4 (1-589) construct. Mutations of Phe596 and Val597 into Ala were introduced using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). pRS314-Mex67-ΔH4-3HA was generated as described previously for pRS314-Mex67-3HA by using a EcoNI–NheI DNA fragment encoding amino acids 494–589 of Mex67. The plasmid was transformed into the MEX67 shuffle strain (Strasser et al., 2000) before selection on 5-fluoroorotic acid plates. Expression levels of Mex67-3HA and Mex67-ΔH4-3HA in the shuffled strains analyzed by Western blotting with anti-hemagglutinin (HA) antibodies were found to be similar.
Protein Purification
Recombinant proteins were expressed in Escherichia coli BL21(DE3). Strains transformed with pGEX4T-1-Mex67, pGEX4T-1-Mex67ΔUBA, or pGEX4T-1-Mex67ΔH4 were grown at 37°C up to OD600 = 0.6. Protein expression was then induced for 48 h at 18°C with 0.5 mM isopropyl β-d-thiogalactoside (IPTG) after cold and chemical shocks. GST-UBA-Mex67 and GST-UBA-Rad23 fusion proteins were expressed and induced as described previously (Gwizdek et al., 2006). GST-UBA-Mex67 was cleaved with thrombin before nuclear magnetic resonance (NMR) analysis. GST-UBAΔH4-Mex67 or His-UBA-Mex67 were expressed in E. coli grown at 37°C to an OD600 of 0.6 and induced with 0.5 mM IPTG for 24 h at 23°C. GST- or His-fusion proteins were then purified on glutathione-Sepharose 4B beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) or nickel-agarose beads (QIAGEN, Hilden, Germany), respectively, according to the manufacturer's instructions in the presence of 0.1% Triton X-100. The His-UBA-Mex67/GST-Hpr1 complex was formed by mixing purified recombinant His-UBA-Mex67 and GST-Hpr1 (Gwizdek et al., 2005) proteins followed by overnight dialysis at 4°C against 50 mM Tris, pH 7.5, 150 mM NaCl, and 10% glycerol and purification on glutathione-Sepharose beads.
Solution Structure of the Mex67 UBA Domain
NMR-spectra were acquired at 27°C on a Bruker Avance 500 spectrometer equipped with triple resonance cryoprobe. All experiments were performed on a uniformly 15N- and 13C-labeled sample containing 0.55 mM protein, 25 mM NaH2PO4, and 50 mM NaCl at pH 7.4 in 90% H2O, 10% D2O. All spectra were processed using the program XWINNMR (Bruker BioSpin, Rheinstetten, Germany). Backbone resonance assignment was carried out using the pair of triple resonance experiments CBCA(CO)NH/CBCANH (Grzesiek and Bax, 1992a,b). Side-chain resonances were identified using HBHA(CO)NH, HCCH-total correlation spectroscopy (TOCSY), and HCCH-correlation spectroscopy (COSY) (Kay et al., 2002) experiments. Interproton distance information for structure calculation was derived from a 15N-nuclear Overhauser effect spectroscopy (NOESY)-heteronuclear single quantum correlation (HSQC) spectrum and a pair of 13C-NOESY-HSQC spectra recorded for both aliphatic and aromatic resonances (Clore and Gronenborn, 1992). All NOESY spectra were acquired using a mixing time of 100 ms. The program CCPNMR Analysis (Vranken et al., 2005; Hajduk et al., 1997) was used for resonance assignment of the protein.
Secondary structure predictions were made using the chemical shift-based dihedral angle prediction software TALOS (Cornilescu et al., 1999), and they were confirmed by manual analysis of 15N- and 13C-3D NOESY spectra. Distance restraints were manually refined by iterative assignment corrections/structure calculations by using XPLOR-NIH (Schwieters et al., 2003). The assigned distance restraints were categorized in four classes for these calculations: I, <3.0 Å; II, <4.0 Å; III, <5.0 Å; and IV, <6.0 Å. Final structure calculations were based on a total of 1206 nuclear Overhauser effect (NOE)-derived interresidue distance restraints (class I, 10; II, 129; III, 318; and IV, 749), and 26 distance restraints mimicking H-bonds identified using characteristic short range NOE patterns (Table 1). All structures were subjected to a refinement in water by using XPLOR-NIH (Schwieters et al., 2003) and the parallhdg5.3 force fields (Linge et al., 2003). The final ensemble of 20 structures selected by lowest NOE energy showed the following distribution in the Ramachandran plot: 82.5% most favored, 12.8% additionally allowed, 2.3% generously allowed, and 2.5% disallowed. For analysis of the refined ensemble MOLMOL (Koradi et al., 1996), Procheck-NMR (Laskowski et al., 1996) were used. Structural information derived from an ensemble of 20 structures is shown in Table 1. Structural coordinates have been registered in Protein Data Bank (accession code: 2jp7).
Table 1.
NMR and refinement statistics for protein structures
| Protein | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 1206 |
| Intraresidue | 4 |
| Interresidue | 1202 |
| Sequential (|i-j|= 1) | 432 |
| Medium range (|i-j|<4) | 429 |
| Long range (|i-j| >4) | 294 |
| Ambiguous | 47 |
| Hydrogen bonds | 26 |
| Total dihedral angle restraints | 0 |
| Structure statistics | |
| Violations (mean ± SD) | |
| Distance constraints (Å) | 0.0354 ± 0.0007 |
| Maximum distance constraint violation (Å) | 0.437 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0171 ± 0.0003 |
| Bond angles (°) | 2.04 ± 0.04 |
| Impropers (°) | 2.3 ± 0.1 |
| Avg. rmsd of 20 structures to the mean (Å) | |
| Heavy (ordered regions) | 1.19 ± 0.16 (0.83 ± 0.13) |
| Backbone (ordered regions) | 0.47 ± 0.11 (0.32 ± 0.10) |
Chromatin immunoprecipitation analysis, fluorescence in situ hybridization (FISH), and fluorescence titration experiments were performed as described in Gwizdek et al. (2006).
Surface Plasmon Resonance (SPR)
Quantitative interaction analyses were performed on a BIAcore 2000 instrument (BIAcore, Uppsala, Sweden). GST and GST-Hpr1 were amine-coupled to CM5 sensor chip to a density (RL) of 2500 and 7000 resonance units (RUs), respectively. A mock-immobilized surface was used as a control. Coupling was performed using N-hydroxysuccinimide/N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide reagents, ethanolamine, and HBS-EP buffer (10 mM HEPES, pH 7.5, 150 mM NaCl, 3 mM EDTA, and 0005% surfactant P20) (BIAcore). Binding experiments were performed in HSP-EP buffer at 25°C with 5 μM His-UBA-Mex67 as analyte in the mobile phase. The analyte was injected at a flow rate of 5 μl/min during 6 min alone or together with different concentrations of K48-linked tetraubiquitin. Specific sensorgrams were obtained after subtracting binding of the analyte to the mock-immobilized surface. After each run, surfaces were regenerated using 1 M NaCl, 50 mM NaOH. A control binding assay conducted after each round of regeneration shows a reproducible response. Experimental equilibrium responses and Rmax values were obtained using a “two-state reaction model with conformation change” (BIAevaluation software; BIAcore). Each experiment has been repeated at least twice.
RESULTS
Solution Structure of the Mex67UBA Domain
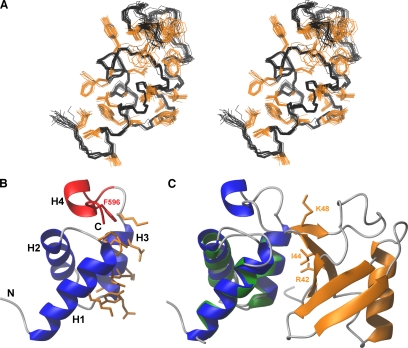
To elucidate the basis for the interaction of the Mex67 UBA domain with its partners, we obtained the solution NMR structure of the Mex67 UBA domain (Figure 1A) by using a set of 1232 structural restraints (Table 1), from which a well-defined ensemble of 20 structures was obtained that showed an average root mean square deviation (rmsd) to the mean structure of 0.47 Å for all backbone-heavy atoms, and an average rmsd to the mean structure of 1.19 Å when all heavy atoms were included. The structure contained four α-helices (residues 546–559 [H1], 563–572[H2], 578–586[H3], and 593–596[H4]) and overall, the fold is very similar to that of the TAP UBA domain (Grant et al., 2002; 2003). In contrast to other UBA folds, the Mex67 UBA domain has an additional C-terminal helix (H4) that helps stabilize the core by burying hydrophobic side chains of helix H1 and loop 1 (Figure 1). The solution structures of the Mex67 and TAP UBA domains can be superimposed with rmsd of 1.4 Å, consistent with their sequence homology (Supplemental Figure 1).
Figure 1.
NMR structure of the Mex67 UBA domain and a model of its likely interaction interface with ubiquitin. (A) Stereo view of the ensemble of 20 NMR structures having the lowest total energy. Positions of the backbone-heavy atoms are given in black, whereas those of hydrophobic and aromatic side chains are in orange. (B) Representation of the secondary structure elements of the UBA-Mex67 closest to the mean of the NMR ensemble. The residues of UBA-Mex67 that may interact with monoubiquitin are in orange. The region of the structure deleted in UBAΔH4-Mex67 and Phe596 are highlighted in red. (C) Prediction of the likely docking geometry of the Mex67 UBA domain onto ubiquitin by analogy with the structure of the Ede1:ubiquitin complex (PDB:2G3Q). Ubiquitin is orange, and the Ede1 UBA domain is green. Helices H1-H3 of the UBA-Mex67 domain (blue) were superimposed on the corresponding helices in the Ede1 structure. The side chains of the key residues Lys48, Ile44, and Arg42 on ubiquitin are also shown in orange.
UBA-Mex67 retains several features that in other UBA domains are involved in the interaction with ubiquitin. Thus, residues in both helix H1, loop1, and helix H3 have been reported to interact with ubiquitin in UBA:monoubiquitin complexes (Kang et al., 2003; Mueller et al., 2004; Ohno et al., 2005). Analogous residues are present in Mex67, and they are exposed on the surface. These include the hydrophobic residues Tyr577, Glu578, Val579, Ile581, Lys582, Phe584, and Gln585 of helix H3 and Lys554 and Glu558 of the helix H1 (Figure 1C). Mex67 and TAP show a high degree of structural homology with the UBA domain of yeast Ede1 (Swanson et al., 2006). The structures of the Mex67 and Ede1 UBA domains can be superimposed with rmsd of 1.13 Å, and many residues in either the hydrophobic core or binding to ubiquitin are conserved between these UBA domains (Figure 1C). However, in both Mex67 and TAP, loop 1 is shorter than in other UBA folds and likely forms hydrophobic interactions with H4, suggesting that interaction with ubiquitin might be altered in the Mex67 UBA domain.
Helix H4 Influences the UBA-Mex67:Ubiquitin Interaction
Titrations using fluorescently labeled ubiquitin and recombinant UBA-Mex67 (Table 2) showed that, in contrast to UBA2-Rad23, UBA-Mex67 had low affinity (KD = 400 μM) for monoubiquitin, regardless the tag used. However, deletion of H4 (UBAΔH4-Mex67) increased the affinity of UBA-Mex67 for monoubiquitin substantially (KD = 63 μM), suggesting that the presence of helix H4 may impede the interaction with ubiquitin. Deletion of helix H4 allowed UBA-Mex67 to bind Lys48-linked tetraubiquitin with a slightly higher affinity than monoubiquitin (KD = 18 μM). It should be noted that full-length Mex67 was able to interact with ubiquitin via its UBA domain, suggesting a conformational control of the UBA-Mex67 and H4 by other domains of the protein in vitro. However, deletion of H4 in the context of full-length Mex67 still increased by twofold the affinity of Mex67 for tetraubiquitin without affecting the KD for monoubiquitin (Table 2). Consistently, pull-down experiments using GST-Mex67 or GST-Mex67ΔH4 and extracts from Δerg6 cells revealed that deletion of H4 improves to some extent the ability of Mex67 to interact with polyubiquitylated proteins (data not shown). We also investigated the effect of point mutations in helix H4 on the UBA-Mex67:ubiquitin interaction and found that, whereas UBA-Mex67(V597A) was indistinguishable from wild type, mutation of F596A (that faces and probably interacts with residues in loop 1 allowed binding to ubiquitin (KD = 68 μM). Because the corresponding mutation in the TAP UBA domain (F617A) causes a conformational change in helix H4, these data should not be interpreted as indicating a direct involvement of Phe596 in preventing the interaction with ubiquitin but rather as a regulatory role on residues involved in ubiquitin binding (Grant et al., 2003).
Table 2.
In vitro measurement of dissociation constants of Mex67 and Rad23-derived GST and His6 fusion proteins of the respective UBA domains for mono- and Lys48-linked tetraubiquitin (K48-Ub4)
| Recombinant protein | KD for mono-Ub (μM) | KD for K48-Ub4 (μM) |
|---|---|---|
| GST-UBA2-Rad23 | 19 ± 2 | 7.8 ± 1.2 |
| His-UBA2-Rad23 | 12.9 ± 3.2 | 6.1 ± 0.2 |
| GST-UBA-Mex67 | 400 | >300 |
| His-UBA-Mex67 | 400 | >400 |
| UBA-Mex67 | Not determined | >300 |
| GST-UBAΔH4-Mex67 | 63 ± 4 | 18 ± 2.4 |
| UBA-Mex67(V597A) | Not determined | >300 |
| UBA-Mex67(F596A) | Not determined | 68 ± 5 |
| His-UBA-Mex67:GST-Hpr1 | Not detectable | 7.5 ± 3 |
| GST-Hpr1 | No binding | No binding |
| GST-Mex67 | 12 ± 3 | 6.2 ± 0.6 |
| GST-Mex67ΔUBA | 300–400 | 300 |
| GST-Mex67ΔH4 | 13 ± 2.1 | 3.7 ± 1.2 |
The association with ubiquitin was measured in vitro by monitoring the fluorescence enhancement of fluorescently labeled ubiquitin.
Role of Helix H4 in the Interaction with Hpr1
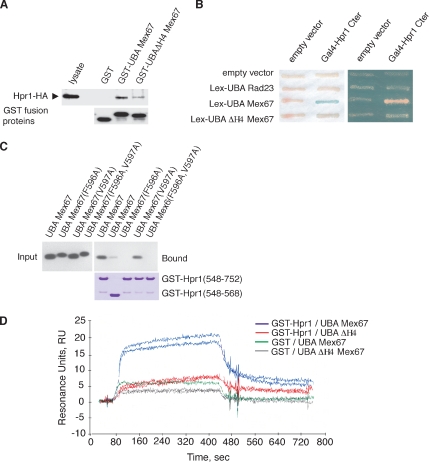
Besides binding ubiquitin, the UBA-Mex67 UBA also interacts specifically with the C-terminal domain of Hpr1 (Gwizdek et al., 2006). GST-pull-down and two-hybrid assays both showed that the strength of the interaction of UBA-Mex67 with Hpr1 was greatly reduced by deletion of helix H4 (Figure 2, A and B) or by the F596A point mutation (Figure 2C), indicating that helix H4 is also important for the interaction with Hpr1. It was unlikely that this loss of function was associated with UBA-Mex67 being denatured by the removal of helix H4, because these mutants showed a gain of function in terms of affinity for ubiquitin.
Figure 2.
Role of helix H4 of the Mex67 UBA domain in its interaction with Hpr1. (A) Pull-down assays using indicated GST fusion recombinant proteins and lysates from cells expressing HA-tagged versions of Hpr1. (B) Two-hybrid analysis of the indicated LexA-fusion proteins versus Gal4-Hpr1 C terminus. Expression levels of wild-type and deletion mutant of UBA-Mex67 were analyzed by Western blotting and found to be similar. (C) Purified recombinant wild-type and mutant forms of UBA-Mex67 and indicated GST-Hpr1 fusion proteins were mixed (input) and complexes analyzed after purification on glutathione-Sepharose beads (bound) by Western blotting by using an anti-Mex67 antibody. (D) Duplicate SPR sensorgrams of the interaction between 2.5 μM His-UBA-Mex67 or UBAΔH4-Mex67 as analyte on amine-coupled GST or GST–Hpr1 surfaces. The kinetic fit was obtained using BIAcore 2000 BIAevaluation software (two-state reaction with conformation change).
The interaction between UBA-Mex67 and Hpr1 was also analyzed by SPR by using GST-Hpr1 as ligand and recombinant His-UBA-Mex67 or UBAΔH4-Mex67 as the mobile phase. Specific association and dissociation could be observed between GST-Hpr1 and UBA-Mex67 (2.5 μM) but not with GST alone (Figure 2D). Consistent with the GST-pull-down and two-hybrid assays, UBAΔH4-Mex67 bound GST-Hpr1 with much lower affinity. The best fits for UBA-Mex67 required a two-state reaction model that included a conformation change and not the classical 1:1 Langmuir model (BIAevaluation software; BIAcore). The kinetically determined KD value of the UBA-Mex67:Hpr1 interaction was ∼4 μM, whereas k2 and k−2 corresponding to the conformation change were 6.5 × 10−3 and 10−3 s−1, respectively (Table 3), consistent with there being a conformational change in the UBA-Mex67:Hpr1 complex following the initial binding.
Table 3.
SPR measurements of the interaction of immobilized GST-Hpr1 with His-UBA-Mex67 as an analyte in the absence or in the presence of Lys48-linked tetraubiquitin (K48-Ub4)
| Analyte | Apparent KD (M) | Rmax (RU) | k2 (s−1) | k−2 (s−1) |
|---|---|---|---|---|
| UBA (5 μM)+ | ||||
| 0 | 3.7 × 10−6 | 31 | 6.5 × 10−3 | 9.9 × 10−4 |
| K48-Ub4 (1 μM) | 4.3 × 10−6 | 83 | 6.7 × 10−3 | 9.8 × 10−4 |
| K48-Ub4 (3.5 μM) | 5.0 × 10−6 | 133 | 7.1 × 10−3 | 12 × 10−4 |
| K48-Ub4 (7 μM) | 6.1 × 10−6 | 146 | 6.6 × 10−3 | 9.0 × 10−4 |
Apparent KD corresponds to KD of the interaction between GST-Hpr1 and UBA-Mex67 ± K48-Ub4 without considering the conformational change whose k2 and k−2 are indicated separately. Data correspond to mean of at least two independent experiments.
Hpr1 Binding Facilitates the UBA-Mex67:Ubiquitin Interaction
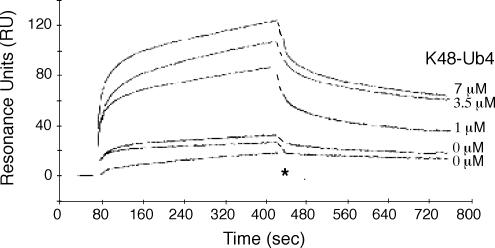
These results prompted us to investigate whether Hpr1 binding altered the affinity of UBA-Mex67 for ubiquitin. Although the purified recombinant GST-Hpr1:His-UBA-Mex67 complex did not have measurable affinity for monoubiquitin, it clearly interacted with Lys48-linked tetraubiquitin with a KD of 7.5 ± 3 μM, comparable with that observed for Rad23-UBA (Table 2). Consistently, we did not detect an interaction between GST-Hpr1:UBA-Mex67 neither with 1.4 nor 10 μM monoubiquitin by SPR (data not shown). However, when increasing concentrations of K48-linked tetraubiquitin (0–7 μM) were used in SPR studies together with 5 μM His-UBA-Mex67 for binding to immobililized GST-Hpr1, the signal increased fivefold in the presence of 7 μM ubiquitin, consistent with the formation of a GST-Hpr1:UBA-Mex67:tetraubiquitin tripartite complex and with every GST-Hpr1:UBA-Mex67 complex binding ubiquitin (Figure 3). Both the equilibrium RU values as well as the kinetically determined value of global KD indicated that the KD for the interaction between the GST-Hpr1:UBA-Mex67 complex and tetraubiquitin was in the micromolar range and so in reasonable agreement with the fluorescence titration data (Tables 2 and 3). In addition, k2 and k−2, corresponding to the putative conformation change after formation of the GST-Hpr1:UBA-Mex67 complex, were not altered by the presence of tetraubiquitin (Table 3). These results indicate that Hpr1 binding, likely followed by a conformational change, influences the binding of UBA-Mex67 to ubiquitin.
Figure 3.
Tetraubiquitin binding to UBA-Mex67 requires interaction of the Mex67 UBA domain with Hpr1. SPR sensorgrams of interaction of 5 μM His-UBA-Mex67 with immobilized GST-Hpr1 in the absence or in the presence of increasing concentrations of Lys48-linked tetraubiquitin (K48-Ub4). The association of 1 μM Lys48-Ub4 with the GST–Hpr1 surface is shown as control (*). Interruption of association and dissociation curves due to alterations in refractive index during buffer changes were artificially filled with dashed lines.
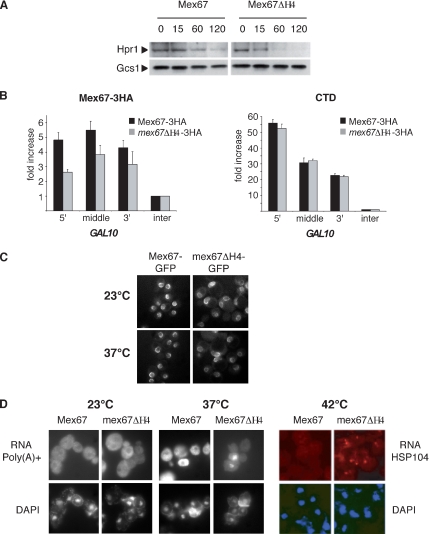
Deletion of Helix H4 Reduces Recruitment of Mex67 to mRNA In Vivo and Produces an mRNA Nuclear Export Defect
To confirm the contribution of H4 to the function of the UBA-Mex67 domain in vivo in the context of the full-length protein, we analyzed the consequences of deleting helix H4, by using yeast strains expressing HA-tagged-wild type Mex67 (Mex67-HA) or ΔH4-Mex67 (mex67-ΔH4-HA) that were generated from the MEX67 shuffle strain. Because binding of UBA-Mex67 results in the transient protection of ubiquitylated Hpr1 from proteasome-dependent degradation (Gwizdek et al., 2006), the stability of Hpr1 was analyzed in both strains. Heat-induced degradation of Hpr1 was accelerated in mex67-ΔH4-HA compared with Mex67-HA cells (Figure 4A) consistent with the weak interaction observed between ΔH4-UBA-Mex67 and Hpr1 in vivo (Figure 2). Because the Mex67 UBA domain is required for both its recruitment to transcribing genes and mRNA nuclear export (Gwizdek et al., 2006), we used chromatin immunoprecipitation analysis to assess the consequences of deleting helix H4 on Mex67 recruitment to active genes. Deletion of helix H4 in mex67-ΔH4-HA cells resulted in decreased cotranscriptional recruitment of Mex67 on GAL10 (Figure 4B). As shown previously for deletion of the entire UBA domain, deletion of helix H4 did not influence recruitment of RNA polymerase II (CTD) to GAL10, consistent with helix H4 contributing to the recruitment of Mex67 to actively transcribing genes. Deletion of helix H4 did not affect the ability of Mex67-GFP to localize at the nuclear pore complex in vivo (Figure 4C). However, absence of helix H4 resulted in lower levels of mRNA export when the distributions of poly(A)+ RNA or the specific HSP104 transcript were examined in mex67-ΔH4-HA cells by FISH and compared with wild-type Mex67-HA cells (Figure 4D). In agreement with observations in mex67-ΔUBA-HA cells (Gwizdek et al., 2006), no poly(A)+ RNA export defect was detected at 23°C. However, 20% of mex67-ΔH4-HA cells accumulated poly(A)+ RNA in the nucleus after a 3-h shift at 37°C, whereas mRNA export was unaffected in wild-type cells. The export defect was more pronounced for HSP104, as these transcripts accumulated within a nuclear dot in 70% of mutant cells after a 30-min shift to 42°C. Thus, the weaker Mex67 cotranscriptional binding observed in mex67-ΔH4-HA cells was paralleled by an mRNA export defect. Overall, both deletion of helix H4 alone or deletion of the entire UBA domain produce similar mRNA nuclear export phenotypes consistent with helix H4 regulating at least some functions of the Mex67 UBA domain.
Figure 4.
Role of helix H4 of the Mex67 UBA domain in cotranscriptional recruitment of Mex67 and mRNA export. (A) Steady-state level of Hpr1 expression was analyzed in Mex67-3HA and mex67-ΔH4-3HA cells shifted to 37°C for different incubation times. The Gcs1 protein was used as a loading control. (B) Chromatin immunoprecipitation experiments on GAL10 were performed with extracts prepared from Mex67-3HA or mex67-ΔH4-3HA strains shifted to galactose for 2 h by using the anti-HA or anti-CTD antibodies. Significance of the differences observed for Mex67 recruitment between wild-type and mutant cells was evaluated using Student's t test. Values were found to be p = 0.004 for the 5′, p = 0.02 for the middle, and p = 0.07 for the 3′. (C) Subcellular localization of Mex67 was analyzed by direct green fluorescent protein (GFP) fluorescence on Mex67-GFP and mex67ΔH4-GFP cells. (D) Subcellular localization of poly(A)+ RNA was analyzed by FISH by using oligo(dT) Cy3 as probe in cells kept at 23°C or shifted to 37°C for 3 h (left). Alternatively, HSP104 mRNA was analyzed by FISH by using a specific probe in Mex67-3HA or in mex67ΔH4-3HA shuffle strains after a 30-min shift to 42°C (right).
DISCUSSION
Our data show that helix H4 of the UBA domain of Mex67 influences its interaction with other partners in different ways. Thus, whereas in vitro deletion of helix H4 or the F596A mutation increases the affinity of the UBA domain for ubiquitin and ubiquitin multimers, it reduces the affinity for Hpr1. Moreover, deletion of helix H4 reduces both cotranscriptional recruitment of Mex67 to mRNPs and generates an mRNA export defect at 37°C.
It is unlikely that deletion of helix H4 results in a complete loss of structure of the UBA domain, because these constructs retain the ability to bind ubiquitin. Instead, helix H4 seems to be involved directly in Hpr1 binding, and the pronounced mRNA export defect seen in mex67-ΔH4-HA cells would be consistent with the Hpr1:Mex67UBA interaction being an important step in the overall export pathway. However, it seems unlikely that helix H4 alone can account for all of the effects seen with deletion of the entire UBA domain, because previous studies have, for example, shown a role for the intact domain in interacting with phenylalanine-glycine nucleoporins to facilitate translocation through nuclear pore complexes (Grant et al., 2003). Our results indicate that the core of the UBA domain (helices H1–H3) is responsible for ubiquitin binding, whereas H4 participates to Hpr1 binding and also seems to regulate the interaction of the Mex67 UBA domain with ubiquitin. It should be noted that although H4 is necessary for Hpr1 binding, it is likely not sufficient for this interaction as suggested by the residues within the UBA domain that are affected upon Hpr1 binding in NMR spectra (our unpublished observations). Because both ubiquitin binding and Hpr1 stabilization are important for cotranscriptional recruitment and mRNA export, interfering with either UBA domain or H4 results in both cotranscriptional recruitment and mRNA export defects.
Based on these results, we hypothesize that, in vivo, helix H4 may function to coordinate the ubiquitin binding activity of the Mex67 UBA domain with recognition of specific substrates, in particular Hpr1, and also prevent binding to nonspecific substrates. Such a function would probably require a two-stage mechanism for the recognition of ubiquitinated Hpr1 by UBA-Mex67, with an initial helix H4-dependent binding of Hpr1 followed by an interaction with ubiquitins conjugated to Hpr1. This type of UBA-Mex67:ubiquitin interaction might also occur with other partners. Substrate specificity and ubiquitin linkage selectivity of UBA domains seem to be interdependent and do not exclusively rely on intrinsic properties of UBA domains, but rather they are precisely controlled by their molecular environment, as we show here for UBA-Mex67. For example, a specific interaction of the HIV-1 accessory protein Vpr with hHR23A-UBA1 domain involves loop 1 (Withers-Ward et al., 2000), whereas interaction of the N-terminal Ubiquitin-like domain of hHR23A with hHR23A-UBA1 modulates its selectivity for polyubiquitin linkage.
Comparison of the structure of the Mex67 UBA domain with the structures of other UBA:ubiquitin conjugates indicates that much of the interaction surface has been retained. For example, when the structure of the Mex67 UBA domain is superimposed on the UBA domain in the Ede1:ubiquitin complex (Swanson et al., 2006), much of the general character and structure of the interaction interface is retained for helix H3 (Figure 1). How might helix H4 influence the interactions? Steric hindrance seems unlikely, because in our model for the UBA-Mex67:monoubiquitin complex, helix H4 does not contact ubiquitin, although steric hindrance may possibly be important for binding of Lys48-linked ubiquitin multimers. Alternatively, previous studies have shown that the structure of the TAP-UBA domain is relatively plastic and mutations can introduce movements of helices H1, H2, and H3 that significantly alter interactions with other partners. Structural studies revealed that helix H4 in TAP-UBA is highly mobile and undergoes major conformational changes upon binding FXFG repeats or in the F617A mutant (Grant et al., 2003). This result, together with the involvement of helix H4 in the interaction with Hpr1, would be consistent with the conformational change observed by SPR being associated with a change in helix H4 being triggered by binding to Hpr1. However, a change in the overall structure of the UBA domain associated with its binding Hpr1cannot be formally excluded. In either case, the conformational change induced by Hpr1 would facilitate ubiquitin binding by the UBA domain.
Although further work will be required to define the precise manner in which helix H4 modulates the interactions of the Mex67 UBA domain, our present studies demonstrate how the binding of Hpr1 can influence the affinity of this domain for ubiquitin and thereby provide a novel mechanism by which interactions between different components of the mRNP processing and export machinery can be modulated.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Hurt (Heidelberg University, Germany) and A. Jacquier (Institut Pasteur, Paris, France) for reagents and J. M. Camadro and B. Gontero-Meunier for help with SPR. This study was funded by grants from the Ministèrede la Recherche (ACI BCMS), Agence Nationale pour la Recherche grant BLAN06-1_134099 (to C.D.), the Ligue contre le Cancer (Equipe labelisée), Swiss National Science Foundation grant 102235 (to F.S.), and the Canton de Genève. M.H. is supported by the Ministèredélégué à la Recherche and holds a European Molecular Biology Organization short-term fellowship. C.B. holds a Federation of European Biochemical Societies postdoctoral fellowship.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0153) on May 2, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Clore G. M., Gronenborn A. M. Application of three- and four-dimensional heteronuclear NMR spectroscopy to protein structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 1992;23:43–92. [Google Scholar]
- Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Duncan K., Umen J. G., Guthrie C. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 2000;10:687–696. doi: 10.1016/s0960-9822(00)00527-3. [DOI] [PubMed] [Google Scholar]
- Grant R. P., Hurt E., Neuhaus D., Stewart M. Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat. Struct. Biol. 2002;9:247–251. doi: 10.1038/nsb773. [DOI] [PubMed] [Google Scholar]
- Grant R. P., Neuhaus D., Stewart M. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J. Mol. Biol. 2003;326:849–858. doi: 10.1016/s0022-2836(02)01474-2. [DOI] [PubMed] [Google Scholar]
- Grzesiek S., Bax A. An efficient experiment for sequential backbone amide assignment of medium sized isotopically enriched proteins. J. Magn. Reson. 1992a;99:201–207. [Google Scholar]
- Grzesiek S., Bax A. Correlating backbone amide and sidechain resonances in proteins by multiple triple resonance NMR. J. Am. Chem. Soc. 1992b;115:11620–11621. [Google Scholar]
- Gwizdek C., Hobeika M., Kus B., Ossareh-Nazari B., Dargemont C., Rodriguez M. S. The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J. Biol. Chem. 2005;280:13401–13405. doi: 10.1074/jbc.C500040200. [DOI] [PubMed] [Google Scholar]
- Gwizdek C., Iglesias N., Rodriguez M. S., Ossareh-Nazari B., Hobeika M., Divita G., Stutz F., Dargemont C. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc. Natl. Acad. Sci. USA. 2006;103:16376–16381. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk P. J., Meadows R. P., Fesik S. W. Discovering high-affinity ligands for proteins. Science. 1997;278:497–499. doi: 10.1126/science.278.5337.497. [DOI] [PubMed] [Google Scholar]
- Harper J. W., Schulman B. A. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Hicke L., Schubert H. L., Hill C. P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Kang R. S., Daniels C. M., Francis S. A., Shih S. C., Salerno W. J., Hicke L., Radhakrishnan I. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell. 2003;113:621–630. doi: 10.1016/s0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- Kay L. E., Ikura M., Bax A. Proton-proton correlation via carbon-carbon coupling: a three-dimensional NMR approach for the assignment of aliphatic resonances in proteins labeled with carbon 13. J. Am. Chem. Soc. 2002;112:888–889. [Google Scholar]
- Koradi R., Billeter M., Wüthrich K. MOLMOL a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Liker E., Fernandez E., Izaurralde E., Conti E. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- Mueller T. D., Kamionka M., Feigon J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J. Biol. Chem. 2004;279:11926–11936. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- Neumann S., Petfalski E., Brugger B., Grosshans H., Wieland F., Tollervey D., Hurt E. Formation and nuclear export of tRNA, rRNA and mRNA is regulated by the ubiquitin ligase Rsp5p. EMBO Rep. 2003;4:1156–1162. doi: 10.1038/sj.embor.7400026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno A., Jee J., Fujiwara K., Tenno T., Goda N., Tochio H., Kobayashi H., Hiroaki H., Shirakawa M. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure. 2005;13:521–532. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Eddins M. J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Raasi S., Varadan R., Fushman D., Pickart C. M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Rodriguez M. S., Dargemont C., Stutz F. Nuclear export of RNA. Biol. Cell. 2004;96:639–655. doi: 10.1016/j.biolcel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Rodriguez M. S., Gwizdek C., Haguenauer-Tsapis R., Dargemont C. The HECT ubiquitin ligase Rsp5p is required for proper nuclear export of mRNA in Saccharomyces cerevisiae. Traffic. 2003;4:566–575. doi: 10.1034/j.1600-0854.2003.00115.x. [DOI] [PubMed] [Google Scholar]
- Schwieters C. D., Kuszewski J. J., Tjandra N., Marius C. G. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Strasser K., Bassler J., Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell. Biol. 2000;150(4):695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K. A., Hicke L., Radhakrishnan I. Structural basis for monoubiquitin recognition by the Ede1 UBA domain. J. Mol. Biol. 2006;358:713–724. doi: 10.1016/j.jmb.2006.02.059. [DOI] [PubMed] [Google Scholar]
- Trempe J. F., Brown N. R., Lowe E. D., Gordon C., Campbell I. D., Noble M. E., Endicott J. A. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005;24:3178–3189. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan R., Assfalg M., Haririnia A., Raasi S., Pickart C., Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- Varadan R., Assfalg M., Raasi S., Pickart C., Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol. Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Vranken W. F., et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Withers-Ward E. S., Mueller T. D., Chen I. S., Feigon J. Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr. Biochemistry. 2000;39:14103–14112. doi: 10.1021/bi0017071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.