Abstract
The inhibition of the ubiquitin-dependent proteasome system (UPS) via specific drugs is one type of approach used to combat cancer. Although it has been suggested that UPS inhibition prevents the rapid decay of AU-rich element (ARE)-containing messages, very little is known about the cellular mechanisms leading to this effect. Here we establish a link between the inhibition of UPS activity, the formation of cytoplasmic stress granules (SGs), and mRNA metabolism. The assembly of the SGs requires the phosphorylation of the translation initiation factor eIF2α by a mechanism involving the stress kinase GCN2. On prolonged UPS inhibition and despite the maintenance of eIF2α phosphorylation, SGs disassemble and translation recovers in an Hsp72 protein-dependent manner. The formation of these SGs coincides with the disassembly of processing bodies (PBs), known as mRNA decay entities. As soon as the SGs assemble, they recruit ARE-containing messages such as p21cip1 mRNA, which are stabilized under these conditions. Hence, our findings suggest that SGs could be considered as one of the players that mediate the early response of the cell to proteasome inhibitors by interfering temporarily with mRNA decay pathways.
INTRODUCTION
In eukaryotic cells, the ubiquitin-dependent proteasome system (UPS) is the main nonlysosomal pathway responsible for selective intracellular protein degradation (Nandi et al., 2006). The UPS is composed of the ubiquitin-conjugating system and the multicatalytic protease complex known as the proteasome (Vabulas and Hartl, 2005). The activity of the UPS is not only essential for protein quality control but also for sustaining protein synthesis by supplying the translational machinery with amino acids (Hershko and Ciechanover, 1998; Hershko, 2005). Interfering with the UPS activity by pharmacological agents reduces general translation initiation, followed by the induction of the expression of heat shock proteins such as Hsp72; two main features of cell stress response (Glickman and Maytal, 2002; Pritts et al., 2002; Takimoto and Diggikar, 2002; Vabulas and Hartl, 2005). Conversely, prolonged inhibition of the UPS leads to a partial recovery of general translation via an unknown mechanism (Baugh and Pilipenko, 2004; Ding et al., 2006).
The UPS has also been implicated in RNA metabolism. It was shown that interfering with UPS activity prevents the rapid decay of a reporter message containing AU-rich elements (AREs) in the 3′ untranslated region (3′UTR; Laroia et al., 1999). The effect, however, of UPS inhibition on endogenous ARE-containing messages is not known. The AREs consist of multiple copies of the destabilizing sequence AUUUA, responsible for the rapid decay of many short-lived mRNAs including cytokines, growth factors, and proto-oncogenes (Bevilacqua et al., 2003; Barreau et al., 2005). In addition to the AREs, the AU-rich–mediated decay (AUMD) pathway involves trans-acting factors known as AU-binding proteins (AUBPs) that positively or negatively affect the AUMD process. AUBPs such as HuR and CUG-BP1 protect ARE-messages from the decay machinery, whereas TTP, KSRP, BRF1, CUG-BP2, and AUF1 target them for rapid degradation (Fan and Steitz, 1998; Stoecklin et al., 2003; Iakova et al., 2004; Raineri et al., 2004; Barreau et al., 2005; Briata et al., 2005).
It is well established that under stress conditions the stabilization of ARE-containing messages correlates with the recruitment of some of the AUBPs (HuR and TTP) to cytoplasmic foci called stress granules (SGs; Andrews et al., 1987; Gallouzi et al., 2000; Kedersha et al., 2000; Stoecklin et al., 2004). Likewise, other RNA-binding proteins (RBPs) known to repress translation, such as TIA/TIAR and FMRP (Fragile X Mental Retardation protein), or to induce mRNA turnover, such as G3BP (ras-GAP SH3 binding protein), also localize to SGs in response to various stresses (Gallouzi et al., 1998; Mazroui et al., 2002; Tourriere et al., 2002; Kedersha et al., 2005; Anderson and Kedersha, 2006). Under these conditions, RBPs and their associated mRNAs are recruited from translating ribosomes to accumulate into SGs as untranslated mRNPs. Because SGs contain proteins involved in both the stabilization and degradation of mRNAs, their assembly might be important for the regulation of mRNA decay during stress.
Recently it has been suggested that under normal growth conditions, mRNA decay occurs in cytoplasmic entities referred to as processing bodies (PBs; Kedersha et al., 2005; Sheth and Parker, 2006). These are dynamic foci where factors involved in different mRNA decay pathways are concentrated (Fenger-Gron et al., 2005; Lykke-Andersen and Wagner, 2005). Examples include factors involved in AUMD, such as TTP and BRF1, in nonsense-mediated decay (NMD) such as Smg (5, 2) and Upf1 and in basic mRNA decay such as the decapping enzymes Dcp1/2 and the exonuclease Xrn1. Therefore, SGs and PBs share several common proteins (Kedersha et al., 2005), raising the possibility that a cross-talk between these two structures is required to ensure cellular protection from harmful conditions and rapid recovery from stress. It is well known that once the stress is relieved, SGs disassemble, and the majority of released mRNPs are recruited back to the translation machinery (Kedersha and Anderson, 2002). These observations indicate that the assembly/disassembly of SGs might constitute an important mechanism to regulate gene expression in response to stress. Because proteasome inhibition induces a typical stress response such as reduction in translation activity (Jiang and Wek, 2005) as well as the overexpression of the Hsp72 protein (Meriin et al., 1998), it is possible that under these conditions the cell forms SGs and directly or indirectly affects the function of PBs, which could explain in part the block in the decay of ARE-containing messages (Laroia et al., 1999).
The mechanisms that regulate the formation of PBs are largely unknown, although their size and numbers can increase when 5′ to 3′ mRNA decay is prevented or when translation initiation is inhibited by stress (Kedersha et al., 2005; Anderson and Kedersha, 2006). The formation of PBs can be blocked in response to treatments with a variety of translation elongation inhibitors known to trap mRNPs within polysomes, as well as during mitosis (Brengues et al., 2005). In contrast to PBs, two different mechanisms have been described to induce the formation of SGs. The first relies on the phosphorylation of the translation initiation factor 2 (eIF2α; Kedersha et al., 1999; Anderson and Kedersha, 2006). The phosphorylation of eIF2α is known to be mediated by four different stress kinases: heme-regulated inhibitor kinase (HRI), protein kinase RNA (PKR), PKR-like endoplasmic reticulum (ER) kinase (PERK), and general control nonderepressible-2 (GCN2), depending on the type of stress applied (Holcik and Sonenberg, 2005; Wek et al., 2006). The second mechanism is independent of eIF2α phosphorylation and does not involve the activation of any of these kinases. Indeed, we and others have showed that interfering with the activity of the translation initiation factor eIF4A (Dang et al., 2006; Mazroui et al., 2006) or eIF4G (Mazroui et al., 2006) triggers SG formation independently of eIF2α phosphorylation. Under these conditions, however, we did not observe any effect on the shape and number of PBs. Although these studies identified some of the molecular mechanisms by which SGs are formed, the effect of these treatments on mRNA turnover is still elusive.
Although the observations mentioned above describe some of the molecular mechanisms by which SGs are formed, the signaling events involved in their disassembly are not well defined. It has been suggested that Hsp72 protein could be involved in the disassembly of cytoplasmic aggregates induced by the prion-like domain (PRD) of the TIA protein (Gilks et al., 2004). The fact that the overexpression of PRD prevents SG formation and stimulates a massive production of the Hsp72 protein raised the possibility that this heat shock protein (HSP) could be one of the factors that triggers SGs disassembly. HSPs are a group of proteins that act as chaperones or regulate protein translocation into organelles (Beere, 2004). Hsp72 is the major inducible HSP during stress (Gabai and Sherman, 2002). Interestingly, the inhibition of the UPS pathway is known to induce a high expression level of Hsp72 (Meriin et al., 1998). However, the link between Hsp72 expression and the translation-recovery observed during prolonged proteasome inhibition is not known. Under other stresses, however, the expression of Hsp72 is induced during the phase of translation recovery (Rylander et al., 2005a,b), suggesting a potential link between Hsp72 expression, translation reinitiation, and SG disassembly.
Here we investigated whether the inhibition of UPS affects the localization and the fate of endogenous ARE messages. We found that interfering with the activity of UPS induces the formation of SGs and disrupts PB assembly, leading to the stabilization of ARE mRNAs. We also show that the assembly of SG in response to the inhibition of the proteasome requires GCN2-mediated phosphorylation of eIF2α. Finally, we observed that the disassembly of SGs as well as the recovery of translation that occurred upon prolonged inhibition of UPS correlates with the cytoplasmic accumulation of the Hsp72 protein.
MATERIALS AND METHODS
Cell Lines and Cultures
HeLa cells and 293 cells stably transfected with green fluorescent protein ubiquitin (GFPu) (Bence et al., 2005; Salomons et al., 2005) were obtained from American Type Culture Collection (Manassas, VA; ATCC). PERK KO and GCN2 KO mouse embryonic fibroblasts (MEFs) are a gift from Dr. D. Ron (New York University). HeLa cells stably transfected with TCR-β WT or TCR-β containing a PTC were a generous gift from Dr. M. Wilkinson (University of Texas, M. D. Anderson Cancer Center, Houston, TX). Wild-type (WT) MEFs, and MEFs harboring the mutation eIF2αS51A/S51A were previously described (Scheuner et al., 2001). Unless specified, all cell lines were maintained in DMEM (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS; Sigma), and penicillin and streptomycin (Sigma). All drug treatments were done when cells reached 60–80% confluency.
Reagents
Two proteosome inhibitors MG132 (Z-Leu-Leu-Leu-al, purity ∼80% HPLC) and lactacystin (purity ∼90% thin layer chromatography) were acquired from Sigma. Both drugs were dissolved in DMSO (Sigma) at 100 mM stock solution and stored at −20°C.
Antibodies
Anti-HuR and anti-G3BP antibodies were previously described (Gallouzi et al., 2000; Mazroui et al., 2006). Anti-eIF4E and anti-TIA antibodies were provided by Dr. J. Pelletier (McGill University, Canada) and Dr. P. Anderson (Brigham and Women's Hospital, Boston, MA), respectively. Anti-FMRP and anti-FXR1 antibodies were a generous gift from Dr. E. Khandijian (St-Francois d'Assise, Quebec, Canada). Anti-Dcp1a antibody was obtained from Dr. J. Lykke Anderson (Boulder University, Colorado). Anti-EDD antibody was a gift from Dr. Simon Wing (McGill University, Canada), Anti-Hsp72 (Stressgene), anti- p21cip1 (Chemicon, Temecula, CA), anti-actin (Sigma). Anti-caspase-3, anti- eIF2α, and anti-phospho-eIF2α, (Cell Signaling, Beverly, MA) antibodies were obtained from the indicated companies. Anti-p53 was obtained from Dr. P. Branton (McGill University, Canada). Secondary antibodies were from Biocan Jackson (West Grove, PA).
Plasmids and Transfections
The plasmid encoding Flag-Hsp72 was a generous gift from Dr. P. Anderson and the plasmid encoding HA-eIF4ET was provided by Dr. N. Sonenberg (McGill University, Canada). DNA transfections were performed using Lipofectamine Plus Reagent (Invitrogen, Carlsbad, CA) as recommended by the manufacturer. For a six-well plate, 0.5 μg of DNA was used. Hsp72 small interfering RNA (siRNA), purchased from Santa Cruz Biotechnology (Santa Cruz, CA), was transfected into HeLa cells as described previously (Di Marco et al., 2005). Twenty-four hours before transfection, cells were trypsinized to obtain 50–60% confluency on the day of transfection. For a six-well plate, 1.5 μl of siRNA duplex (80 μM annealed duplex from Dharmacon [Boulder, CO] or Santa Cruz) was mixed with 100 μl of OPTI-MEM medium and 3.5 μl of Plus Reagent (Invitrogen) and incubated for 15 min at room temperature. A mixture of 4 μl of Lipofectamine (Invitrogen) and 100 μl of OPTI-MEM was then added to the precomplexed RNA mix and incubated for an additional 15 min before adding to cells. EDD siRNA was obtained from Dr. Simon Wing (McGill University) and used as described (Yoshida et al., 2006).
Proteins Extraction and Western Blotting
For total extract, cells (4 × 106) were washed with cold PBS, lysed directly in 1 ml of buffer A (20 mM Tris-HCl, pH 7.4, 150 mM KCl, 1.5 mM MgCl2, 100 mM NaF, 10 ng/ml aprotinin, 1 mM phenylmethyl-sulfonylfluoride, 1 mM dithiothreitol (DTT), 10 U/ml RNasin; Pharmacia, Arlington Heights, IL) and 0.5% Nonidet P-40. The lysates were scraped from the plates, and proteins were quantified with the Bradford assay. An equal volume of 2× Laemmli loading dye was then added to the extract before boiling. For soluble-insoluble fractionation of lysates, cells were washed twice on ice with cold PBS and then lysed in 1 ml ice-cold buffer A. The lysates were scraped from the plates, incubated on ice for 10 min, and passed through a hypodermic needle (Becton Dickinson, Lincoln Park, NJ) 15 times. Lysates were then clarified by centrifugation at 12,000 × g for 30 min at 4°C to separate the soluble from the pellet insoluble fraction. The soluble fraction was then removed and mixed with one volume of 2× Laemmli loading dye. The pellet was dissolved in 1 ml of buffer A and mixed with 1 ml of 2× Laemmli loading dye. Proteins were boiled, resolved in 10% SDS-polyacrylamide gel, transferred to nitrocellulose membrane, and incubated with the indicated antibodies.
Fluorescence Microscopy
Immunofluorescence was performed as previously described (Mazroui et al., 2006). Essentially, cells were fixed in 3% paraformaldehyde and permeabilized with 0.1% Triton X-100/phosphate-buffered saline (PBS). Slides were incubated with primary antibodies diluted in 0.1% normal goat serum for 1 h at room temperature (RT). After washing, slides were incubated with goat anti-mouse/-rabbit IgG (H+L) secondary antibodies coupled to Alexa Fluor 488/594 (Molecular Probes, Eugene, OR; Invitrogen). Fluorescence microscopy was performed using a Zeiss Axiovision 3.1 microscope equipped with Axiocam HR (Zeiss, Thornwood, NY) digital camera. Images were compiled using Adobe Photoshop software (San Jose, CA).
Annexin V-Fluorescein Isothiocyanate/Propidium Iodide Assay
HeLa cells were treated with 10 μM of MG132 for different time points or with 3% formaldehyde containing PBS for 30 min on ice. We harvested both adherent and detached cells. First the media was removed and collected in a 15-ml Falcon tube (Becton Dickinson) and stored on cold ice. Second the remained attached cells were scraped off the plate than added to the Falcon tube. On centrifugation for 5 min at 1500 rpm, cells were washed with ice-cold PBS, then pelleted again at 1500 rpm for 10 min at 4°C, and resuspended in ice-cold binding buffer (10 mM HEPES/NaOH, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2). The cells were subsequently stained with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) for 15 min in the dark. A total of 50,000 cells were counted, and dead cells were examined by flow cytometry.
Northern Blot Analysis
Northern blot analysis was performed as was previously reported (Di Marco et al., 2005) using 10 μg of RNA isolated by TRIZOL reagent (Invitrogen). RNA was transferred to a Hybond-N membrane, and after cross-linking the blots were hybridized with 32P-dCTP-labeled human p21cip1, TCR-β, and GABDH probes. After hybridization and washing, blots were exposed to Biomax films (Perkin Elmer, ON, Canada).
In Vitro Transcription and RNA Fluorescence In Situ Hybridization
A DNA fragment corresponding to the 3′UTR of p21cip1 was amplified by PCR using the following primers fused either to the T7 or T3 minimal promoter sequence: T3- p21cip1 Forward: 5′ AATTAACCCTCACTAAAGGGCTCAGAGGAGGCGCCATGTCA 3′ and T7- p21cip1 Reverse: 5′ TAATACGACTCACTATAGGGGCCGTTTTCGACCCTGAGAGTG 3′.
The amplified fragment was used as a template for in vitro transcription to produce p21cip1 sense RNA from T3 promoter or p21cip1 anti-sense RNA from T7 promoter, using the fluorescence in situ hybridization (FISH) tag RNA Green Kit with Alexa fluor 488 (Invitrogen). In vitro–transcribed RNA was quantified, denatured, and incubated with fixed, permeabilized cells at 37°C for 16 h in the hybridization buffer (50% formamide, 5× SSC, 50 mM phosphate buffer, pH 7.4, 5× Denhardt's, 1 mM EDTA, and 250 ng/μl salmon sperm DNA). After hybridization, cells were processed for immunofluorescence as described above.
RESULTS
The Inhibition of the Ubiquitin-Proteasome Activity Induces the Formation of Stress Granules
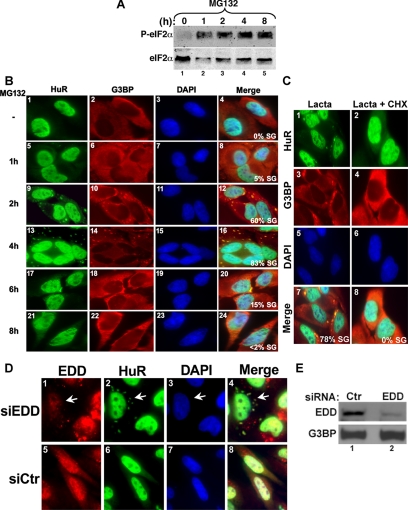
The inactivation of the UPS is known to induce the phosphorylation of eIF2α, leading to the transient inhibition of translation initiation (Baugh and Pilipenko, 2004; Jiang and Wek, 2005; Ding et al., 2006). Because phosphorylation of eIF2α was also linked to SG formation triggered by several stresses (Anderson and Kedersha, 2006), we asked whether UPS inactivation might also result in the formation of these cytoplasmic foci. We confirmed the effect of UPS inhibition on eIF2α phosphorylation (Figure 1A) and found that treatment of HeLa cells with MG132 or lactacystin (two well-characterized inhibitors of the UPS; Voorhees and Orlowski, 2006) induced the formation of SGs, as illustrated by the distribution of the SG markers, HuR and G3BP (Figure 1, B and C). The involvement of the proteasome in the formation of SGs was further confirmed by the depletion of the E3 ubiquitin ligase EDD using RNA interference (RNAi) duplexes against EDD mRNA (Yoshida et al., 2006). A significant reduction in EDD expression (Figure 1E) correlated with the formation of SGs (Figure 1D, compare panels 1 and 2). However, cells that did not take up EDD siRNA duplexes did not form SGs (Figure 1D).
Figure 1.
Inhibition of the proteasome activity induces SGs formation. (A) Treatment of HeLa cells with MG132 induces eIF2α phosphorylation. HeLa cells were treated with 10 μM MG132 for the indicated amounts of time, and protein extracts were prepared and analyzed by Western blotting using an anti-phospho eIF2α (top panel) or pan anti-eIF2α (bottom panel) antibodies. (B) The SGs induced by MG132 disassemble 6 h after treatment. Untreated HeLa cells or cells treated with 10 μM MG132 for different time points were permeabilized and fixed. The presence of stress granules was subsequently verified by immunofluorescence using antibodies against two surrogate markers of SGs: HuR and G3BP. The primary antibodies used were a monoclonal anti-HuR antibody and a polyclonal anti-G3BP antibody. (C) Lactacystin-induced SGs are dynamic. HeLa cells were incubated with lactacystin (10 μM) for 4 h (Lacta) or for 3 h before adding cycloheximide (50 μg/ml) for an additional hour (Lacta + CHX), and then the distribution of HuR and G3BP was analyzed as described above. In B and C the percentage of cells harboring SGs (>5 granules/cell), from three different fields and three different experiments containing a total of 450 cells, is indicated to the right bottom of 4, 8, 12, and 16. (D and E) The RNAi-mediated depletion of the E3 ubiquitin ligase EDD induces SG formation. HeLa cells transfected with siRNA against EDD or control siRNA (Ctr) for 48 h were either fixed on coverslips and used for immunofluorescence with anti-EDD and anti-HuR antibodies (D) or used to prepare total cell extracts to perform Western blot analysis using the indicated antibodies (E). (D) Cells containing SGs are indicated with arrowheads. Representative images of two independent experiments are shown.
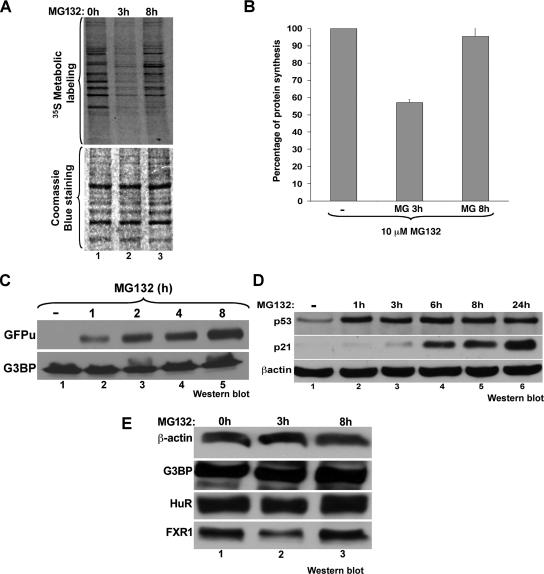
Our observations described above indicated that the assembly of SGs in response to proteasome inhibition is time dependent and reversible; this assembly started 1 h after treatment with 10 μM MG132 and coincided with the phosphorylation of eIF2α. The granules then disassembled after 6 h of treatment (Figure 1, compare A and B). It was previously shown that prolonged exposure of cells to MG132 (>6 h) leads to a partial recovery of translation (Baugh and Pilipenko, 2004; Ding et al., 2006). To determine whether the disassembly of SGs correlates with this phenomenon, we treated HeLa cells with MG132 for different times followed by 30 min of incubation with [35S]methionine and analyzed the synthesis of de novo protein using a 10% SDS-polyacrylamide gel (Figure 2A). The results showed that translation recovered, albeit partially, when SGs disassembled (Figure 2, A and B). Of note, we observed that the translation pattern after 8-h exposure to MG132 is slightly different from untreated cells (Figure 2A). This could indicate that some of the population of mRNAs that recover their translation after prolonged UPS inhibition could be different from the ones expressed before the MG132 treatment. To ensure that the UPS is still inhibited at a time when SGs disassembled, we used 293 cells stably transfected with a reporter plasmid expressing GFP fused to a proteasome-targeting sequence (GFPu). In these cells and in the absence of any UPS inhibitors, the GFPu protein is constitutively degraded by the ubiquitin-dependent proteasome pathway (Bence et al., 2005; Salomons et al., 2005). The addition of 10 μM MG132 to these cells prevented the degradation of GFPu protein even after the disassembly of SGs (Figure 2C), demonstrating that UPS inhibition is maintained beyond the 6-h time point. This result was confirmed in HeLa cells by following the expression of p53 protein, which is known to be rapidly degraded by UPS in this cell system (Hougardy et al., 2006; Kuballa et al., 2007). Western blot analysis on total HeLa cell extracts prepared upon treatment with 10 μM MG132 for various periods of time showed a significant increase in the levels of p53 compared with untreated cells (Figure 2D). These results indicated that the disassembly of SGs occurred without reactivation of the ubiquitin-proteasome system.
Figure 2.
The proteasome inhibitor MG132 induces a transient reduction in translation activity that correlates with SG assembly. (A and B) MG132 triggers a transient translation inhibition. (A) HeLa cells were treated with 10 μM MG132 for the indicated times and were then labeled for 30 min with [35S]methionine (50 μCi/ml). Proteins were resolved by SDS-polyacrylamide gel, stained with Coomassie blue (bottom panel), and detected by autoradiography (top panel). (B) The intensity of the signal in each lane was measured using ImageQuant software. The results were plotted as bar graphs. Error bars, SD of three independent experiments. The percentage of each signal was defined as the intensity of each sample minus the background level and normalized against the signal for the untreated cells. (C and D) Treatment with MG132 for more than 8 h is still effective. (C) GFPu cells (293 cells stably transfected with a GFP reporter construct fused to a proteasome targeting sequence) were treated with 10 μM MG132 for different periods of time. The expression of GFP protein was monitored by Western blot using the anti-GFP antibody to assess the inhibition of the proteasome activity. (D) HeLa cells were treated with 10 μM MG132 for different periods of time (0–24 h). The expression of p53 protein was monitored by Western blot using the anti-p53 antibody. (E) The expression levels of β-actin, G3BP, HuR, and FXR1 proteins are not affected during MG132 treatment. HeLa cells were treated with MG132 as in A. The steady state level of the indicated proteins was analyzed by Western blot using specific antibodies as indicated.
Previous observations showed that the overexpression of some RBPs such as G3BP, FXR1, and FMRP induce spontaneous SGs assembly (Mazroui et al., 2002; Tourriere et al., 2002). Thus it is possible that the MG132-induced SGs could simply be the result of the stabilization and the accumulation of SG-promoting proteins. To test this possibility, we performed a Western blot analysis with total cell extracts treated for different periods of time with 10 μM MG132, and then the membrane was probed with the anti-G3BP, -FXR1, and -FMRP antibodies. We observed that the expression levels of G3BP, FXR1, FMRP, and HuR proteins remained unchanged throughout the treatment (Figure 2E and Supplementary Figure 4). Additionally, the knockdown of the EDD protein did not affect the expression of G3BP (Figure 1, D and E). Thus, these observations suggested that during MG132-mediated proteasome inhibition the assembly of SGs does not correlate with the accumulation of some SG-promoting proteins (G3BP, FXR1, and FMRP).
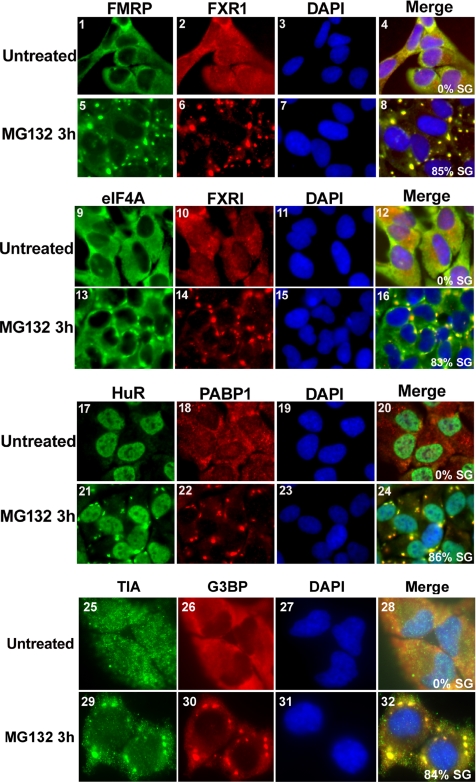
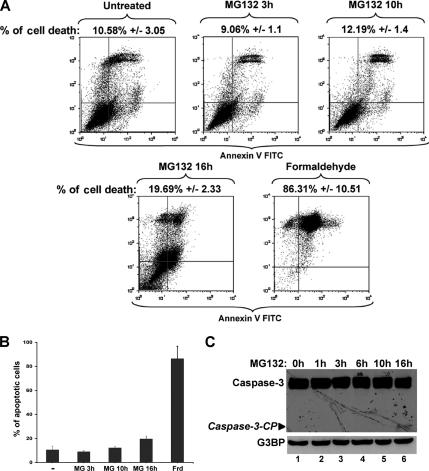
Our results indicated that the SGs induced by MG132 could be similar to those triggered by arsenite or heat shock treatments (Andrews et al., 1987; Gallouzi et al., 2000; Kedersha et al., 2000; Stoecklin et al., 2004). It has been shown that during biochemical fractionation of cells treated with various stresses the levels of SG-associated proteins significantly increases in the insoluble fraction (Mazroui et al., 2002; Gilks et al., 2004). When we performed the same biochemical fractionation on HeLa cells treated with MG132 for 3 h, we observed a threefold increase in the levels of G3BP and FMRP proteins in the insoluble fraction compared with untreated cells (Supplementary Figure 4). This indicated that MG132-induced SGs display the same biochemical characteristics found in SGs that are induced by other stresses. This conclusion was confirmed when we followed the cellular distribution of other SG markers. Using immunofluorescence, we observed that in addition to HuR and G3BP (Figures 1B), SGs in cells treated with MG132 for 3 h recruit FMRP, FXR1, PABP, eIF4A, and TIA-1 (Figure 3). Addition of cycloheximide (a drug that inhibits the elongation step of translation by trapping mRNPs into polysomes; Sonenberg and Dever, 2003) to cells pretreated with lactacystin (Figure 1C, panels 2 and 4) or with MG132 (data not shown) disrupted preformed SGs, attesting to their dynamic nature. This conclusion was further supported by the fact that MG132-induced SGs disassemble after prolonged exposure to 10 μM MG132 (Figure 1B). However, this disassembly of SGs could simply be the result of massive cell death. To test this possibility we treated HeLa cells for various periods of time (0–16 h) with 10 μM MG132 and measured cell death by quantifying the percentage of annexin V/PI-positive cells using flow cytometry (Martin et al., 1995) or by following the activation by cleavage of the apoptotic effector caspase-3 (Zou et al., 1997). Annexin V is known to associate with externalized phosphatidylserines exposed at the surface of cell dying by either apoptosis or necrosis (Sabourin et al., 2000; Suzuki et al., 2001; Green and Steinmetz, 2002; Cobb et al., 2004). As shown in Figure 4A, the maximum levels of apoptotic cells was ∼ 20% upon 16 h of MG132 treatment compared with the 86% of cell death reached after only 30 min of exposure to formaldehyde (Figure 4, A and B). Additionally, a low level of cleaved caspase-3 was only detected after 16 h of exposure to MG132 (Figure 4C). These observations clearly indicated that the MG132-induced SG assembly and disassembly are not due to a massive cell death. Therefore, together these data suggested that the inhibition of the UPS induced the formation of dynamic and reversible SGs.
Figure 3.
MG132-induced SGs recruit a variety of RNA-binding proteins. Untreated HeLa cells or cells treated with 10 μM MG132 for 3 h were subjected to immunofluorescence using antibodies specific to FMRP, FXR1, eIF4A, HuR, PABP, TIA, and G3BP. The Merge represents a staining for the two indicated proteins and the DAPI. The percentage of cells harboring SGs (>5 granules/cell), from three different fields and three different experiments containing a total of 450 cells, is indicated to the right bottom of 4, 8, 12, and 16. Representative pictures are shown for each protein.
Figure 4.
A small percentage of apoptotic cells are detected only after prolonged exposure (>15 h) to 10 μM MG132. (A and B) The effect of MG132 treatment on the expression of annexin V. (A) Untreated HeLa cells or cells that were treated with MG132 for the indicated times were collected, stained with annexin V-FITC and PI, and analyzed by flow cytometry. Formaldehyde treatment for 30 min was used as a positive control for apoptosis. The percentage of total dead or dying cells is indicated on the top of each box (% of cell death) and was defined as the sum of early (lower right box) and late (upper right box) apoptosis. The values were normalized to control, untreated cells. (B) Histogram presenting the results from A as the means ± SEM, from two independent experiments. (C) The cleavage of the apoptotic protein caspase-3 is detected only upon 16 h of exposure to MG132. HeLa cells were treated for the various indicated time points with 10 μM MG132 and then used to prepare total cell extracts. Thirty micrograms from each extract were used for Western blot, and the membrane was probed for caspase-3 and G3BP. The molecular weight (MW) of the caspase-3 cleavage product (Caspase-3-CP) is ∼14 kDa, whereas the precursor is 35 kDa. Shown are representatives of two independent experiments.
The Formation of SGs in Response to MG132 Requires the Phosphorylation of eIF2α
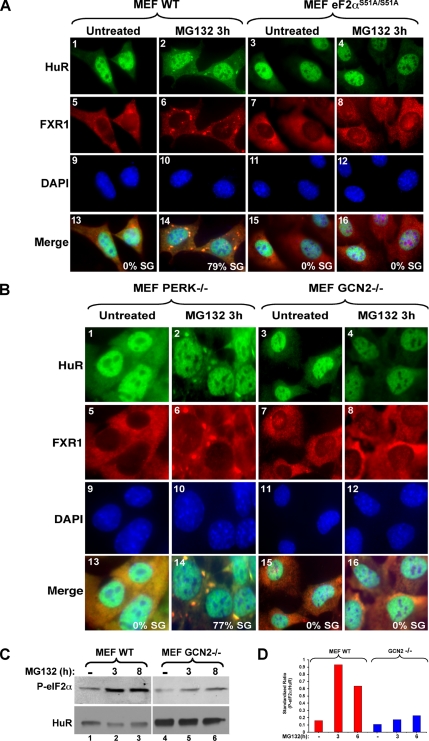
The assembly of SGs under different stresses can be triggered by phosphorylation of eIF2α at serine 51 (Kedersha and Anderson, 2002). Recently, we reported that SGs can also form via a mechanism unrelated to this modification (Mazroui et al., 2006). Thus, we investigated the requirement of eIF2α phosphorylation in UPS-inhibition–mediated SG formation. For this, we analyzed the ability of MEF cells expressing the nonphosphorylatable eIF2α mutant (eIF2αS51A/S51A), obtained from knockin mice (Scheuner et al., 2001), to form SGs in response to MG132. Treating these cells with 10 μM MG132 for 3 h did not induce SG formation (Figure 5A, panels 3, 4, 7, and 8). In contrast, and under the same conditions, SGs were efficiently formed in WT MEFs (Figure 5A, panels 1, 2, 5, and 6). This result established that SG assembly triggered by UPS-inhibition requires the phosphorylation of eIF2α.
Figure 5.
The formation of stress granules in response to the inhibition of the proteasome requires activation of the stress-induced kinase GCN2 and phosphorylation of eIF2α. (A) The phosphorylatable isoform of eIF2α is required for MG132-induced SGs. Wild-type cells as well as cells expressing a nonphosphorylatable eIF2α isoform (S51A/S51A) were treated with 10 μM MG132 for 3 h. The formation of SGs was then verified by immunofluorescence using antibodies against HuR and FXR1. (B) MG132-mediated SGs require the normal expression of the GCN2 kinase. WT MEFs or MEFs lacking GCN2 or PERK were treated with 10 μM MG132 for 3 h. The formation of SGs was verified by immunofluorescence as indicated above. (C and D) MG132-mediated eIF2α phosphorylation requires the GCN2 kinase. (C) Western blot analysis of phospho-eIF2α in wild-type as well as in GCN2−/− cells treated with 10 μM MG132 for the indicated periods of time. HuR was used as a loading control. (D) The results of Western blot were quantified and normalized against HuR. The bar graph represents the results of two independent experiments.
Recent reports have suggested that during UPS-inhibition the phosphorylation of eIF2α is mediated by the GCN2 kinase (Jiang and Wek, 2005). To test the implication of GCN2 in MG132-induced SG assembly, we analyzed the formation of these granules in MEF cells isolated from GCN2 knockout mice. As a control, we followed the formation of these SGs in PERK−/− cells (PERK is a stress kinase known to phosphorylate eIF2α under ER stress; Kimball et al., 2003). We observed that in contrast to PERK−/−, GCN2−/− cells do not form SGs upon MG132 treatment for 3 h (Figure 5B, compare panels 2 and 6 with 4 and 8). The role of GCN2 in this process seems to be specific to proteasome inhibition, because arsenite-induced oxidative stress or pateamine-mediated inhibition of eIF4A (a drug that forms SGs in an eIF2α-phosphorylation-independent manner; Mazroui et al., 2006) is able to induce the formation of SGs in both GCN2−/− and PERK−/− cells (Supplementary Figure 2). Thus, the lack of SG induction in GCN2−/− cells in the presence of MG132 is not due to a general failure of these cells to assemble SGs, but rather to a defect of a single pathway that requires GCN2 activity. Consistent with the reported role of GCN2 kinase in phosphorylating eIF2α during the proteasome inhibition (Jiang and Wek, 2005), our Western blot analysis showed that this posttranslational modification is weakly induced in GCN2−/− compared with PERK−/− or WT MEFs in response to MG132 (Figure 5, C and D). Altogether these results suggested that GCN2 activity is part of the stress-activated signaling pathway leading to SG assembly in response to the inhibition of the UPS through phosphorylation of eIF2α. The disassembly of SGs that occurs upon prolonged exposure to MG132, however, seems to be independent of the phosphorylation state of eIF2α (Figure 1).
The Activity of the HSP Hsp72 Is Involved in Both SG Disassembly and Translational Recovery during the Inhibition of the UPS
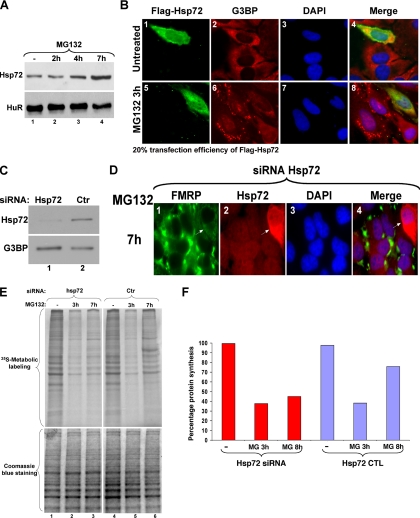
Hsp72 is the major HSP produced in cells recovering from stresses that induce SGs, such as heat shock or arsenite (Gabai and Sherman, 2002). It was also shown that Hsp72 is the main HSP induced during proteasome inhibition (Meriin et al., 1998), and we have found that in NIH-3T3 cells, the induction of Hsp72 in response to MG132 precedes SG disassembly (Supplementary Figure 1). In HeLa cells, although Hsp72 is constitutively expressed (Figure 6A, lane 1), its production significantly increased during prolonged proteasome inhibition when SGs disassemble (Figure 6A, compare lanes 1 and 4). Thus, we hypothesized that Hsp72 activity might play a role in SG disassembly. First we analyzed the effect of overexpressing Flag-tagged Hsp72 on SGs in HeLa cells and observed that only cells expressing high levels of Flag-Hsp72 (∼20% of total cells), compared with nontransfected cells, failed to assemble SGs upon treatment with 10 μM MG132 (Figure 6B, compare panels 1, 2, 5, and 6). The same result was obtained using other cell types such as NIH-3T3 (known to express Hsp72 only during a recovery from stress; Supplementary Figure 1C). Of note, we observed that only cells expressing very high levels of Flag-tagged-Hsp72 (50% of total Flag-tagged Hsp72-positive cells) failed completely to form SG in response to MG132. Thus, it is possible that a high level of Hsp72 protein is required to prevent MG132-induced SGs. To confirm the role of Hsp72 in SG disassembly during prolonged inhibition of the UPS, we depleted Hsp72 from HeLa cells by RNAi using specific siRNA duplexes. Western blot analysis showed that Hsp72 protein levels were significantly reduced in HeLa cells treated with siRNA-Hsp72 compared with cells treated with control siRNA (siRNA-Ctr; Figure 6C). Immunofluorescent experiments using antibodies against FMRP (Mazroui et al., 2003) or Hsp72 proteins showed that treatment with 10 μM MG132 for 7 h failed to induce SG disassembly in cells transfected with siRNA-Hsp72 duplexes compared with nontransfected cells (Figure 6D) or those treated with siRNA-Ctr (data not shown). These observations suggested that the overexpression of the Hsp72 protein could be implicated in the disassembly of MG132-induced SGs.
Figure 6.
Link between the cellular localization of the Hsp72 protein, the disassembly of SGs and translation reactivation during prolonged UPS inhibition. (A) The expression level of Hsp72 protein is enhanced during UPS inhibition. HeLa cells were treated with or without 10 μM MG132 for the indicated periods of time. The expression of Hsp72 was then analyzed by Western blot. (B) The overexpression of Hsp72 prevents SG formation. HeLa cells transfected with a construct encoding Flag-Hsp72 were treated with or without 10 μM MG132 for 3 h as indicated. The localization of Flag-Hsp72 was verified by immunofluorescence using the anti-Flag antibody. The formation of SGs was verified by immunofluorescence using antibodies against G3BP. Shown are representative images of three independent experiments. Cells overexpressing Flag-Hsp72 and did not form SGs are indicated with arrowheads. (C and D) RNAi-mediated Hsp72 depletion prevents SG disassembly after 7 h of treatment with MG132. HeLa cells incubated with an siRNA targeting Hsp72 or a control siRNA (Ctr) for 48 h were subsequently treated with 10 μM MG132 for 7 h and then lysed, and protein extracts were prepared and analyzed by Western blot for Hsp72 expression (C, HuR levels are included as a loading control). (D) The same experiment as in C was performed, and cells were fixed and used for immunofluorescence experiment with anti-FMRP and anti-Hsp72 antibodies. Cells that did not uptake the Hsp72-siRNA and lacking SGs are indicated with arrowheads. Representative images of two independent experiments are shown. (E and F) The knockdown of Hsp72 correlates with the absence of translation recovery upon treatment with MG132 for 7 h. (E) HeLa cells incubated with an siRNA targeting Hsp72, or a control siRNA, were treated with or without 10 μM MG132 for 3 or 7 h (h) and then subjected to 35S-methionine labeling for 30 min to monitor de novo protein translation. Proteins were resolved by SDS-polyacrylamide gel, stained with Coomassie blue (bottom panel), and detected by autoradiography (top panel). (F) The results were plotted as a bar graph. The intensity of the signals was defined as described in Figure 2B.
Our data described above established a link between MG132-induced SG formation and reduction in general translation (Figure 1). To investigate the role of Hsp72 in translation recovery during proteasome inhibition, we depleted the expression of Hsp72 from HeLa cells with siRNA as described above and analyzed the effect on the synthesis of de novo proteins during MG132 treatment using [35S]methionine in vivo labeling. Our results showed that the siRNA-mediated depletion of Hsp72 protein did not affect the inhibition of translation triggered by MG132, but significantly impaired translation recovery (Figures 6, E and F), which is normally observed 6 h upon MG132 treatment (Figure 1). Together, these results suggested that Hsp72 protein could be involved in both SG disassembly and in the partial recovery of translation that occurs during prolonged inhibition of the UPS.
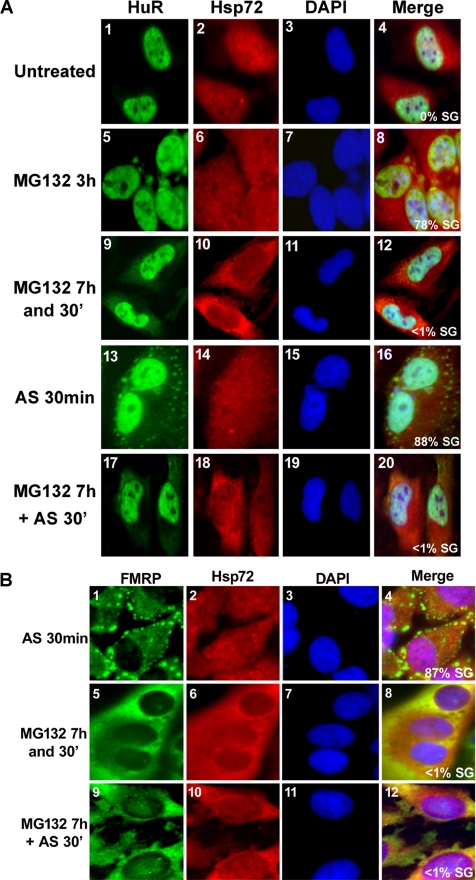
Previous studies have shown that the localization of HSPs such as Hsp27 and Hsc70 changes during the stress response of eukaryotic cells (Cuesta et al., 2000; Kodiha et al., 2005). Hence, we examined the cellular distribution of Hsp72 in HeLa cells in response to MG132. Immunofluorescence experiments showed that Hsp72 was more concentrated in the nucleus of untreated cells, as well as in cells treated with 10 μM MG132 for 3 h (Figure 7A, panel 2). In contrast, Hsp72 became more cytoplasmic upon prolonged exposure to MG132 (>6 h), which correlated with the time during which SG disassembly occurred (Figure 7A, compare panels 9 and 10). These results suggested that the cytoplasmic accumulation of Hsp72 correlated with SGs disassembly. If this assumption is true, cells in which Hsp72 accumulates in the cytoplasm might fail to assemble SGs if treated with another stress. To test this possibility, we first exposed HeLa cells to 10 μM MG132 for 7h and 30min to allow cytoplasmic accumulation of Hsp72, then assessed SG formation upon treatment with different stresses known to induce SGs in an eIF2α phosphorylation-dependent manner. Because as expected these cells retained the cytoplasmic accumulation of Hsp72 (Figure 7A, panel 18), they failed to assemble SGs upon exposure to 0.5 mM arsenite for 30 min (Figure 7A, panel 17) or to 10 μM MG132 for 3 h (data not shown). In contrast, cells that were not pretreated with MG132 had nuclear localization of Hsp72 and formed SGs upon arsenite treatment (Figure 7A, panels 13 and 14). Additionally, the same result has been obtained when we followed the cellular distribution of the FMRP protein in cells treated as described above (Figure 7B). Subsequently, we tested the effect of arsenite on eIF2α phosphorylation in cells pretreated with MG132 for 7 h. We performed a time course of MG132 treatment and followed eIF2α phosphorylation by Western blot analysis. We observed that the level of eIF2α phosphorylation increases by twofold in the presence of arsenite when cells were pretreated with MG132 for 7 h (Supplementary Figure 3, compare lane 7 to lanes 1–5). However this effect is fivefold less than the phosphorylation of eIF2α observed in cells exposed to only arsenite for 30 min (Supplementary Figure 3, compare lane 6 and 7). This observation could argue that MG132 only needs a low level of eIF2α phosphorylation to induce SGs compared with arsenite. However these cells preserved their ability to induce eIF2α-phosphorylation-independent SGs. We observed that exposing pretreated HeLa cells with 10 μM MG132 for 7 h to Pateamine A or Hippuristanol (Mazroui et al., 2006) for 30 min triggered SG formation (Supplementary Figure 3B). Together our results suggested that the accumulation of Hsp72 in the cytoplasm not only correlated with SGs disassembly, but also with the inhibition of their reassembly and this effect is not due to a general defect in SG formation.
Figure 7.
Prolonged inhibition of UPS induces cytoplasmic accumulation of Hsp72 and renders the cell resistant to SG formation. (A) Inhibition of arsenite-induced HuR recruitment to SGs upon exposure to MG132 for 7 h. HeLa cells were either untreated or treated with 10 μM MG132 for 3 h, 7 h and 30 min, or 7 h with MG132 followed by 30-min exposure to 0.5 mM of arsenite (AS) and then analyzed by immunostaining with antibodies against HuR and Hsp72. HeLa cells treated with only 0.5 mM arsenite were used as a positive control for AS-induced SGs. Shown are representative images from four independent experiments. (B) Inhibition of arsenite-induced FMRP recruitment to SGs upon exposure to MG132 for 7 h. HeLa cells were treated as described in A except that SGs were visualized by following the cellular distribution of FMRP protein. Shown are representative images from three independent experiments. In both panels, the percentage of cells harboring SGs were determined as described in Figure 1.
The Inhibition of the UPS Prevents Both ARE- and Nonsense-mediated Decay
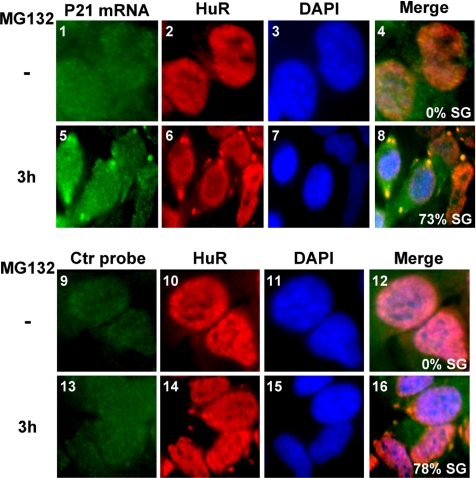
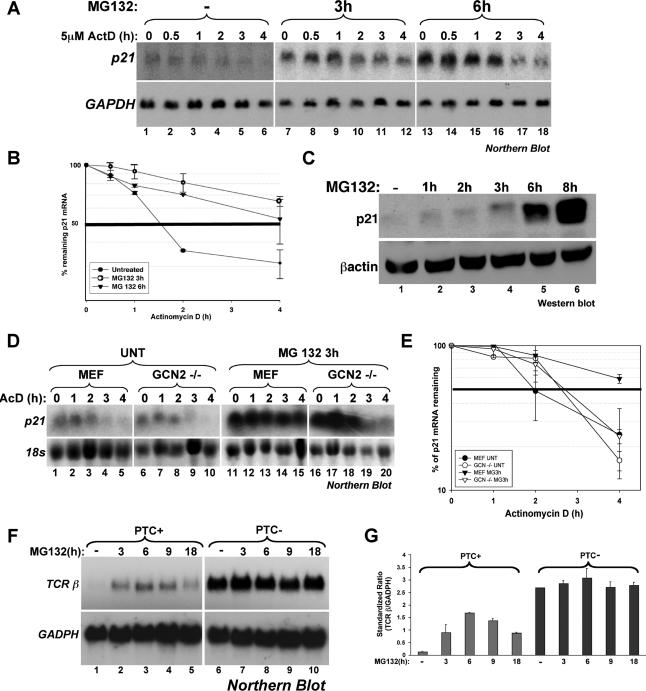
Previous studies have reported that the inhibition of the UPS leads to the stabilization of ARE-containing messages via a mechanism that involves Hsp72 protein (Laroia et al., 1999). Stabilization of ARE messages has also been observed in response to other stresses known to induce SG formation such as heat shock, UV, or arsenite (Andrews et al., 1987; Wang et al., 2000; Stoecklin et al., 2004). Therefore, it is possible that the SGs induced by UPS inhibition could play a direct role in protecting mRNA from the degradation machinery. To test this possibility, we assessed whether exposing HeLa cells to 10 μM MG132 for 3 h induces the recruitment of mRNAs to SGs. Using RNA FISH, we observed that the endogenous ARE-containing mRNA p21cip1, which is known to be stabilized under stress conditions (Wang et al., 2000), was recruited to MG132-induced SGs (Figure 8). These observations suggested that SGs could play a direct role in protecting labile mRNAs from rapid decay. This conclusion was further confirmed by measuring the half-life of p21cip1 mRNA during UPS inhibition. HeLa cells untreated or incubated with 10 μM MG132 for 3 or 6 h were subsequently treated with actinomycin D (ActD) for various periods of time to block synthesis of de novo pol-II transcripts. Total RNA was collected and used for Northern blot analysis to assess the levels of p21cip1 mRNA. We observed that treatment with MG132 for 3 h, conditions under which SGs assemble (Figure 1B), resulted in a significant shift of the p21cip1 mRNA half-life from 1 h 30 min (untreated cells) to >4 h (Figure 9, A and B). Because under normal conditions p21cip1 mRNA is an unstable message, we had to overexpose the blot containing mRNA isolated from untreated cells about five times longer than the one containing mRNA extracted from MG132-treated cells. However, upon prolonged treatment of HeLa cells with MG132 (>6 h) we observed that the half-life of p21cip1 mRNA was <4 h (Figure 9, A and B). Although this increase in the decay rate of p21cip1 mRNA is modest, it could indicate that as soon as SG disassembly starts, the ARE-containing messages that were recruited to SGs become accessible to the decay machinery. This could be the case because after the 6-h MG132 treatment and during the 4-h exposure to ActD the MG132, although still active (Figure 2C and data not shown), did not induce cell death (Figure 4). Therefore, these observations suggested that during the inhibition of the UPS, the stabilization of p21cip1 message could be due in part to its recruitment to SGs.
Figure 8.
MG132-induced SGs recruit the ARE-containing message p21cip1. On treatment with MG132 for 3 h, cells were fixed, permeabilized, and then incubated with a Cy3-labeled in vitro–transcribed antisense RNA probe to detect p21cip1 mRNA (p21 mRNA probe, panels 1–8) and as a control with sense RNA probe (Ctr probe, panels 9–16). SGs were visualized using anti-HuR antibodies. Shown are representative images from two independent experiments.
Figure 9.
The inhibition of the proteasome blocks both ARE-mediated decay and NMD. (A and B) Effect of MG132 treatment on the half-life of p21cip1 mRNA in HeLa cells. (A) HeLa cells were first treated with or without MG132 for 3 h and then incubated with 5 μg/ml actinomycin D (ActD) for the indicated times. Total RNA was prepared and the expression levels of p21cip1 mRNA was determined using Northern blot analysis. Endogenous GAPDH mRNA was used as a loading control. (B) The expression of p21cip1 mRNA was quantified using the ImageQuant software program. The expression levels of p21cip1 mRNA were then standardized against GAPDH message and plotted as the percentage of remaining mRNA compared with message levels at the 0 time point (where there is 100% maximum mRNA level). Error bars, SD of three independent experiments. (C) Effect of MG132-treatment on the synthesis of the p21cip1 protein in HeLa cells. Total cell extracts prepared from HeLa cells treated with 10 μM MG132 for the indicated time periods were harvested and used for Western blot analysis. The membrane was probed for p21cip1 and β-actin proteins. (D and E) The GCN2 kinase seems to be required for the MG132-mediated stabilization of the p21cip1 mRNA. (D) Wild-type MEFs and MEFs derived from GCN2−/− mice were treated as in A, and the expression levels of mouse p21cip1 mRNA was determined using Northern blot analysis. (E) The expression of p21cip1 mRNA was quantified as in A. Levels were standardized against 18S rRNA. Error bars, SD of two independent experiments. (F and G) UPS inhibition by MG132 interferes with the nonsense-mediated decay pathway. (F) HeLa cells stably expressing T-cell receptor beta minigene wild type (TCR-β-wt) or a TCR-β minigene containing a PTC were treated with 10 μM MG132 for different time points, and the levels of TCR-β mRNA were analyzed by Northern blot. (G) TCR-β mRNA levels were standardized as described in A and B. Error bars, SD of two independent experiments.
Interestingly, our data suggested that p21cip1 mRNA colocalizes in SGs with the RBP HuR (Figure 8). This colocalization is consistent with the fact that HuR is known to stabilize p21cip1 mRNA under stress conditions (Wang et al., 2000). Moreover, SGs are also known as translation repression sites. Thus, it is possible that SG-mediated stabilization of p21cip1 mRNA coincides with a reduction/inhibition in its translation. To examine this possibility, we followed the expression of p21cip1 protein under UPS inhibition. Cells were treated with MG132 for different periods of time, and total extracts were prepared and used for Western blot analysis using the anti- p21cip1 antibody. We observed that the level of p21cip1 protein remained low during the time when SGs were induced by MG132 and when p21cip1 message was highly stabilized (3–6 h), compared with its level in untreated cells (Figure 9C). This result is consistent with the fact that p21cip1 mRNA is recruited to SGs where its translation is repressed. However, p21cip1 protein becomes highly abundant after prolonged treatment with MG132 (>6 h), which correlates with SG disassembly (Figure 9C). Of note, our Northern blot analysis showed that the steady state levels of p21cip1 mRNA at 3 and 6 h after MG132 treatment are almost identical (Figure 9A, compare lanes 7 and 13), indicating that the observed increase in protein expression at 6 h is not due to an increase in the transcription rate of p21cip1 message. Our data indicated that although the inhibition of the UPS prevented the decay of ARE-messages, it does not necessarily lead to the immediate translation of these messages due to their sequestration into SGs.
If MG132-induced SGs play a role in the stabilization of p21cip1 mRNA under conditions of UPS inhibition, this stabilization effect should be prevented in the GCN2−/− cells that are not able to form SGs in response to MG132 (Figure 3). To test this possibility, we performed the same ActD pulse-chase experiment on GCN2−/− cells as described above and measured the half-life of the endogenous p21cip1 mRNA. We observed that although exposure to MG132 for 3 h enhanced the steady state levels of p21cip1 mRNA by fivefold in GCN2−/− cells (Figure 9D, compare lane 16 to lane 6), the rate of its decay was the same than in untreated cells (MEF or GCN2−/−; Figure 9D, compare lanes 1, 10, 16, and 20, and 9E). As expected, however, and similarly to HeLa cells (Figure 9A), exposure of wt-MEFs to 10 μM MG132 for 3 h induced SG formation (data not shown) and increased the half-life of p21cip1 mRNA (from 2 h in untreated cells to >4 h; Figure 9E). Thus, the stabilization of p21cip1 mRNA in response to MG132 seems to require the activity of GCN2 kinase and SG formation. Altogether, our observations strengthen the correlation between MG132-induced SG formation and the stabilization of ARE-containing messages such as the p21cip1 mRNA.
The role of UPS-inhibition in ARE-mRNA stabilization prompted us to question whether this inhibition could interfere with other mRNA degradation pathways. To address this question, we tested the effect of MG132 treatment on the decay of another class of mRNA that contains a premature-termination codon (PTC) and is normally eliminated by the NMD pathway (Lykke-Andersen, 2004). HeLa cells stably expressing the WT T-cell receptor beta minigene (TCR-β-wt) or a TCR-β minigene containing a PTC (Li et al., 1997) were treated with 10 μM MG132 for different times and the levels of TCR-β mRNA were analyzed by Northern blot (Figure 9, F and G). As expected, the level of the PTC-containing mRNA was very low in untreated cells (Figure 9F, lane 1), confirming its rapid decay by NMD (Li et al., 1997). In contrast, NMD was impaired upon MG132 treatment (Figure 9F, compare lanes 2–5 with lane 1), which correlated with SG formation (data not shown). Thus, our data suggested that the role of UPS inhibition-induced SGs in mRNA metabolism is not restricted to ARE-containing messages.
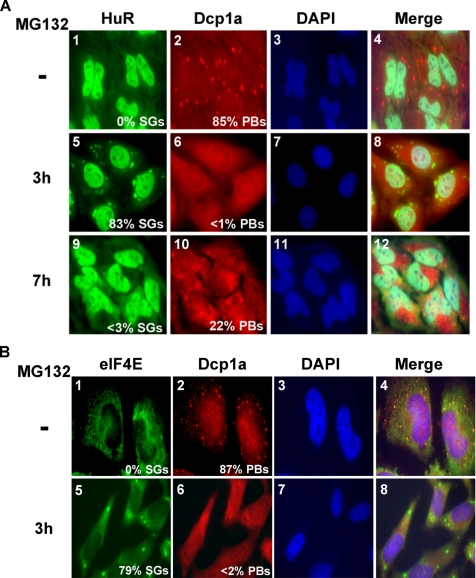
Recent studies have shown that both AUMD and NMD occur in specific cytoplasmic foci called PBs (Fenger-Gron et al., 2005; Sheth and Parker, 2006). Although stress treatment is not required for PB assembly, it does increase their size and number (Kedersha et al., 2005; Anderson and Kedersha, 2006). Similar to SGs, PBs have been described as sites for storage of translationally repressed mRNAs (Sheth and Parker, 2006). Thus we investigated the effect of UPS inhibition on the integrity of PBs. HeLa cells were treated with 10 μM MG132 for different periods of time, and PBs were visualized by immunofluorescence experiments using a polyclonal antibody against a marker of PBs, Dcp1a (Lykke-Andersen, 2002). As expected, we observed that under normal conditions, Dcp1a was detected in PBs (Figure 10A, panel 2). In contrast, upon treatment with MG132 for 3 h, the number of Dcp1a-containing PBs was significantly reduced (Figure 10A, panel 6). These results indicated that exposure to MG132 resulted in the disruption of PBs. We also analyzed the distribution of other markers of PBs, such as the cap-binding protein eIF4E (Ferraiuolo et al., 2005). We found that similar to Dcp1a, treatment with 10 μM MG132 for 3 h prevented the localization of eIF4E in PBs and targeted it to SGs (Figure 10B). These data are consistent with previous reports demonstrating that arsenite treatment targeted eIF4E to SGs (Kedersha et al., 2002). Altogether, these observations indicated that although the inhibition of the UPS induces SGs, it disrupts PBs.
Figure 10.
The inhibition of the UPS disrupts PBs. (A) After incubation with 10 μM MG132 for 3 or 7 h, HeLa cells were fixed and used for immunofluorescence to detect SGs with anti-HuR antibodies and to detect PBs with anti-Dcp1a antibodies. (B) Untreated HeLa cells or cells treated with MG132 for 3 h were analyzed by immunostaining using anti-eIF4E and anti-Dcp1a antibodies. (A and B) Shown are representative images for three independent experiments. The percentage of SGs and PBs in each cell has been determined as described in Figure 1 for SGs.
DISCUSSION
It is well established that interfering with the normal function of the UPS induces a stress characterized by the accumulation of proteins that are normally targeted for decay (Zimmermann et al., 2000; Lee et al., 2003). UPS inhibition also stabilizes reporter ARE-containing messages (Laroia et al., 1999). Very little is known, however, about the mechanisms linking these stress conditions to the block in mRNA decay. In this study, we show that UPS inhibition triggers SG assembly through a process that requires the GCN2-mediated phosphorylation of eIF2α as well as a significant reduction in protein synthesis. The assembly of these SGs is reversible since they disassemble during proteasome inhibition allowing for a partial recovery in translation. Under these circumstances, both SG disassembly and translation recovery involves the Hsp72 protein. We provide evidence suggesting that these stress conditions induce the stabilization of the ARE-containing mRNA p21cip1, which correlates with the recruitment of this message to SGs. This effect is not restricted to ARE messages because PTC-containing mRNAs are also protected from the degradation machinery during UPS inhibition. While assessing the status of the cytoplasmic mRNA decay bodies (PBs) under these conditions, we observed that the inhibition of both ARE-mediated decay and NMD correlated with PB disassembly. Together, these findings suggest that SGs could be involved in protecting mRNAs from decay during UPS inhibition, which preserves the ability of the cell to recover from this stress.
One of the main features of cell stress response is to first activate survival mechanisms to prevent the damage of vital proteins and mRNAs that are needed for the recovery process. If the stress is unsustainable, the cell will be irreversibly damaged and death pathways are activated (Beere, 2004). Because prolonged treatment of cells with proteasome inhibitors such as MG132 has been shown to activate apoptosis in an eIF2α-phosphorylation–dependent manner (Jiang and Wek, 2005), it is possible that the assembly and the disassembly of SGs observed in our experiments (Figure 1) are part of the mechanisms involved in the sequential activation of anti- and pro-death pathways. In fact, eIF2α phosphorylation has been associated with both pro- and anti-apoptotic activities (Adams, 2004). The fact that SGs have been associated with a reduction in translation activity and have been shown to recruit poly A+ mRNAs and key cellular proteins raised the possibility that their assembly could be part of stress-induced survival pathways (Anderson and Kedersha, 2002, 2006). Our data showing that the assembly of SGs upon MG132 treatment depends on the phosphorylation of eIF2α (Figure 5A) supports this possibility. A recent study by Jiang and Wek (2005) has shown that GCN2 is the primary kinase that mediates eIF2α phosphorylation-dependent apoptosis upon treatment with MG132. However, with other stresses such as UV treatment, GCN2 plays the opposite role by activating the anti-apoptotic NFκB pathways (Ghosh and Karin, 2002). The high level of cell death mediated by GCN2 and eIF2α-phosphorylation occurs mainly after prolonged exposure to MG132 (Jiang and Wek, 2005). While examining cell death in our system, we observed that a modest level of apoptosis was detected only after exposure to MG132 for >15 h (Figure 4). Therefore, it is possible that by inducing the assembly of SGs during the early time points of MG132 treatment, GCN2 participates in the first step of the cell stress response, which is activation of survival pathways. It is still intriguing, however, as to why and how after 6 h of proteasome inhibition and despite the maintenance of eIF2α phosphorylation, the cell partially recovers its translation activity and disassembles SGs (Figures 1 and 6). These observations could simply indicate that at this stage the MG132 stress becomes unsustainable, signaling to the cell that death is imminent. Consequently and in response to these signals the cell slowly starts preparing itself for the activation of suicide mechanisms by increasing the levels of proapoptotic proteins. Therefore, defining the exact factors involved in MG132-mediated SG assembly could help identify better ways to accelerate the transition from the survival to the apoptotic step, which will help improve anticancer treatments.
In this study, we provide evidence suggesting that the HSP Hsp72 (also called Hsp70; Nylandsted et al., 2004) plays a positive role in the disassembly of MG132-induced SGs (Figures 6 and 7). Previous experiments have indicated that the level of Hsp72 protein increases significantly in response to proteasome inhibition in mice fibroblasts (Jiang and Wek, 2005). Although Hsp72 is constitutively expressed in HeLa cells (Gallouzi et al., 2001; Nylandsted et al., 2004), our results suggest that the increase in Hsp72 expression levels upon prolonged treatment with MG132 is concomitant with its accumulation in the cytoplasm (Figure 6). The nuclear localization of Hsp72 that is seen at early time points of MG132 treatment (<6 h; Figure 7A, panels 2 and 6) could indicate a need for keeping this chaperone away from the area where SGs assemble. This conclusion was further supported by the fact that overexpressing Hsp72 prevented SG formation, whereas its depletion led to the maintenance of SGs beyond 6 h of MG132 treatment (Figure 6). Although these observations could argue that Hsp72 plays a role in SG disassembly, we still do not know the molecular mechanisms leading to this effect. However, it is also possible that this MG132-induced SG assembly and disassembly could simply be the result of a shift from cap-dependent to cap-independent translation as has been recently observed for some viral infection systems (McInerney et al., 2005; MacCallum et al., 2006; Lin et al., 2007). Further studies are needed to define the exact function of Hsp72 during SG disassembly as well as during the recovery from stress in general.
Previous studies have reported that the inhibition of the UPS leads to the stabilization of reporter ARE-mRNAs, and suggested that this effect is due to the sequestration of AUF1 by Hsp72 in perinuclear aggregates (Laroia et al., 1999). It is unlikely that these aggregates represent SGs because our experiments show that Hsp72 is excluded from SGs (Figure 7A, panel 6). AUF1 is a member of a family of factors that include TTP, KSRP, BRF1, G3BP, and CUGBP2—all known to induce the decay of ARE messages (Barreau et al., 2005; Anderson and Kedersha, 2006). Thus, it is plausible to assume that the defect in ARE-mRNA decay during UPS inhibition involves a negative effect on the function of these factors and the activation of a general mRNA stabilization processes. Recent reports have shown that the recruitment of TTP to SGs correlates with the inhibition of its ability to induce ARE-mRNA decay (Stoecklin et al., 2004). Our data suggest that MG132-induced SGs could be the cellular entities in which ARE-containing messages are recruited (Figure 8) and most likely stabilized (Figure 9). The presence of p21cip1 mRNA in SGs (Figure 8) correlates with a significant increase in its half-life and an inhibition of its translation (Figures 9C). These data indicate a dual function of MG132-induced SGs in the metabolism of a given message; protection from degradation and translation repression. Thus, mRNAs that are stabilized during stress are not necessarily accompanied by an increase in the level of protein that they encode. Previous studies have reported that the inhibition of UPS induces accumulation of proteins involved in cell cycle arrest such as p21cip1, p27, and p53 (Adams, 2004). This effect was attributed to protein stabilization because these factors are normally degraded by the proteasome. Our data provides more mechanistic detail regarding the steps leading to the expression of proteins such as p21cip1 under condition of UPS inhibition. However, we still do not know how p21cip1 mRNA is recruited to SGs. A recent study has shown that although the RBP ZBP1 is not required for targeting β-actin mRNA to SGs under stress, it plays a key role in its stabilization (Stohr et al., 2006). Defining the protein factor(s) responsible for the recruitment of p21cip1 to SGs under UPS inhibition will help in the understanding of how and why this message is first stabilized and then released to the translation machinery.
In absence of stress, both translation repression and mRNA degradation are known to occur within PBs. Because the number and size of PBs seem to be induced during stress (Kedersha et al., 2005), it is possible that many of the translationally repressed mRNAs are still targeted to PBs rather than to SGs. However, under stress, SGs and PBs can be detected in close proximity in single cells (Kedersha et al., 2005), suggesting a direct communication between both entities. Indeed, a model for a functional link between PBs and SGs was recently proposed (Kedersha et al., 2005); mRNAs that are recruited to SGs are either stored or delivered (e.g., via the destabilizing factor TTP) to adjacent PBs for degradation. According to this model, the two entities are distinct. It has also been shown that under certain conditions SGs can recruit PB-markers such as Dcp1a and GW182 (Kedersha et al., 2005). Our data shows that during the early time points of UPS inhibition, SG assembly coincides with the disruption of PBs (Figure 10). The disappearance of PBs is not due to their fusion with SGs because Dcp1a was excluded from SGs during the inactivation of the UPS (Figure 10). It is possible that UPS inactivation prevents the decay of proteins that are antagonistic to PBs assembly. Previous studies have suggested that interfering with UPS activity by overexpressing the deubiquitinating enzyme prevents ARE-mediated decay (Laroia et al., 2002). It will be interesting to investigate whether the activity of these enzymes can also affect PB and/or SG formation, and how they affect mRNA decay in general. In fact, our experiments show that treating cells with MG132 not only blocks the decay of ARE-mRNAs, but also that of PTC-containing messages which are normally targeted for NMD (Figure 9). Because PBs are known to be the site of nonsense-mediated mRNA decay (Sheth and Parker, 2006), their disassembly during UPS inhibition in addition to the observed translation inhibition explains in part the impairment of the NMD machinery. However, whether PTC-containing messages are recruited to SGs remains unknown. Thus the inhibition of the UPS seems to affect more than one mRNA decay pathway, and it will be interesting to investigate whether other mRNA processes such as miRNA-mediated translation repression and mRNA degradation can also be linked to the activity of the UPS. Although SGs, PBs, and UPS are separate compartments, they appear to be functionally connected. Because drugs known to block the proteasome activity are now used in cancer therapy (Adams, 2004), defining the exact pathways by which they affect cell metabolism will help improve our strategy to combat this disease. Our work opens the door to further investigate the role of SGs in mediating the effect of these drugs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christopher von Roretz for critical reading of the manuscript as well as all the members of the Gallouzi laboratory for their helpful discussion and suggestions. R.M. is a recipient of an National Cancer Institute of Canada (NCIC) Postdoctoral Terry Fox Fellowship and a CIHR Postdoctoral Fellowship. This work was supported by National Institutes of Health Grant DK42394 to R.J.K. and an NCIC operating grant (NCIC016247) to I.G. I.G. is a recipient of TierII Canada Research Chair.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1079) on May 2, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews G. K., Harding M. A., Calvet J. P., Adamson E. D. The heat shock response in HeLa cells is accompanied by elevated expression of the c-fos proto-oncogene. Mol. Cell. Biol. 1987;7:3452–3458. doi: 10.1128/mcb.7.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C., Paillard L., Osborne H. B. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh J. M., Pilipenko E. V. 20S proteasome differentially alters translation of different mRNAs via the cleavage of eIF4F and eIF3. Mol. Cell. 2004;16:575–586. doi: 10.1016/j.molcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Beere H. M. “The stress of dying”: the role of heat shock proteins in the regulation of apoptosis. J. Cell Sci. 2004;117:2641–2651. doi: 10.1242/jcs.01284. [DOI] [PubMed] [Google Scholar]
- Bence N. F., Bennett E. J., Kopito R. R. Application and analysis of the GFP(u) family of ubiquitin-proteasome system reporters. Methods Enzymol. 2005;399:481–490. doi: 10.1016/S0076-6879(05)99033-2. [DOI] [PubMed] [Google Scholar]
- Bevilacqua A., Ceriani M. C., Capaccioli S., Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P., Forcales S. V., Ponassi M., Corte G., Chen C. Y., Karin M., Puri P. L., Gherzi R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Cobb L. J., Salih D. A., Gonzalez I., Tripathi G., Carter E. J., Lovett F., Holding C., Pell J. M. Partitioning of IGFBP-5 actions in myogenesis: IGF-independent anti-apoptotic function. J. Cell Sci. 2004;117:1737–1746. doi: 10.1242/jcs.01028. [DOI] [PubMed] [Google Scholar]
- Cuesta R., Laroia G., Schneider R. J. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W. K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J. O. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- Di Marco S., Mazroui R., Dallaire P., Chittur S., Tenenbaum S. A., Radzioch D., Marette A., Gallouzi I. E. NF-(kappa)B-mediated MyoD decay during muscle wasting requires nitric oxide synthase mRNA stabilization, HuR protein, and nitric oxide release. Mol. Cell. Biol. 2005;25:6533–6545. doi: 10.1128/MCB.25.15.6533-6545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Dimayuga E., Markesbery W. R., Keller J. N. Proteasome inhibition induces reversible impairments in protein synthesis. FASEB J. 2006;20:1055–1063. doi: 10.1096/fj.05-5495com. [DOI] [PubMed] [Google Scholar]
- Fan X. C., Steitz J. A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M., Fillman C., Norrild B., Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo M. A., Basak S., Dostie J., Murray E. L., Schoenberg D. R., Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabai V. L., Sherman M. Y. Invited review: Interplay between molecular chaperones and signaling pathways in survival of heat shock. J. Appl. Physiol. 2002;92:1743–1748. doi: 10.1152/japplphysiol.01101.2001. [DOI] [PubMed] [Google Scholar]
- Gallouzi I. E., Brennan C. M., Steitz J. A. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7:1348–1361. doi: 10.1017/s1355838201016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi I. E., Brennan C. M., Stenberg M. G., Swanson M. S., Eversole A., Maizels N., Steitz J. A. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi I. E., Parker F., Chebli K., Maurier F., Labourier E., Barlat I., Capony J. P., Tocque B., Tazi J. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol. Cell. Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Karin M. Missing pieces in the NF-kappaB puzzle. Cell 109. 2002;(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M. H., Maytal V. Regulating the 26S proteasome. Curr. Top. Microbiol. Immunol. 2002;268:43–72. doi: 10.1007/978-3-642-59414-4_3. [DOI] [PubMed] [Google Scholar]
- Green A. M., Steinmetz N. D. Monitoring apoptosis in real time. Cancer J. 2002;8:82–92. doi: 10.1097/00130404-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Hougardy B. M., Maduro J. H., van der Zee A. G., de Groot D. J., van den Heuvel F. A., de Vries E. G., de Jong S. Proteasome inhibitor MG132 sensitizes HPV-positive human cervical cancer cells to rhTRAIL-induced apoptosis. Int. J. Cancer. 2006;118:1892–1900. doi: 10.1002/ijc.21580. [DOI] [PubMed] [Google Scholar]
- Iakova P., Wang G. L., Timchenko L., Michalak M., Pereira-Smith O. M., Smith J. R., Timchenko N. A. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. Y., Wek R. C. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 2005;280:14189–14202. doi: 10.1074/jbc.M413660200. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I. J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E., Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fitzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S. R., Horetsky R. L., Ron D., Jefferson L. S., Harding H. P. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- Kodiha M., Chu A., Lazrak O., Stochaj U. Stress inhibits nucleocytoplasmic shuttling of heat shock protein hsc70. Am. J. Physiol. Cell Physiol. 2005;289:C1034–C1041. doi: 10.1152/ajpcell.00590.2004. [DOI] [PubMed] [Google Scholar]
- Kuballa P., Matentzoglu K., Scheffner M. The role of the ubiquitin ligase E6-AP in human papillomavirus E6-mediated degradation of PDZ domain-containing proteins. J. Biol. Chem. 2007;282:65–71. doi: 10.1074/jbc.M605117200. [DOI] [PubMed] [Google Scholar]
- Laroia G., Cuesta R., Brewer G., Schneider R. J. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- Laroia G., Sarkar B., Schneider R. J. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl. Acad. Sci. USA. 2002;99:1842–1846. doi: 10.1073/pnas.042575699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. H., Iwakoshi N. N., Anderson K. C., Glimcher L. H. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. USA. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Leonard D., Wilkinson M. F. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J. Exp. Med. 1997;185:985–992. doi: 10.1084/jem.185.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. C., Hsu M., Tarn W. Y. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc. Natl. Acad. Sci. USA. 2007;104:2235–2240. doi: 10.1073/pnas.0611015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. Making structural sense of nonsense-mediated decay. Nat. Struct. Mol. Biol. 2004;11:305–306. doi: 10.1038/nsmb0404-305. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum P. R., Jack S. C., Egan P. A., McDermott B. T., Elliott R. M., Chan S. W. Cap-dependent and hepatitis C virus internal ribosome entry site-mediated translation are modulated by phosphorylation of eIF2alpha under oxidative stress. J. Gen. Virol. 2006;87:3251–3262. doi: 10.1099/vir.0.82051-0. [DOI] [PubMed] [Google Scholar]
- Martin S. J., Reutelingsperger C. P., McGahon A. J., Rader J. A., van Schie R. C., LaFace D. M., Green D. R. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., Huot M. E., Tremblay S., Boilard N., Labelle Y., Khandjian E. W. Fragile X Mental Retardation protein determinants required for its association with polyribosomal mRNPs. Hum. Mol. Genet. 2003;12:3087–3096. doi: 10.1093/hmg/ddg335. [DOI] [PubMed] [Google Scholar]
- Mazroui R., Huot M. E., Tremblay S., Filion C., Labelle Y., Khandjian E. W. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- Mazroui R., Sukarieh R., Bordeleau M. E., Kaufman R. J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney G. M., Kedersha N. L., Kaufman R. J., Anderson P., Liljestrom P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell. 2005;16:3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriin A. B., Gabai V. L., Yaglom J., Shifrin V. I., Sherman M. Y. Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J. Biol. Chem. 1998;273:6373–6379. doi: 10.1074/jbc.273.11.6373. [DOI] [PubMed] [Google Scholar]
- Nandi D., Tahiliani P., Kumar A., Chandu D. The ubiquitin-proteasome system. J. Biosci. 2006;31:137–155. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- Nylandsted J., Gyrd-Hansen M., Danielewicz A., Fehrenbacher N., Lademann U., Hoyer-Hansen M., Weber E., Multhoff G., Rohde M., Jaattela M. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J. Exp. Med. 2004;200:425–435. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritts T. A., Hungness E. S., Hershko D. D., Robb B. W., Sun X., Luo G. J., Fischer J. E., Wong H. R., Hasselgren P. O. Proteasome inhibitors induce heat shock response and increase IL-6 expression in human intestinal epithelial cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;282:R1016–R1026. doi: 10.1152/ajpregu.00492.2001. [DOI] [PubMed] [Google Scholar]
- Raineri I., Wegmueller D., Gross B., Certa U., Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylander M. N., Diller K. R., Wang S., Aggarwal S. J. Correlation of HSP70 expression and cell viability following thermal stimulation of bovine aortic endothelial cells. J. Biomech. Eng. 2005a;127:751–757. doi: 10.1115/1.1993661. [DOI] [PubMed] [Google Scholar]
- Rylander M. N., Feng Y., Bass J., Diller K. R. Thermally induced injury and heat-shock protein expression in cells and tissues. Ann. NY Acad. Sci. 2005b;1066:222–242. doi: 10.1196/annals.1363.009. [DOI] [PubMed] [Google Scholar]
- Sabourin L. A., Tamai K., Seale P., Wagner J., Rudnicki M. A. Caspase 3 cleavage of the Ste20-related kinase SLK releases and activates an apoptosis-inducing kinase domain and an actin-disassembling region. Mol. Cell. Biol. 2000;20:684–696. doi: 10.1128/mcb.20.2.684-696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomons F. A., Verhoef L. G., Dantuma N. P. Fluorescent reporters for the ubiquitin-proteasome system. Essays Biochem. 2005;41:113–128. doi: 10.1042/EB0410113. [DOI] [PubMed] [Google Scholar]
- Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Dever T. E. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Stoecklin G., Gross B., Ming X. F., Moroni C. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22:3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W. F., Blackwell T. K., Anderson P. MK2-induced tristetraprolin:14–3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr N., Lederer M., Reinke C., Meyer S., Hatzfeld M., Singer R. H., Huttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kostin S., Person V., Elsasser A., Schaper J. Time course of the apoptotic cascade and effects of caspase inhibitors in adult rat ventricular cardiomyocytes. J. Mol. Cell. Cardiol. 2001;33:983–994. doi: 10.1006/jmcc.2001.1364. [DOI] [PubMed] [Google Scholar]
- Takimoto C. H., Diggikar S. Heat shock protein and proteasome targeting agents. Hematol. Oncol. Clin. North Am. 2002;16:1269–1285. doi: 10.1016/s0889-8588(02)00048-5. [DOI] [PubMed] [Google Scholar]
- Tourriere H., Chebli K., Tazi J. mRNA degradation machines in eukaryotic cells. Biochimie. 2002;84:821–837. doi: 10.1016/s0300-9084(02)01445-1. [DOI] [PubMed] [Google Scholar]
- Vabulas R. M., Hartl F. U. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- Voorhees P. M., Orlowski R. Z. The proteasome and proteasome inhibitors in cancer therapy. Annu. Rev. Pharmacol. Toxicol. 2006;46:189–213. doi: 10.1146/annurev.pharmtox.46.120604.141300. [DOI] [PubMed] [Google Scholar]
- Wang W., Furneaux H., Cheng H., Caldwell M. C., Hutter D., Liu Y., Holbrook N., Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Jiang H. Y., Anthony T. G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Yoshida K., Kozlov G., Lim N. S., De Crescenzo G., Pang Z., Berlanga J. J., Kahvejian A., Gehring K., Wing S. S., Sonenberg N. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006;25:1934–1944. doi: 10.1038/sj.emboj.7601079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J., Erdmann D., Lalande I., Grossenbacher R., Noorani M., Furst P. Proteasome inhibitor induced gene expression profiles reveal overexpression of transcriptional regulators ATF3, GADD153 and MAD1. Oncogene. 2000;19:2913–2920. doi: 10.1038/sj.onc.1203606. [DOI] [PubMed] [Google Scholar]
- Zou H., Henzel W. J., Liu X., Lutschg A., Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.