Abstract
Background
It is estimated that 5–10% of all breast cancer are hereditary and attributable to mutations in the highly penetrance susceptibility genes BRCA1 and BRCA2. The genetic analysis of these genes is complex and expensive essentially because their length. Nevertheless, the presence of recurrent and founder mutations allows a pre-screening for the identification of the most frequent mutations found in each geographical region. In Spain, five mutations in BRCA1 and other five in BRCA2 account for approximately 50% of the mutations detected in Spanish families.
Methods
We have developed a novel PCR multiplex SNaPshot reaction that targets all ten recurrent and founder mutations identified in BRCA1 and BRCA2 in Spain to date.
Results
The SNaPshot reaction was performed on samples previously analyzed by direct sequencing and all mutations were concordant. This strategy permits the analysis of approximately 50% of all mutations observed to be responsible for breast/ovarian cancer in Spanish families using a single reaction per patient sample.
Conclusion
The SNaPshot assay developed is sensitive, rapid, with minimum cost per sample and additionally can be automated for high-throughput genotyping. The SNaPshot assay outlined here is not only useful for analysis of Spanish breast/ovarian cancer families, but also e.g. for populations with Spanish ancestry, such as those in Latin America.
Background
Germline mutations in BRCA1 (OMIM 113705) and BRCA2 (OMIM 600185) account for familial clustering in the majority of families with both breast and ovarian cancer and in approximately one-half of families with site-specific breast cancer [1,2]. BRCA1 and BRCA2 are both large genes containing 5,592 and 11,385 nucleotides spread over ~100,000 bases of genomic DNA each. There are more than 1,000 BRCA mutations reported to the Breast Cancer Information Core (BIC) database [3]; many of them are unique but there are also numerous examples of recurrent and founder mutations. Founder mutations have been reported in genetic isolate populations such as Ashkenazi Jewish [4,5], Icelandic [6], Finnish [7], Dutch [8,9] and French Canadian [10], as well as in Slavic countries, including Poland [11].
In Spain, a compilation of BRCA test results from different laboratories shows that five mutations in the BRCA1 gene (c.187-188delAG [p.Q23fs], c.330A > G, [p.64-71del], c.5236G > A [p.G1706A], c.5242C > A [p.A1708E], c.589-590delCT [p.S157X]; numbered GeneBank U14680) account for 46.6 % of BRCA1 mutations and four in BRCA2 (c.3036-3039delACAA [p.K936fs], c.6857-6858delAA [p.E2210fs], c.9254-9258del5 [p.Y3009fs], c.9538-3539delAA [p.K3104fs]; numbered GenBank U43746) account for 56.6 % of the BRCA2 mutations [12]. Moreover, an additional founder mutation in the Spanish population, located in BRCA2 has recently been observed: c.5374-5377delTATG [p.Y1716fs] (Comunicated by Infante M, et al. at the XXIII Congreso Nacional de Genética Humana, Valladolid, Spain, 2006).
Automated sequencing using standardized procedures represents the "gold standard" system of analysis (complemented by an assay that can detect copy number changes as well). However this procedure is time-consuming and expensive. The identification of fully characterized, recurrent mutations could reduce the time and cost of the analysis. Thus, for instance, Revillion et al. [13] developed a multiplex single-nucleotide primer extension analysis to simultaneous detects 36% of French mutations and 32 % of international ones.
In the present work we describe a rapid, sensitive and low cost assay based on a multiplexed single nucleotide primer extension reaction using SNaPshot dye labeled terminators and capillary electrophoresis to allow rapid detection of approximately 50% of the mutations detected in BRCA1 and BRCA2 in Spanish families.
Methods
Samples and Extraction method
The 48 DNA patient samples included in this study were sent to our laboratory for BRCA1 and BRCA2 diagnosis. Written informed consent was obtained from each patient. Twenty-four of the 48 DNA samples carried one of the ten recurring germline mutations in the BRCA1 and BRCA2 genes identified in Spain (BRCA1: c.187-188delAG, c.330A > G, c.5236G > A, c.5242C > A, c.589-590delCT; numbered GeneBank U14680; BRCA2: c.3036-3039delACAA, c.5374-5377delTATG, c.6857-6858delAA, c.9254-9258del5, c.9538-3539delAA numbered GeneBank U43746). In the remaining 24 samples, no BRCA1/2 mutations were present. DNA was extracted from peripheral blood leucocytes by standard procedures.
SNaPshot multiplex primer design
The primers for PCR amplification and SNaPshot extension reactions (Tables 1 and 2) were both designed to have an annealing temperature around 60°C using Primer3 software [14]. To test for possible repetitive sequences, primers were aligned with the NCBI sequence databases using BLAST [15]. AutoDimer Software [16] was used to test for potential hairpin structures and primer-dimer problems. Primer synthesis was performed by Sigma-Genosys Ltd. (Pampisford, Cambridgeshire, UK). All minisequencing primers were purified by HPLC to remove incomplete primer synthesis products. Co-amplification in the same amplicon was used to detect c.5236 G > C and c.589-590delCT closely positioned in exon 18 of BRCA1. The PCR product sizes ranged from 151 bp to 250 bp.
Table 1.
PCR primer sequences (5'→3')
| Mutation (Predicted effect) | PCR primers | Size bp | Exon |
| BRCA1 c.187-188delAG (p.Q23fs) | F ttcgcgttgaagaagtacaaaa R ttccctagtatgtaaggtcaattctg |
151 | 2 |
| BRCA1 c.330 A > G (p.64-71del) | F ggctcttaagggcagttgtg R cctactgtggttgcttccaa |
235 | 5 |
| BRCA1 c.589-590delCT (p.S157X) | F ctcttcaggaggaaaagcaca R cctgagacccttacccaattc |
186 | 8 |
|
BRCA1 c.5236 G > C (p.G1706A) BRCA1 c.5242 C > A (p.A1708E) |
F ggacagcacttcctgattttg R taaagggaggaggggagaaa |
174 | 18 |
| BRCA2 c.3036-3039delACAA (p.K936fs) | F ttcatgaaacagacttgacttgtg R ttcaaggagatgtccgatttt |
208 | 11 |
| BRCA2 c.5374-5377delTATG (p.Y1716fs) | F tggcttagagaaggaatatttgatg R ctggctcaataccagaatcaag |
250 | 11 |
| BRCA2 c.6857-6858delAA (p.E2210fs) | F tgggaaaagaacaggcttca R gaatgtgtggcatgacttgg |
217 | 11 |
| BRCA2 c.9254-9258del5 (p.Y3009fs) | F tcttccattgcatctttctca R ggtttgtaccggtagttgttga |
209 | 23 |
| BRCA2 c.9538-9539delAA (p.K3104fs) | F gagtttcctttcttgcatcttaaa R gcctgatttggattctggtc |
221 | 25 |
Table 2.
Extension primer sequences
| Mutation | Extension Primer | Size bp | Change |
| BRCA1 c.187-188delAG | F gactcattaatgctatgcagaaaatcttag | 30 | A/T |
| BRCA1 c.5236 G > C | F gacttgaacggacactgaaatattttctag | 30 | G/C |
| BRCA1 c.330 A > G | F gactgatgtcctttatgtaagaatgatataaccaaa | 36 | A/G |
| BRCA2 c.5374-5377delTATG | F gactgactgactaatactgcagattatgtaggaaattatttg | 42 | T/A |
| BRCA1 c.589-590delCT | F gactgactgactgactgactgactaaaccagtctcagtgtccaactct | 48 | C/A |
| BRCA2 c.3036-3039delACAA | F gacgactgactgactgactgactgactggttttatatggagacacaggtgataa | 54 | A/G |
| BRCA1 c.5242C > A | R acgactgactgactgactgactgactgactgactgactgctaactacccattttcctccc | 60 | T/G |
| BRCA2 c.9254-9258del5 | F gacgactgactgactgactgactgactgactgttaacagaaggaaagagatacagaattt | 60 | A/C |
| BRCA2 c.6857-6858delAA | F gacgactgactgactgactgactgactgactgactgactctgatgttcctgtgaaaacaaatatag | 66 | A/G |
| BRCA2 c.9538-9539delAA | F gagactgactgactgactgactgactgactgactgactgactgactacgaatgttacaatttactggcaata | 72 | A/G |
* Poly(dGACT) tail
The extension primer was designed to anneal immediately adjacent to the nucleotide at the mutation site, on either the sense or anti-sense DNA strand, for the detection of BRCA1 c.5242C > A mutation we used anti-sense primer. The primer orientation allowed a multiplexed SNaPshot reaction encompassing all the loci to be detected in the absence of intra- and inter-primer complementarities.
PCR multiplex amplification
The PCR multiplex co-amplified 9 fragments for the 10 BRCA1/BRCA2 mutations, with sizes ranging from 174 to 250 bp with a single amplicon encompassing the two exon 18 mutations: c.5242 G > A, and c.589-590delCT of BRCA1 gene. Amplification was performed using 5 ng of DNA template, 2 × PCR master mix from QIAGEN Multiplex PCR kit (Qiagen Hilden Germany), 10 × amplification primer mix (2 μM each primer with final concentration of 0.2 μM, optimal for most primer-template systems). Cycling was carried out using a 9700 thermocycler (Applied Biosystems, Foster City, CA, USA). Following a 95°C initial activation step for HotStar Taq DNA polymerase of 15 minutes, a total of 35 cycles were made using the following conditions: 94°C denaturation for 30 seconds, primer annealing at 60°C for 90 seconds, followed by 15 minutes of final extension at 72°C and a 4°C holding step. PCR quality and yield were checked using an Agilent 2100 Expert Bioanalyzer DNA 500/DNA 1000 (Agilent Technologies Inc. Santa Clara, CA USA).
Minisequencing SNaPshot reaction
After amplification, PCR products required purification to remove primers and un-incorporated dNTPs. This post-PCR purification was performed as follows: 1 μl of PCR product was incubated with 0.5 μl of ExoSapIT (Amershan Pharmacia Biotech) for 15 min at 37°C followed by 15 min at 80°C for enzyme inactivation. The minisequencing reaction was performed using the SNaPshot™ kit in a 9700 thermocycler with 2 μl of SNaPshot ready reaction mix, 0.2 μM of extension primer for each mutation and 1.5 μl of purified PCR products in a 5 μl total volume. Extension used 25 cycles of denaturation at 96°C for 10 seconds, annealing at 50°C for 5 seconds and extension at 60°C for 30 seconds. Detection of extension products is based on four different fluorochromes to assay each base and controlled extension primer sizes (the length of a primer regulated using different sized non-homologous Poly-dGACT tails at the 5' end). Final extension product sizes ranged between 30 and 72 bp (see Table 2). After the minisequencing reaction, a post-extension treatment to remove the 50-phosphoryl group of the ddNTPs helps to prevent unincorporated terminators from comigrating with the extended primers and producing high background signal. For this a 10 μl final volume was treated with 1 μl of SAP (Amersham Biosciences) for 60 min at 37°C, followed by 15 min at 80°C for enzyme inactivation.
Electroforetic SNaPshot conditions
The minisequencing products (1.5 μl) were mixed with 10 μl of HiDi™ formamide and 0.5 μl of GeneScan-120 LIZ size standard (Applied Biosystems) and denatured at 95°C for 5 minutes. The fluorescently labeled fragments were resolved by capillary electrophoresis on an ABI PRISM 3730 × l Genetic Analyzer (Applied Biosystems). Minisequencing products were injected electrokinetically for 10 seconds at 15 kV and electrophorezed for 20 min at 15 kV and 9 mA at 60°C (default module) in a 36 cm length capillary using POP-7™ polymer with the laser set at a constant power of 9.9 mW. The resulting data was analyzed with GeneMapper™ 3.7 Software (Applied Biosystems).
Results
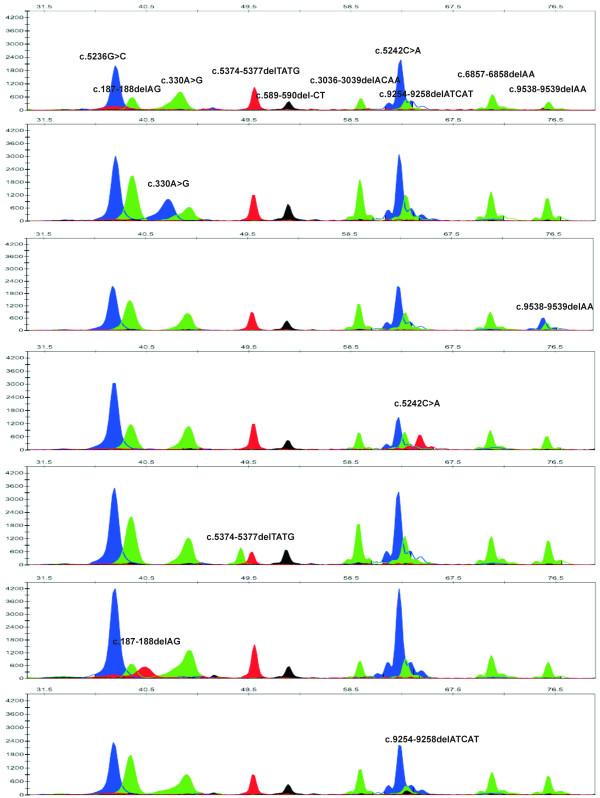
The SNaPshot reaction was performed on samples previously analyzed by direct sequencing and all mutations were concordant. Mutations are usually present in the heterozygous state yielding two peaks, one with the expected size and color and a "mutant" peak with a different color according to the base incorporated (Fig. 1).
Figure 1.
Electropherograms of Genscan Analysis of the SNaPshot reaction. On the top, a control DNA sample with the position and nucleotide of the wild type allele indicated. Following, electropherograms for the most frequents BRCA1/2 mutations.
The electrophoretic mobility of the extension products depends not only on their lengths, but also on the neighboring nucleotide sequences and on the fluorescent dye used in the reaction.
Therefore, as expected, the detected size determined by the automated sequencer and the real size of certain products were slightly different. It can be tentatively said that to ensure correct differentiation of the extended primer, amplicons should differ by at least six nucleotides in length. Some fragments display stronger fluorescent signal than others in the electropherogram (even after colour compensation with the appropriate matrix) due to variation in minisequencing chemistry. Therefore, mutations are detected by the automated sequencer at different peak heights depending on the terminator and dye label. In general, guanine labeled with dR100 produces a higher signal than the other ddNTPs. However, this fact does not unduly affect the readability of the electropherogram [see also [17]
No false positive results were observed in control samples without mutations, demonstrating the reliability of the procedure and repetitive assays proved the reproducibility of the method.
Discussion
The genetic analysis of BRCA1 and BRCA2 is complex and expensive essentially because their length. Nevertheless, the presence of recurrent and founder mutations allows a pre-screening for the identification of the most frequent mutations found in each geographical region. This is the case of the single BRCA2 mutation found in the Icelandic population (c.999del5), and the 3 founder Ashkenazi Jews mutations that comprise the genetic test targeted to this population. In contrast to isolated populations the number of mutations that need to be included in the pre-screening process in Spanish population is substantially higher: ten recurrent and founder mutations represent approximately 50 % of the identified familial mutations, and are good candidates to be included in the pre-screening [12, and communication of Infante M, et al. at the XXIII Congreso Nacional de Genética Humana, Spain]. Based on this high-frequency-mutation group, we have developed a SNaPshot reaction to analyze all ten mutations in a single assay. This strategy has numerous advantages. Firstly, a single tube reaction is used for each patient that implies the reduction in term of number of steps and handling. Secondly, high-throughput genotyping is possible as the process can be automated, and the assay is carried out in 96 wells plates. In addition, the interpretation of the peak patterns is very simple and the method is sensitive [see other forensic applications in 18 and 19] and low cost. Lastly, the design is highly flexible: (i) more mutations can be added to the multiplex and (ii) SNaPshot can detect point mutations but also indels, as in our design (some point mutations: c.330A > G or c.5236G > A, and some indels: c.187-188delAG or c.3036-3039delACAA).
The SNaPshot design we propose here represents an advance in the genetic diagnosis of BRCA1 and BRCA2 Spanish families, since permits the identification of the 46.6 % of BRCA1 and more than 56.6 % of the BRCA2 families in few hours. Moreover it is not only useful for Spanish patients, but also for those populations with Spanish ancestry such as those from Latin American countries. Many families from Latin America can trace their European ancestry to the period of Spanish colonization, and, in the last century, to migratory movements from diverse regions of Spain. An example of this is the occurrence of two Spanish mutations c.330A > G in BRCA1 and c.6857-6858delAA in BRCA2 in populations with Spanish ancestry: the former was found in Latin/South American/Caribbean families with Galician ancestors [20,21], and the later mutation was described in one Chilean family with Catalan origins [12].
The utility of our SNaPshot design in Latin American population also applies to recent admixture populations. Thus, for instance, the study of Torres et al. [22] shows that three mutations account for almost 80% of deleterious mutations identified in a Hispanic American cohort from Colombia. Two of these three mutations are frequent in Spanish population and included in our SNaPshot multiplex design we present here, supporting the utility of our design for the initial screening of Latino American breast cancer families. Five of the nine different mutations observed in Chile [23] are in the Top 20 Mutation Frequency of the BIC database [3]. This suggest that a modified SNaPshot considering these Top 20 mutation (some already included in our SNaPshot design) could yield good results for screening in Chile as in other recently admixture populations.
Conclusion
In summary, here we propose a fast, flexible and cost-effective multiplex assay of the ten most frequent BRCA1 and BRCA2 mutations in Spain, for the initial screening of Spanish and Spanish ancestry population's breast/ovarian cancer families.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
SF performed the genetic design, carried out the genetic analysis and drafted the manuscript. AB and AFM helped with the genetic analysis of the samples and drafted the manuscript. VAI help in the methodological design. CRP and AC helped to draft and revised the manuscript. AV conceived the study, participated in its design and coordination, and wrote the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
The Spanish grants of the Ministerio de Sanidad y Consumo (PI052275), Xunta de Galicia (PGIDIT06BTF910101PR) and Fundación de Investigación Médica Mutua Madrileña given to AV supported this project. AB is a Fellowship from Instituto de Salud Carlos III (Ministerio de Sanidad, Spain). We thank Dr. Antonio Salas for useful comments on the article.
Contributor Information
Sandra Filippini, Email: sandraefilip@yahoo.com.ar.
Ana Blanco, Email: anablancop@hotmail.com.
Ana Fernández-Marmiesse, Email: ana.fernandez.marmiesse@sergas.es.
Vanesa Álvarez-Iglesias, Email: vaneiml@usc.es.
Clara Ruíz-Ponte, Email: crponte@usc.es.
Ángel Carracedo, Email: apimlang@usc.es.
Ana Vega, Email: apimlapv@usc.es.
References
- Narod SA, Ford D, Devilee P, Barkardottir RB, Lynch HT, Smith SA, Ponder BAJ, Weber BL, Garber JE, Birch JM, Cornelis RS, Kelsell DP, Spurr NK, Smyth E, Haites N, Sobol H, Bignon Y-J, Chang-Claude J, Hamann U, Lindblom A, Borg A, Piver MS, Gallion HH, Struewing JP, Whittemore A, Tonin P, Goldgar DE, Easton DF. An evaluation of genetic heterogeneity in 145 breast ovarian cancer families. Am J Hum Genet. 1995;56:254–264. [PMC free article] [PubMed] [Google Scholar]
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BAJ, Gayther SA, Birch JM, Lindblom A, Stoppa-Lyonnet D, Bignon Y, Borg A, Hamann U, Haites N, Scott RJ, Maugard CM, Vasen H, Seitz S, Cannon-Albright LA, Schofield A, Zelada-Hedman M, the Breast Cancer Linkage Consortium Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breast Cancer Information Core internet web site http://www.nhgri.nih.gov/Intramural_research/Lab_transfer/Bic/index.html
- Tonin P, Weber B, Offit K, Couch F, Rebbeck TR, Neuhausen S, Godwin AK, Daly M, Wagner-Costalos J, Berman D, Grana G, Fox E, Kane MF, Kolodner RD, Krainer M, Haber DA, Struewing JP, Warner E, Rosen B, Lerman C, Peshkin B, Norton L, Serova O, Foulkes WD, Lynch HT, Lenoir GM, Narod SA, Garber JE. Frequency of recurrent BRCA1 and BRCA2 mutations in Ashkenazi Jewish breast cancer families. Nat Med. 1996;2:1179–1183. doi: 10.1038/nm1196-1179. [DOI] [PubMed] [Google Scholar]
- Neuhausen S, Gilewski T, Norton L, Tran T, McGuire P, Swensen J, Hampel H, Borgen P, Brown K, Skolnick M, Shattuck-Eidens D, Jhanwar S, Goldgar D, Offit K. Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet. 1996;13:126–128. doi: 10.1038/ng0596-126. [DOI] [PubMed] [Google Scholar]
- Johannesdottir G, Gudmundsson J, Bergthorsson JT, Arason A, Agnarsson BA, Eiriksdottir G, Johannsson OT, Borg A, Ingvarsson S, Easton DF, Egilsson V, Barkardottir RB. High prevalence of the 999del5 mutation in Icelandic breast and ovarian cancer patients. Cancer Res. 1996;56:3663–3665. [PubMed] [Google Scholar]
- Huusko P, Paakkonen K, Launonen V, Poyhonen M, Blanco G, Kauppila A, Puistola U, Kiviniemi H, Kujala M, Leisti J, Winqvist R. Evidence of founder mutations in Finnish BRCA1 and BRCA2 families. Am J Hum Genet. 1998;62:1544–1548. doi: 10.1086/301880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peelen T, van Vliet M, Petrij-Bosch A, Mieremet R, Szabo C, van den Ouweland AMW, Hogervorst F, Brohet R, Ligtenberg MJL, Teugels E, van der Luijt R, van der Hout AH, Gille JJP, Pals G, Jedema I, Olmer R, van Leeuwen I, Newman B, Plandsoen M, van der Est M, Brink G, Hageman S, Arts PJW, Bakker MM, Willems HW, van der Looij E, Neyns B, Bonduelle M, Jansen R, Oosterwijk JC, Sijmons R, Smeets HJM, van Asperen CJ, Meijers-Heijboer H, Klijn JGM, de Greve J, King M-C, Menko FH, Brunner HG, Halley D, van Ommen G-JB, Vasen HFA, Cornelisse CJ, van 'tVeer LJ, de Knijff P, Bakker E, Devilee P. A high proportion of novel mutations in BRCA1 with strong founder effects among Dutch and Belgian hereditary breast and ovarian cancer families. Am J Hum Genet. 1997;60:1041–1049. [PMC free article] [PubMed] [Google Scholar]
- Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FB, Hageman S, Arts PJ, Ligtenberg MJ, Meijers-Heijboer H, Klijn JG, Vasen HF, Cornelisse CJ, van't Veer LJ, Bakker E, van Ommen GJ, Devilee P. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet. 1997;17:341–345. doi: 10.1038/ng1197-341. [DOI] [PubMed] [Google Scholar]
- Tonin PN, Mes-Masson AM, Futreal PA, Morgan K, Mahon M, Foulkes WD, Cole DE, Provencher D, Ghadirian P, Narod SA. Founder BRCA1 and BRCA2 mutations in French Canadian breast and ovarian cancer families. Am J Hum Genet. 1998;63:1341–1351. doi: 10.1086/302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski B, Byrski T, Huzarski T, Jakubowska A, Menkiszak J, Gronwald J, Pluzanska A, Bebenek M, Fischer-Maliszewska L, Grzybowska E, Narod SA, Lubinski J. Founder mutations in the BRCA1 gene in Polish families with breast-ovarian cancer. Am J Hum Genet. 2000;66:1963–1968. doi: 10.1086/302922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez O, Osorio A, Duran M, Martínez-Ferrandis JI, de la Hoya M, Salazar R, Vega A, Campos B, Rodríguez-López R, Velasco E, Chaves J, Díaz-Rubio E, Jesús Cruz J, Torres M, Esteban E, Cervantes A, Alonso C, San Roman JM, González-Sarmiento R, Miner C, Carracedo A, Eugenia Armengod M, Caldés T, Benítez J, Baiget M. Analysis of BRCA1 and BRCA2 genes in Spanish breast/ovarian cancer patients: a high proportion of mutations unique to Spain and evidence of founder effects. Hum Mut. 2003;22:301–312. doi: 10.1002/humu.10260. [DOI] [PubMed] [Google Scholar]
- Revillion F, Verdiere A, Fournier J, Hornez L, Peyrat JP. Multiplex single-nucleotide primer extension analysis to simultaneously detect eleven BRCA1 mutations in breast cancer families. Clin Chem. 2004;50:203–206. doi: 10.1373/clinchem.2003.023713. [DOI] [PubMed] [Google Scholar]
- Primer3 software http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi
- BLAST (Basic Local Alignment Search Tool) http://www.ncbi.nlm.nih.gov/blast/blast.cgi
- AutoDimer Software http://www.cstl.nist.gov/div831/strbase//AutoDimerHomepage/AutoDimerProgramHomepage.htm
- Álvarez-Iglesias V, Jaime JC, Carracedo Á, Salas A. Coding region mitochondrial DNA SNPs: targeting East Asian and Native American haplogroups. Forensic Sci Int Genet. 2007;1:44–55. doi: 10.1016/j.fsigen.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Quintáns B, Álvarez-Iglesias V, Salas A, Phillips C, Lareu MV, Carracedo A. Typing of mitochondrial DNA coding region SNPs of forensic and anthropological interest using SNaPshot minisequencing. Forensic Sci Int. 2004;140:251–257. doi: 10.1016/j.forsciint.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Crespillo M, Paredes MR, Prieto L, Montesino M, Salas A, Albarran C, Álvarez-Iglesias V, Amorin A, Berniell-Lee G, Brehm A, Carril JC, Corach D, Cuevas N, Di Lonardo AM, Doutremepuich C, Espinheira RM, Espinoza M, Gómez F, González A, Hernandez A, Hidalgo M, Jimenez M, Leite FP, López AM, López-Soto M, Lorente JA, Pagano S, Palacio AM, Pestano JJ, Pinheiro MF, Raimondi E, Ramón MM, Tovar F, Vidal-Rioja L, Vide MC, Whittle MR, Yunis JJ, Garcia-Hirschfel J. Results of the 2003–2004 GEP-ISFG collaborative study on mitochondrial DNA: focus on the mtDNA profile of a mixed semen-saliva stain. Forensic Sci Int. 2006;160:157–167. doi: 10.1016/j.forsciint.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Vega A, Campos B, Bressac-de-Paillerets B, Bond PM, Janin N, Douglas FS, Domenech M, Baena M, Pericay C, Alonso C, Carracedo A, Baiget M, Díez O. The R71G BRCA1 is a founder Spanish mutation and leads to aberrant splicing of the transcript. Hum Mut. 2001;17:520–521. doi: 10.1002/humu.1136. [DOI] [PubMed] [Google Scholar]
- Vega A, Torres M, Martínez JI, Ruíz-Ponte C, Barros F, Carracedo A. Analysis of BRCA1 and BRCA2 in Breast and Breast/Ovarian Cancer Families shows Population Substructure in the Iberian Peninsula. Ann Hum Genet. 2002;63:29–36. doi: 10.1017/S0003480001001014. [DOI] [PubMed] [Google Scholar]
- Torres D, Rashid MU, Gil F, Umana A, Ramelli G, Robledo JF, Tawil M, Torregrosa L, Briceno I, Hamann U. High proportion of BRCA1/2 founder mutations in Hispanic breast/ovarian cancer families from Colombia. Breast Cancer Res Treat. 2006. [DOI] [PubMed]
- Jara L, Ampuero S, Santibanez E, Seccia L, Rodríguez J, Bustamante M, Martínez V, Catenaccio A, Lay-Son G, Blanco R, Reyes JM. BRCA1 and BRCA2 mutations in a South American population. Cancer Genet Cytogenet. 2006;166:36–45. doi: 10.1016/j.cancergencyto.2005.08.019. [DOI] [PubMed] [Google Scholar]