Abstract
The control of alternative pre-mRNA splicing often requires the participation of factors displaying synergistic or antagonistic activities. In the hnRNP A1 pre-mRNA, three elements promote the exclusion of alternative exon 7B, while a fourth intron element (CE9) represses splicing of exon 7B to the downstream exon. We have shown previously that the 5′ portion of the 38-nucleotide-long CE9 element is bound by SRp30c, and that this interaction is important for repression in vitro. To determine whether SRp30c alone can impose repression, we tested a high-affinity SRp30c binding site that we identified using the SELEX protocol. We find that multiple high-affinity SRp30c sites are required to replicate the level of repression obtained with CE9, and that both the 5′ and the 3′ portions of CE9 contribute to SRp30c binding. Performing RNA affinity chromatography with the complete CE9 element recovered hnRNP I/PTB. Surprisingly however, His-tagged PTB reduced the binding of SRp30c to CE9 in a nuclear extract, stimulated splicing to a downstream 3′ splice site, and relieved the CE9-mediated splicing repression in vitro. Our in vivo results are consistent with the notion that increasing PTB levels alleviates the repression imposed by CE9 to a downstream 3′ splice site. Thus, PTB can function as an anti-repressor molecule to counteract the splicing inhibitory activity of SRp30c.
Keywords: SRp30c, hnRNP I/PTB, 3′ splice site selection, alternative splicing, anti-repression
INTRODUCTION
The majority of mammalian pre-messenger RNAs (pre-mRNAs) are alternatively spliced to produce mRNA isoforms that can be translated to increase the protein-coding capacity of genomes. The control of alternative splicing often relies on combinatorial networks involving several cis-acting elements bound by factors that belong to the hnRNP and SR families of RNA binding proteins (Black 2003; Matlin et al. 2005). hnRNP proteins frequently mediate splicing repression, particularly through binding to exonic splicing silencer elements or by sterically interfering with the interaction of generic splicing factors (Lin and Patton 1995; Romano et al. 2002). PTB is a hnRNP protein that represses the inclusion of alternative exons in numerous systems (Wagner and Garcia-Blanco 2001). Nevertheless, hnRNP proteins can also associate with enhancer elements to elicit exon inclusion (Caputi and Zahler 2002), and a generic role for hnRNP A1 and hnRNP F/H proteins in intron definition has been proposed recently (Martinez-Contreras et al. 2006). The situation is somewhat reversed for the SR family of proteins. Mammalian SR proteins were initially described as positive regulators of splicing and mediators of the activity of exonic splicing enhancers (Lavigueur et al. 1993; for review, see Graveley 2000). However, recent results suggest a more complex situation since SR proteins can participate in the negative control of splicing and alternative splicing. For example, SRp38 has been implicated in the cell-cycle repression of splicing at the M phase, and the dephosphorylation of SRp38 promotes an interaction with U1 snRNP that interferes with 5′ splice site recognition (Shin and Manley 2002). The binding of ASF/SF2 to a branch site region has been associated with the steric repression of the 3′splice site of the adenovirus IIIa pre-mRNA (Kanopka et al. 1996). Moreover, we have shown that SRp30c mediates the repressor activity of a conserved intronic element (CE9) located 120 nucleotides (nt) upstream from the 3′ splice site of exon 8 in the hnRNP A1 pre-mRNA (Fig. 1A). In heterologous constructs, the SRp30c/CE9 interaction represses a downstream 3′ splice site (Simard and Chabot 2000, 2002). Thus, intronic binding sites for SR proteins have been associated with splicing inhibition on many occasions (Gallego et al. 1997; Pagani et al. 2000; Ibrahim et al. 2005).
FIGURE 1.
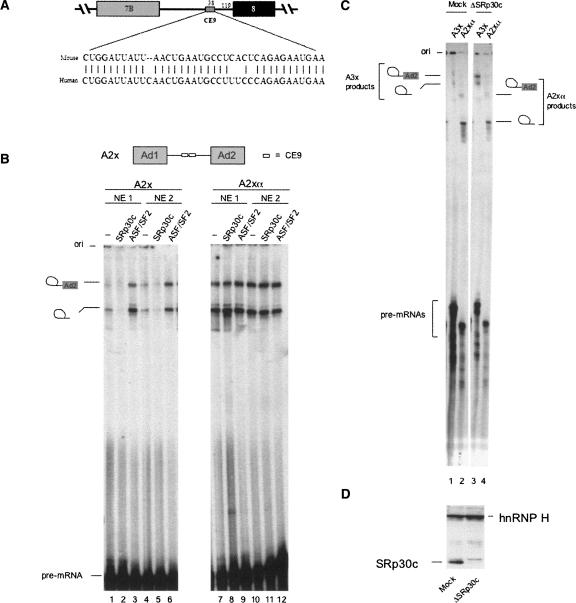
SRp30c mediates the repressor activity of CE9. (A) The CE9 element maps in the intron downstream of alternative exon 7B in the hnRNP A1 pre-mRNA. The sequence of CE9 is conserved in mouse and human hnRNP A1 introns. (B) The A2x pre-mRNA contains two copies of CE9. A2xα contains two copies of the complementary sequence. Pre-mRNAs were incubated in two independent preparations of HeLa nuclear extracts (NE1 and NE2). Some mixtures were supplemented with 5 μM of His-SRp30c or GST-ASF/SF2, as indicated. Splicing products were fractionated on a 6.5% acrylamide/8 M urea gel. (C) The A3x pre-mRNA contains three copies of CE9. Pre-mRNAs were incubated in an extract prepared from HeLa cells that had previously been transfected with a siRNA against SRp30c (ΔSRp30c). Mock-transfected cells were used to prepare the control extract (mock). (D) Western analysis of the nuclear extracts from cells that were depleted of SRp30c by RNA interference (ΔSRp30c) or from mock-treated cells (mock). An anti-SRp30c antibody was used first to determine the position of SRp30c. The membrane was then redecorated with an antibody against hnRNP H (Garneau et al. 2005).
SRp30c has been implicated in the activity of many elements that control splice site selection. SRp30c stimulates the inclusion of SMN exon 7 (Young et al. 2002). This activity apparently requires an interaction with hTra2β since SRp30c does not bind independently to the SMN exon. hTra2β also binds to a splicing enhancer in the tau alternative exon 10 to promote exon inclusion, and overexpression of SRp30c can promote tau exon 10 inclusion (Jiang et al. 2003; Kondo et al. 2004). These observations are in contradiction with a more recent report indicating that SRp30c inhibits the inclusion of both alternative tau exons 2 and 10 (Wang et al. 2005). In the latter study, silencer elements located at the 5′ end of both exons were identified as binding sites for SRp30c and SRp55. Because SRp30c could interact with SRp55 and hTra2β, it was suggested that SRp30c prevents the productive interaction of hTra2β with the purine-rich enhancer (Wang et al. 2005). SRp30c has also been implicated in the alternative splicing of the glucocorticoid receptor β in neutrophils (Xu et al. 2003) and in the gonadotropin-releasing hormone pre-mRNA (Park et al. 2006). In the latter case, SRp30c binds to several exonic enhancer elements, interacts with Tra2α, and collaborates with the SR protein 9G8 to facilitate intron A splicing. Finally, SRp30c can interact with other proteins implicated in alternative splicing: YB-1, a factor capable of altering adenovirus E1A alternative splicing (Raffetseder et al. 2003); rSLM-1 and rSLM2, two RNA binding proteins that influence the alternative splicing of CD44 exon v5 (Stoss et al. 2004); E4-ORF4, an adenoviral protein that activates IIIa splicing (Estmer Nilsson et al. 2001); Nop30, which affects the alternative splicing of preprotachykinin and SRp20 reporter genes (Stoss et al. 1999); and p32, a cellular protein that inhibits ASF/SF2 function (Petersen-Mahrt et al. 1999).
Given the wide range of action of SRp30c and its interaction with many factors to activate or repress splicing, we have pursued our efforts at characterizing the function of SRp30c in the activity of the intronic CE9 element found in the hnRNP A1 pre-mRNA. Using a recombinant SRp30c protein in a SELEX protocol, we have identified an optimal binding site for SRp30c and have defined some of the sequence requirements for high-affinity binding. We show that multiple high-affinity binding sites for SRp30c can replicate the impact of the CE9 element. Moreover, we show that the 3′ portion of CE9 cooperates with the 5′ portion to promote stronger SRp30c binding. Finally, we find that PTB can interact with CE9. However, and in contrast to the numerous examples implicating PTB in splicing repression, PTB counteracts the SRp30c-mediated repressor activity of CE9.
RESULTS
SRp30c and ASF/SF2 modulate the activity of CE9
When placed in between two competing 3′ splice sites, the 38-nt-long CE9 element can shift 3′ splice site selection toward the upstream site (Simard and Chabot 2000). CE9 most likely acts by repressing the downstream 3′ splice site since tandem copies of CE9 inhibit in vitro splicing of a single-intron reporter pre-mRNA (Simard and Chabot 2000, 2002). The repressor activity of CE9 is observed in different preparations of HeLa nuclear extracts using an adenovirus model pre-mRNA that contains two intronic copies of CE9 (Fig. 1B, cf. lanes 1,4 and 7,10). SRp30c can bind to the first 16 nt of CE9 (Simard and Chabot 2002), and the addition of recombinant His-SRp30c to splicing mixtures further inhibits splicing in a CE9-dependent manner (Fig. 1B, cf. lanes 2,5 and 1). Adding SRp30c to the control pre-mRNA that lacks CE9 has no repressive effect (Fig. 1B, cf. lanes 8,11 and 7). ASF/SF2 is the closest homolog of SRp30c, with 74% amino acid identity (Screaton et al. 1995). In contrast to SRp30c, ASF/SF2 stimulates splicing of the CE9-containing pre-mRNA (Fig. 1B, cf. lanes 1,4 and 3,6). This stimulation is not observed with a control pre-mRNA that contains the complementary sequence of two CE9 elements (Fig. 1B, cf. lanes 7,10 and 9,12). Thus, ASF/SF2 can antagonize the repressing activity of SRp30c in the context of CE9. This antagonism may involve a competition for binding, although we have been unable to detect ASF/SF2 binding to CE9 using the conditions of our gel mobility shift assays (Simard and Chabot 2002).
A role for SRp30c in the activity of CE9 was also supported by performing a splicing experiment in a nuclear extract prepared from HeLa cells that had been previously transfected with a siRNA targeting SRp30c (Fig. 1C). This treatment drastically reduces the level of SRp30c, as judged by Western analysis (Fig. 1D). In an extract prepared from mock-treated cells, a pre-mRNA containing three copies of CE9 (A3x) is spliced less efficiently than a control pre-mRNA carrying a complementary insert (A2xα) (Fig. 1C, lanes 1,2). Notably, the splicing efficiency of A3x in the SRp30c-depleted extract was stimulated (Fig. 1C, lanes 3,4). These results support a role for SRp30c in mediating CE9-dependent repression.
Identification of a high-affinity binding site for SRp30c
To assess whether SRp30c is sufficient to impose repression, we wished to test the repression activity of a strong SRp30c binding site. To identify such a site, we relied on the iterative in vitro genetic selection protocol (SELEX) using recombinant GST-SRp30c and a pool of randomized RNA sequences. We used a large molar excess of a pool of synthetic RNA molecules containing a randomized 20-nt region flanked by constant regions for primer annealing. Following six rounds of selection–amplification, the bound RNA material was reverse transcribed, amplified, and cloned. Sequencing 37 individual clones revealed that the sequence AGGAC was the most frequent, although the consensus emerged as AGSAS (S = G or C) (Fig. 2A). Twenty-two clones had at least one copy of this motif and five had two. Ten other clones carried sequences with a four out of five match to AGGAC.
FIGURE 2.
Binding sites for SRp30c as determined by SELEX. (A) In vitro selection–amplification of high-affinity RNA target sequences for SRp30c. The sequences of 37 individual clones after six cycles are shown and arranged according to their fit (in bold) to the consensus AGSAS motif (S = G or C). The diagram below shows the frequency of each nucleotide based on this arrangement. The height of each letter is proportional to its frequency, the percentage being indicated at the right of the letter. (B) Binding of recombinant SRp30c to the S21 RNA oligo. Gel mobility shift assays were set up in the presence of heparin and GST-SRp30c (2.5 and 5 μM), His-SRp30c (2.5 and 5 μM), and ASF/SF2 (5 μM). The sequence of the S21 RNA oligo is shown with the AGGAC sequences underlined. The position of free RNA and the SR protein/RNA complexes is shown. (C) SRp30c binding to mutated versions of S21. The sequence of S21 and derivatives are shown. The positions that have been mutated are boxed. GST-SRp30c was used at 2.5 and 5 μM. (D) Binding of SRp30c to S21 and CE9-derived sequences. The sequences of the RNAs tested are shown above. S21, CE9, and 3xCE9.7 are RNA oligos labeled at their 5′ end while 3xCE9 is a uniformly labeled RNA produced by in vitro transcription. GST-SRp30c was used at 2.5, 3.7, and 5 μM. (E) Binding of SRp30c and ASF/SF2 to S21 and 3xCE9.7. The concentrations of the SR proteins were 2.5 and 5 μM.
Next, we used RNA oligos derived from these sequences to perform binding assays with SRp30c. An oligo containing two AGGAC motifs was bound by SRp30c in a gel mobility shift assay (S21; Fig. 2B, lanes 2,3). Similar results were obtained with a His-tagged SRp30c (Fig. 2B, lanes 4,5). Converting a single AGGAC motif in S21 to AGCAC strongly reduced the efficiency of complex formation (Fig. 2C; S21-CG and S21-GC), while converting both motifs to AGCAC or ACCAC completely eliminated it (Fig. 2C; S21-CC and S21-CCCC). Mutating the sequence adjacent to the AGGAC motifs did not strongly affect SRp30c binding (Fig. 2C; S21-mod6). Thus, two adjacent AGGAC motifs conferred optimal SRp30c binding, as judged by gel mobility shift assays.
SRp30c bound more efficiently to S21 than to the complete CE9 element (Fig. 2D, cf. lanes 13–16 and 9–12). The binding of SRp30c to S21 was equivalent to the binding of SRp30c to an RNA carrying three copies of CE9 (3xCE9) or three copies of the 5′ terminal sequence of CE9 (3xCE9.7) (Fig. 2D, lanes 1–4, 5–8, respectively). We also tested the binding of ASF/SF2 to S21. The preparation of His-ASF/SF2 was as active as the preparation of His-SRp30c at stimulating proximal 5′ splice site selection (not shown). As shown in Figure 2E, ASF/SF2 binds to S21, albeit less efficiently than SRp30c (Fig. 2E, lanes 1–5). In contrast, the binding of ASF/SF2 to 3xCE9.7 could not be detected, even after a longer exposure, consistent with previous observations (Simard and Chabot 2002). Thus, while SRp30c displays more affinity to S21, the CE9 sequence may represent a more specific binding site for SRp30c.
Multiple SRp30c binding sites reproduce the activity of CE9
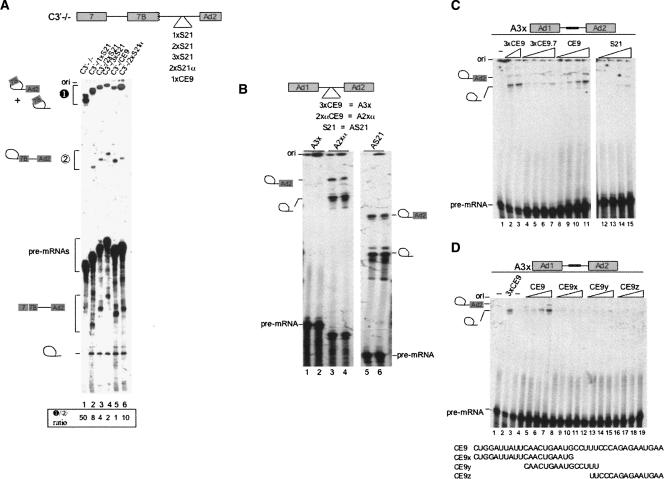
Next, we compared the splicing modulating activity of CE9 to that of the SRp30c binding site represented by S21. As shown previously, one copy of the complete CE9 element inserted in between the two 3′ splice sites of pre-mRNA C3′−/− decreased distal 3′ splice site usage and improved the use of the proximal site (Simard and Chabot 2002; Fig. 3A, cf. lanes 5 and 1). Although one copy of S21 inserted at the same position activated proximal 3′ splice site utilization, the amplitude of the effect was smaller than with CE9 (Fig. 3A, lane 2). Two copies of S21 repressed more efficiently the distal 3′ splice site, while insertion of the complementary sequences had a smaller effect (Fig. 3A, lanes 3,6, respectively). Three copies of S21 affected 3′ splice site selection in a manner that was nearly equivalent to the complete CE9 element (Fig. 3A, lane 4). Thus, although SRp30c binds to CE9 more weakly than to S21, CE9 is more efficient, and multiple copies of S21 are required to reproduce the repression mediated by CE9.
FIGURE 3.
Multiple S21 elements are required to mimic the repressor activity of CE9. (A) 3′ splice site selection assay. Labeled pre-mRNAs containing CE9 or copies of the S21 element were incubated in HeLa extracts for 2 h at 30°C. The structure of the pre-mRNAs is indicated on top. Splicing products were resolved in a 11% acrylamide/8 M urea gel. The position of the pre-mRNA lariat splicing products derived from proximal or distal 3′ splice site use is indicated, as is the mRNA produced from the use of the proximal 3′ splice site. The ratios between both distal lariat products and the intermediate proximal product have been assessed to appreciate the activity of the fragment inserted in between the two 3′ splice sites (values below lane numbers). (B) Splicing repression assay. The structure of the pre-mRNAs is indicated on top. Labeled pre-mRNAs were incubated in HeLa extracts for 2 h at 30°C. Splicing products were fractionated in 6.5% acrylamide/8 M urea gels. The position of the pre-mRNA and lariat molecules is indicated. (C,D) Splicing repression assays were performed in the presence of an excess of competing RNAs. Using A3x in splicing mixtures, increasing amounts of unlabeled competing RNA were incubated in splicing extracts. In panel C, the concentrations of the competing RNAs represent an excess of 250 and 500 for 3xCE9 (relative to the pre-mRNA A3x), whereas they are 5000, 10,000, 25,000 and 50,000 for S21, CE9, 3xCE9.7, CE9x, CE9y and CE9z. The sequences of the oligos representing portions of CE9 are shown.
The reduced repression activity of S21 was confirmed in a splicing inhibition assay. In contrast to a pre-mRNA carrying three intronic copies of CE9 (A3x), one copy of S21 did not inhibit splicing (Fig. 3B, cf. lanes 5,6 and 1,2). Given that S21 and 3xCE9 are bound by recombinant SRp30c with similar affinities, as judged by gel mobility shift assays (Fig. 2D), our results indicate that not all elements bound by recombinant SRp30c display equivalent repressing activity. A similar conclusion was reached when we tested the impact of an excess of various RNA fragments on the CE9-mediated splicing repression. The addition of 3xCE9 RNA, and to a lesser extent of 3xCE9.7 and CE9 RNAs, stimulated the splicing of A3x (Fig. 3C, lanes 2–3, 4–7, 8–11, respectively). Similar amounts of S21 only very weakly stimulated splicing of A3x (Fig. 3C, lanes 12–15). None of the above RNAs affected the splicing efficiency of the control A2xα pre-mRNA (data not shown). We also tested the rescuing ability of RNA oligos corresponding to the 5′, central, and 3′ portions of CE9 (CE9x, CE9y, and CE9z, respectively; Fig. 3D). None of these oligos stimulated splicing of the A3x pre-mRNA.
S21 could be less active than CE9 because other factors may compete with SRp30c for binding. Indeed, ASF/SF2 appears to bind to S21 more efficiently than to CE9 (Fig. 2E). Other elements within CE9 may also improve SRp30c binding to enhance repression. This would be consistent with the observation that the last 11 nt at the 3′ end of CE9 can elicit some repression (Simard and Chabot 2000) and may explain why oligos containing different portions of CE9 were unable to rescue splicing (Fig. 3D). We could not detect binding of SRp30c to the last 11 nt of CE9 (not shown). Nevertheless, we tested the impact of removing these sequences. Recombinant GST-SRp30c and 5′-end-labeled RNA oligonucleotides were incubated with glutathione beads, and the mixtures were washed at increasing salt concentrations. In these conditions, the RNA oligo carrying the first 29 nt of CE9 (CE9-29) was bound by SRp30c less efficiently than the complete CE9 element, consistent with a contribution of the 3′ portion of CE9 to SRp30c binding (Fig. 4C, cf. SRp30c-bound CE9 and CE9-29). Moreover, an oligo containing the first 20 nt of CE9 followed by the last 10 nt of CE9 (CE9-Pyless) was bound by SRp30c as efficiently as CE9. Thus, the last 10 nt at the 3′ end of CE9 contribute to SRp30c binding. Because the 3′ region of CE9 contains AGAAU, a sequence bound by SRp30c in tau exon 2 (Wang et al. 2005), a plausible interpretation of our results is that SRp30c weakly interacts with this sequence to cooperatively improve SRp30c binding to CE9.
FIGURE 4.
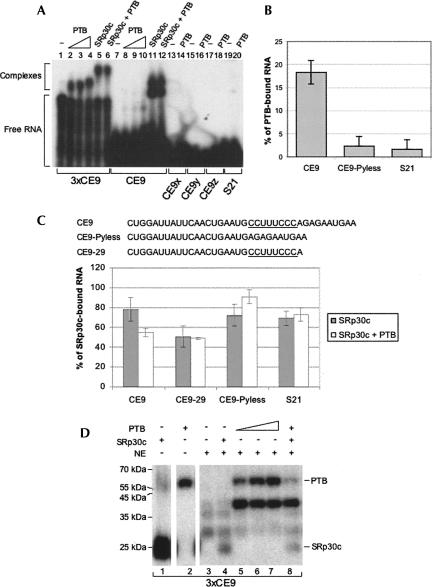
PTB interacts with CE9. (A) Gel mobility shift assay with PTB and SRp30c. His-tagged proteins were incubated with 5′-end-labeled RNA oligos, except for 3xCE9, which was produced by in vitro transcription. (B) His-tagged PTB and various 5′-end-labeled oligonucleotides were mixed and the bound material was recovered using Ni-NTA agarose beads. The experiment was performed in triplicate and standard deviations are indicated. (C) His-tagged PTB and GST-SRp30c were used alone or in combination. The 5′-end-labeled oligonucleotides bound to GST-SRp30c were recovered using glutathione beads. (D) UV cross-linking assay using recombinant PTB and SRp30c incubated in a HeLa nuclear extract with uniformly labeled 3xCE9 RNA. Five micromolar of His-SRp30c was used, whereas 5, 1.0, 2.5, 5, and 5 μM of His-PTB were added in lanes 2, 5, 6, 7, and 8, respectively.
PTB antagonizes the repressor activity of SRp30c
The stronger activity of CE9 relative to S21 can be explained if CE9 provides more specific binding sites for several SRp30c molecules. However, additional factors may interact with or stabilize the CE9/SRp30c complex. To explore this possibility, we carried out RNA affinity chromatography. We had previously recovered SRp30c using the first 16 nt of CE9 (Simard and Chabot 2002). Here, we produced a column with the complete 38-nt-long CE9 element. The column was loaded with a HeLa nuclear extract and washed extensively with a buffer containing 100 mM KCl. The bound material was fractionated on a polyacrylamide gel, and the bands, stained with silver nitrate, were analyzed by nanoliquid chromatography coupled online with tandem mass spectrometry. One component of the eluate was clearly identified as hnRNP I/PTB since six peptides out of 36 were unambiguously assigned to the protein, covering 24% of the sequence.
A gel mobility shift assay using recombinant His-PTB indicates relatively poor binding to a single CE9 element relative to three adjacent CE9 elements (Fig. 4B, lanes 7–10, 1–4, respectively). PTB did not bind to the S21 RNA or to CE9x, CE9y, and CE9z (Fig. 4A, lanes 13–20). While PTB and SRp30c similarly shifted the migration of 3xCE9 RNA (Fig. 4A, lanes 4,5), the binding of PTB to one CE9 element was less efficient than the binding of SRp30c (Fig. 4A, cf. lanes 10 and 11). The superior affinity of SRp30c was also apparent from mixing experiments performed with either CE9 or 3xCE9. In each case, the coincubation of equivalent amounts of PTB and SRp30c produced a complex that comigrated with the complex obtained with SRp30c alone (Fig. 4A, lanes 6,12).
To confirm these observations, we carried out binding assays using His-PTB and labeled oligonucleotides incubated with Ni2+ beads. In this assay, PTB bound specifically to CE9 (Fig. 4B). The central pyrimidine stretch CCUUUCCC was important for binding because removing it considerably reduced PTB binding (CE9-Pyless in Fig. 4B). To ask whether PTB could antagonize SRp30c binding, we tested the impact of adding His-PTB on the binding of GST-SRp30c to CE9 (Fig. 4C). The addition of PTB reduced the binding of SRp30c to CE9. This reduction was not observed if the oligonucleotide lacked the central pyrimidine stretch of CE9 (CE9-Pyless). Notably, PTB also did not affect the binding of SRp30c to an RNA that contains the pyrimidine stretch but not the 3′ portion of CE9 (CE9-29). Thus, PTB does not appear to interfere with the binding of SRp30c to the 5′ portion of CE9. This is expected given that > 10 nt separate this SRp30c binding site from the CU-rich element. Rather, PTB compromises the positive contribution that the 3′ end of CE9 makes to SRp30c binding, and this antagonism requires the CU-rich element, which is located only 2 nt upstream from the putative SRp30c binding sequence AGAAU.
Next, we assessed the interaction of these recombinant proteins in a nuclear extract (Fig. 4D). Using a UV cross-linking assay to monitor the binding of recombinant proteins, we first noted that PTB bound relatively efficiently to CE9 in the extract (Fig. 4D, cf. lanes 2 and 5). PTB also promoted the cross-link of another protein migrating at ∼ 40 kDa. The identity of this protein is unknown and has not been pursued further. In contrast to PTB, the cross-linking signal obtained with recombinant SRp30c decreased dramatically when the assay was performed in a nuclear extract (Fig. 4D, cf. lanes 1 and 4). However, when similar amounts of recombinant PTB and SRp30c (5 μM) were added simultaneously to a nuclear extract, their individual cross-linking signals were both reduced, and more dramatically so for PTB (Fig. 4D, cf. signals for SRp30c and PTB in lanes 4,7 and those in lane 8). Thus, the binding of recombinant SRp30c and PTB to CE9 appears antagonistic.
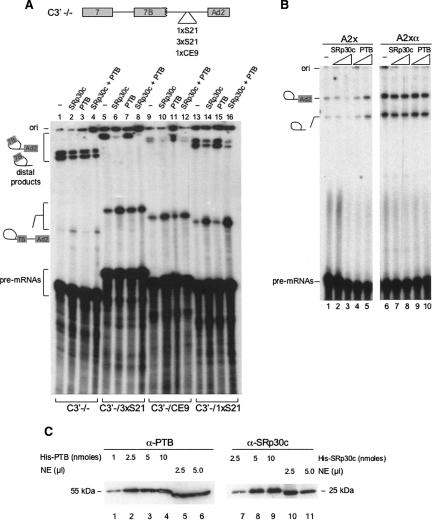
To examine the impact of PTB on the activity of CE9, we performed splicing assays supplemented with recombinant His-PTB (Fig. 5A). As shown previously, one copy of CE9 strongly represses the distal 3′ splice site of the C3′ pre-mRNA (Fig. 5A, cf. lanes 9 and 1). Additional repression at this site can be obtained by adding recombinant SRp30c (Fig. 5A, lane 10). Surprisingly, the addition of recombinant PTB stimulated the distal 3′ splice site in a CE9-dependent manner (Fig. 5A, lane 11). The inclusion of SRp30c in the PTB-supplemented mixture completely neutralized the impact of PTB (lane 12). The addition of PTB to splicing mixtures containing the control C3′ −/− pre-mRNA or a C3′ derivative containing one or three S21 elements did not affect 3′ splice site selection (Fig. 5A, lanes 3,15,7), despite the fact that the addition of SRp30c strongly stimulated splicing to the proximal 3′ splice site of the S21-containing pre-mRNAs (Fig. 5A, lanes 6,14). Thus, PTB appears to antagonize the activity of endogenous SRp30c in the context of CE9, and this situation is reversed by supplementing the extract with recombinant SRp30c.
FIGURE 5.
PTB displays anti-repressor activity. (A) 3′ splice site selection assay. The pre-mRNAs used are described in Figure 3A. Recombinant His-PTB and His-SRp30c proteins were added at a final concentration of 5 μM. The positions and identity of the proximal and distal splicing products are indicated. (B) Splicing repression assay. His-tagged SRp30c and PTB were added to splicing mixtures containing the A2x or A2xα pre-mRNAs. The final concentrations of recombinant proteins were 2.5 and 5 μM. (C) Western analysis of SRp30c and PTB in HeLa nuclear extracts. Titrating amounts of His-PTB and His-SRp30c were added to assess the concentrations of the endogeneous proteins in a HeLa nuclear extract. Anti-PTB and anti-SRp30c antibodies were used.
The anti-repressor activity of PTB was confirmed by investigating the impact of PTB on the splicing of the single-intron A2x pre-mRNA. In this case, while the CE9-mediated repression was exacerbated by adding SRp30c (Fig. 5B, lanes 2,3), the addition of PTB stimulated splicing (Fig. 5A, lanes 4,5). Neither recombinant SRp30c nor PTB affected the splicing efficiency of the control A2xα pre-mRNA (Fig. 5B, lanes 6–10). Given that His-tagged PTB molecules have been used many times to confirm the activity of PTB in other systems, we think that it is unlikely that the anti-repressor activity detected in our system is artifactually conferred by the His-tag.
The repressor activity of CE9 appears to be determined by the relative abundance of SRp30c and PTB. Since CE9 displays repressor activity in a HeLa extract, the activity of endogenous SRp30c appears to be dominant. At equivalent molar concentrations of recombinant SRp30c and PTB (5 μM), SRp30c is also the dominant activity, as judged by the mixing experiment (Fig. 5A, lane 12). To assess the relative abundance of SRp30c and PTB in a HeLa extract, we performed Western analyses with anti-PTB and anti-SRp30c antibodies (Fig. 5C). Based on the signal obtained with titrating amounts of recombinant proteins, we estimate that SRp30c and PTB are present in approximately equimolar concentrations in a nuclear extract. This situation is consistent with the repressing activity displayed by CE9 in cells and in extracts.
PTB affects the activity of CE9 in vivo
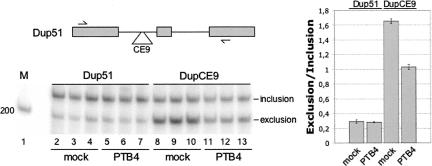
To ascertain the role of PTB in the activity of CE9 in vivo, we altered the concentration of PTB in cells. First, we tested the impact of transiently overexpressing PTB in HeLa cells coexpressing the β-globin-derived mini-gene that contains a CE9 element in the upstream intron of DUP51 (pDUP-CE9). The inclusion of the central alternative exon was monitored by RT-PCR using total RNA extracted 48 h after transfection. Consistent with its ability to repress a downstream 3′ splice site, CE9 reduced the inclusion of the central exon, (Fig. 6A, lanes 8–10). When the PTB-expressing plasmid was cotransfected with pDUP-CE9, we noted a significant improvement (P < 0.002) in the relative frequency of exon inclusion (Fig. 6A, lanes 11–13). Given that PTB had no effect on the alternative splicing of the control transcript lacking CE9 (Fig. 6A, cf. lanes 2–4 and 5–7), these results indicate that upregulating PTB expression can relieve the repression imposed by CE9. We also carried out experiments designed to knock down PTB using specific siRNAs. Despite considerable reductions in the steady-state levels of proteins, we never observed significant CE9-dependent changes in DUP-CE9 alternative splicing (or in the alternative splicing of the endogenous hnRNP A1 exon 7B) (data not shown). A similar result was obtained when both PTB and nPTB were simultaneously knocked down (not shown). However, given that the activity of SRp30c is already dominant over that of PTB, it is somewhat expected that decreasing PTB/nPTB levels should have little impact on a CE9-mediated splicing event.
FIGURE 6.
PTB affects the activity of CE9 in vivo. The human CE9 element was inserted into the upstream intron of the globin DUP51 model β-globin derived mini-gene. DUP splicing was analyzed by RT-PCR in cells cotransfected with a PTB4 expression vector. An experiment performed in triplicate is shown. A two-tailed Student's t-test indicates that the CE9-dependent increase in exon inclusion obtained by overexpressing PTB4 is significant (P < 0.002).
DISCUSSION
Defining an optimal SRp30c binding site
The sequences recovered from a SELEX protocol performed with recombinant SRp30c displayed a strong enrichment for the AGSAS motif (S = G or C). The AGGAC sequence was the most frequent motif and was found in 7 out of the 21 AGSAS-containing clones. Two clones contained two AGGAC motifs. The other most frequent motifs were AGCAG (six occurrences) and AGGAG (four occurrences). Further characterization using RNA oligos carrying specific changes indicated that two AGGAC motifs offered optimal binding affinity for SRp30c in gel mobility shift assays. Moreover, the conversion of any of the two AGGAC motifs into AGCAG produced a strong decrease in SRp30c binding.
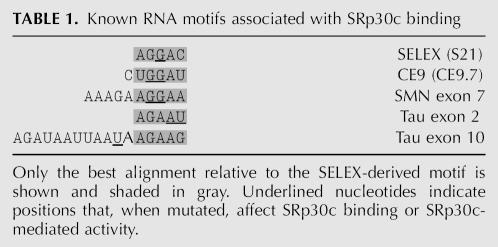
As shown in Table 1, a portion of the SELEX consensus sequence AGGAC is found in the SRp30c-binding portion at the 5′ end of CE9 (CUGGAUU). Consistent with their proposed function, mutating the underlined purines in CE9 compromised SRp30c binding (Simard and Chabot 2002). We have shown that SRp30c binding to CE9 is weaker than to the SELEX-derived oligo carrying two AGGAC (S21). Three CE9 elements were required to duplicate the affinity displayed by SRp30c for S21. Previously identified SRp30c binding sites display varying degrees of homology with the AGGAC motif, suggesting that these sites may be relatively weak. This would appear to be the case at least for the SMN binding site since hTra2β was required to detect the interaction of SRp30c with this element (Young et al. 2002). Our results suggest that the 3′ portion of CE9 also contributes to the binding by SRp30c. Notably, this portion contains the sequence AGAAU, a sequence that matches the SRp30c motif found in tau exon 2 (Table 1). Thus, high-affinity binding of SRp30c may be achieved by using multiple weak binding sites or through participation of a collaborating protein like hTra2β to stabilize the interaction of SRp30c with one weak site.
TABLE 1.
Known RNA motifs associated with SRp30c binding
The AGGAC motif is part of the high-affinity binding site identified by SELEX for ASF/SF2 (AGGACARRAGC; Tacke and Manley 1995). This suggests that ASF/SF2 binding sites that conform to this consensus may also be bound by SRp30c and that the overlapping binding specificities may lead to functional antagonism. However, whether SRp30c can antagonize the activity of splicing enhancers whose activity is mediated by ASF/SF2 remains to be examined. Conversely, ASF/SF2 may interact with a subset of SRp30c binding sites. We have observed that recombinant ASF/SF2 interacted with low affinity to S21 and less so to CE9. However, although the addition of ASF/SF2 to splicing mixtures did counteract the repressor activity of CE9 (Fig. 1B), it did not reduce the repressing activity of S21 elements (not shown).
SRp30c and CE9-mediated repression
Positioning CE9 in between two 3′ splice sites shifts splicing toward the internal 3′ splice site. This switch is thought to be caused by the repression of the downstream 3′ splice site because splicing repression occurs when multiple copies of CE9 are inserted in the intron of a simple model pre-mRNA. We have confirmed the importance of SRp30c in the activity of the CE9 element in both systems. Increasing the concentration of SRp30c in nuclear extracts exacerbates both the CE9-mediated switch in 3′ splice site utilization and the CE9-mediated splicing repression. Importantly, we have shown that multiple copies of the SELEX-derived high-affinity site for SRp30c (S21) could duplicate the impact of one CE9 element on 3′ splice site selection. This is consistent with a previous observation showing that tandem copies of the weaker SRp30c-binding portion of CE9 (2xCE9.7) partially reproduced the shift obtained with a complete CE9 element (Simard and Chabot 2002). Overall, these results suggest that SRp30c is essential for CE9-mediated repression of the downstream 3′ splice site. If the binding of SRp30c is sufficiently strong, no additional RNA binding factors may be required to mediate repression. It remains possible that additional factors help recruit SRp30c or stabilize its interaction with CE9.
An anti-repressor function for PTB
In an attempt to identify factors that collaborate with SRp30c to mediate splicing repression, we recovered hnRNP I/PTB by affinity chromatography using the complete human CE9 element. PTB is well known for its association with numerous splicing silencers (Wagner and Garcia-Blanco 2001). PTB binds to short elements containing mixtures of C and U (Singh et al. 1995; Perez et al. 1997; Simpson et al. 2004). The human CE9 element contains the sequence CCUUUCCC, and we have shown that this element contributes to PTB binding. Expecting to document yet another example of a role for PTB in splicing repression, we were surprised to observe that the addition of recombinant His-PTB counteracted the splicing shift associated with CE9 and stimulated splicing to the distal 3′ splice site. Moreover, PTB improved the splicing of a pre-mRNA that had been repressed by multiple intronic copies of CE9. Increasing the level of SRp30c in extracts supplemented with PTB restored the CE9-dependent repression. Lastly, increasing the levels of PTB in HeLa cells partially neutralized the repressor activity of CE9. Thus, PTB behaves as an anti-repressor that antagonizes the repressing activity of CE9 mediated by SRp30c.
The above observations challenge our view of PTB as a repressor of splicing (Wagner and Garcia-Blanco 2001). The binding of PTB to intron silencers has been described on numerous occasions (Mulligan et al. 1992; Norton 1994; Ashiya and Grabowski 1997; Chan and Black 1997; Gooding et al. 1998; Southby et al. 1999; Carstens et al. 2000; Gromak et al. 2003). The only apparent exception is in the calcitonin/CGRP gene, where PTB enhances exon 4 inclusion (Lou et al. 1999). However, it has been argued that activation may be indirect in this case (Wagner and Garcia-Blanco 2001; Wollerton et al. 2004). PTB has also been found to bind to splicing silencers located in exons (Zheng et al. 1998; Shen et al. 2004; Izquierdo et al. 2005). In cases where it has been examined, PTB associates with other proteins to form a multiprotein complex (Chan and Black 1997; Grossman et al. 1998; Chou et al. 2000; Smith and Valcarcel 2000). In the α-tropomyosin pre-mRNA, raver1 has been identified as an important protein recruited by PTB to elicit repression (Gromak et al. 2003). A paralog of PTB, nPTB, has been implicated in some form of derepression during alternative splicing of the c-src brain-specific N1 exon (Markovtsov et al. 2000). In this case, the nPTB produced from neuronal cells possesses a weaker repressive activity than PTB. nPTB has been proposed to antagonize the assembly of a strong repressor complex by preventing the binding of PTB and by modifying the interactions of auxiliary factors. In the case of CE9, it is currently not known whether additional factors are collaborating with PTB to mediate anti-repression.
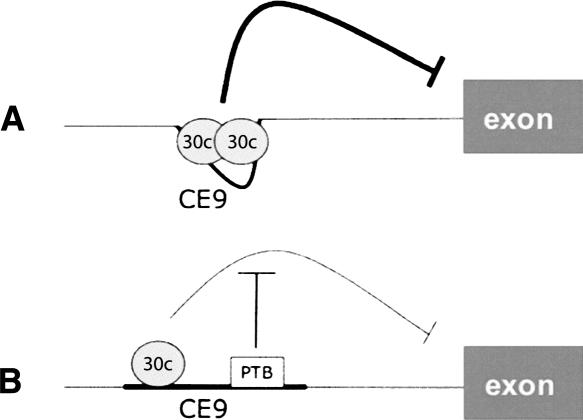
The mechanism by which PTB antagonizes the SRp30c-mediated repression activity of CE9 appears to involve binding interference. The binding site for PTB in CE9 is most probably the pyrimidine-rich sequence located in between two elements that are important for SRp30c binding. The upstream element is bound by SRp30c. Although we have not detected binding of SRp30c to the downstream element, its presence stimulates overall SRp30c binding. PTB does not interfere with SRp30c binding to the upstream element but prevents the improvement normally provided by the downstream element. Thus, PTB may prevent the binding of SRp30c to the downstream element. Alternatively, it may prevent cooperative interactions between SRp30c molecules bound to these sites. We observed previously that the CE9-mediated splicing inhibition does not prevent the assembly of U2- and U4-dependent complexes on a model pre-mRNA, suggesting that SRp30c blocks a late step in spliceosome assembly (Fig. 7A; Simard and Chabot 2000, 2002)). Although PTB appears to antagonize SRp30c binding to the 3′ region of CE9, it may also prevent the upstream bound SRp30c from interacting with components of the spliceosome (Fig. 7B). Such a mechanism would be similar to two recently described molecular mechanisms of action for PTB. In the case of the Fas receptor, the exonic binding of PTB does not alter downstream U1 snRNP binding but compromises exon definition to prevent efficient U2AF binding to the upstream 3′ splice site (Izquierdo et al. 2005). In the case of c-src, PTB interferes with the U1 snRNP–U2AF communication that occurs during intron definition (Sharma et al. 2005). Thus, an interaction between factors bound at distinct sites on a pre-mRNA would be obstructed by one or several intervening PTB molecules. When such interactions are required for splicing stimulation, PTB would act as a repressor. However, if these interactions are required to implement repression, then PTB would display anti-repressor activity, as we have observed in the case of CE9.
FIGURE 7.
A model for the interaction and antagonism of factors binding to CE9. (A) The model proposes that at least two SRp30c proteins interact cooperatively to bind to CE9. This complex promotes repression at the downstream 3′ splice site. (B) PTB binding would antagonize the binding of SRp30c at the downstream site to prevent cooperative interactions. PTB may also prevent interactions between SRp30c and splicing factors bound at the 3′ splice site.
Antagonism between factors that bind to small portions of a pre-mRNA appears to be a common theme in splicing control. The binding of SR proteins to exonic splicing enhancers is important to counteract the repressing effect of a nearby silencers bound by hnRNP A1 (Zhu et al. 2001). Likewise, the binding of hnRNP G to the skeletal-specific exon of the human slow skeletal α-tropomyosin is antagonized by the splicing activator hTra2β (Nasim et al. 2003). PTB has been implicated in several cases of antagonism. It can prevent the binding of U2AF to the 3′ splice site of the α-tropomyosin (Lin and Patton 1995) and can compromise the binding of ETR-3 to intronic elements to prevent exon 5 inclusion in cardiac troponin T (Charlet et al. 2002). Thus, our study adds to the growing list of variations on this theme and underscores the great versatility in the use of combinations of factors to control splicing decisions.
MATERIALS AND METHODS
Plasmids and oligonucleotides
Plasmids C3′−/−, C3′-/CE9, A3x, A2x, and A2xα were described previously (Simard and Chabot 2000, 2002). Plasmid AS21 was constructed by inserting annealed oligonucleotides S21s/S21a into the StuI site of pSPAdStu (Lavigueur et al. 1993). Because this insert contains an EcoRV site, we used it to reinsert two or three copies of S21s/S21a. Plasmids C3′-/1xS21, C3′-/2xS21, and C3′-/3xS21 were constructed by inserting annealed oligonucleotides S21s/S21a at the EcoRV site of pC3′ −/− or the EcoRV site of pC3′-/1xS21 and C3′-/2xS21. The same procedure was followed to produce C3′-/2xS21α, except that the insert was in the reverse orientation.
pGEX-SRp30c and pGEX-ASF/SF2 were obtained by performing RT-PCR using Pwo DNA polymerase on total RNA from HeLa cells using oligos SRp30c-1/SRp30c-2 and ASF/SF2-1/ASF/SF2-B, respectively. The SRp30c fragment was cut with EcoRI and XhoI and was inserted into pGEX. The ASF/SF2 fragment was cut with EcoRI and HindIII and was cloned into pGEX. The BamHI–HindIII fragment of pGEX-SRp30c was inserted in the 6x-His tagged vector pQE80L to generate pQE80L-SRp30c. The MfeI–ScaI fragment of pQE80L-SRp30c was inserted into the EcoRI–SmaI sites of the baculovirus expression vector pVL1392 (BD Biosciences) to produce pVL1392-SRp30c. The pET28a-based plasmid encoding His-PTB was kindly provided by Doug Black (University of California, Los Angeles). The CMV expression plasmid encoding PTB4 was kindly provided by Chris Smith (Cambridge University).
The 38-bp CE9 element was inserted into the ApaI site in the upstream intron of pDUP51 to generate pDUP-CE9.
DNA and RNA oligonucleotides were purchased from Dharmacon Inc. and Integrated DNA Technologies Inc. and were used as cloning adaptors, PCR primers, substrate for RNA binding proteins, splicing assays, and RNAi experiments.
DNA oligos
S21: ACAAAGGACGGCACAGGACA
S21s: ACAAAGGACGGCACAGGACAGATATC
S21a: GATATCTGTCCTGTGCCGTCCTTTGT
CE9: CTGGATTATTCAACTGAATGCCTTTCCCAGAGAATGAA
CE9s: CTGGATTATTCAACTGAATGCCTTTCCCAGAGAATGAAGATATC
CE9a: GATATCTTCATTCTCTGGGAAAGGCATTCAGTTGAATAATCGAC
SRp30c-1 (EcoRI): TAGGAATTCTAATGTCGGGCTGGGCGGA
SRp30c-2 (XhoI): TACCGCTCGAGTCAGTAGGGCCTGAAAG
ASF/SF2-1 (EcoRI): TAGGAATTCTAATGTCGGGAGGTGGTGT
ASF/SF2-B (HindIII): TATCCCAAGCTTTTATGTACGAGAGCGAG
Ge1: ACACAACTGTGTTCACTAGC
Ge2: AGTGGACAGATCCCCAAAGG
RNA oligos
CE9: CUGGAUUAUUCAACUGAAUGCCUUUCCCAGAGAAUGAA
CE9x: CUGGAUUAUUCAACUGAAUG
CE9y: UCAACUGAAUGCCUU
CE9z: UUCCCAGAGAAUGAA
CE9-Pyless: CUGGAUUAUUCAACUGAAUGAGAGAAUGAA
3xCE9.7: CUGGAUUAAGCUGCUGGAUUAAAGCUGCUGGAUUAAGGUG
S21: ACCAAGGACGGCACAGGACA
S21-CG: ACCAAGCACGGCACAGGACA
S21-GC: ACCAAGGACGGCACAGCACA
S21-CC: ACCAAGCACGGCACAGCACA
S21-CCCC: ACCAACCACGGCACACCACA
S21mod6: ACCCAGGACAUUUUAGGACA
S23: CUCAGCCACAGUAGGACACA
Protein purification and RNA chromatography
GST-SRp30c and GST-ASF/SF2 were produced from bacteria as described (Simard and Chabot 2002). His-PTB was purified from bacteria and His-SRp30c was produced from baculovirus-infected cells. Both proteins were purified using the Qiagen purification system. Protein concentration was evaluated using the Bio-Rad protein assay. RNA affinity chromatography using CE9 was carried out as follows: 25 nmol of RNA were coupled to 250 μL of agarose adipic acid hydrazide resin according to the manufacturer's instructions (Amersham Biosciences). One hundred twenty-five microliters of HeLa nuclear extract were incubated with 25 μL of CE9-resin for 10 min at 30°C. The resin was spun and washed four times with D buffer (60 mM HEPES at pH 7.9, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, and 20% glycerol). 2×-Laemmli dye (10% glycerol, 5% β-mercaptoethanol, 2.3% SDS, 62.5 mM Tris-HCl at pH 6.8, and 0.1% bromophenol blue) was added to the beads, and the mixture was boiled and loaded onto an acrylamide/SDS gel. After staining with silver nitrate, protein bands were excised and digested in the gel with trypsin. All peptide analyses were performed on a Q-TOF-2 (Waters/Micromass Corp.) Peptide mass searches were performed with the MASCOT algorithm (http://www.matrixscience.com) and the latest version of the NCBI protein database. Peak lists were prepared from raw tandem mass spectra after smoothing, background-subtraction, and centroiding using the MassLynx 3.5 software, as input data. The MASCOT software default settings were used for protein identification with tandem mass spectrometry-derived data.
SELEX
In vitro selection of SRp30c RNA ligands was performed with GST-SRp30c immobilized on glutathione agarose beads as described (Tacke and Manley 1995). The random oligonucleotide pool had the following sequence: ATACCGTCGACCCGAGGGG-N20 CCCGGTACCCAGCTTTTGTT. The PCR products obtained using SELREV (TTACTCGACAGGCGCGGATCCATACCGTCGACCTGGAGGGG) and SELT7 (TATCCGGAATTCTAATACGACTCACTATAG AACAAAAGCTGGGTACCGGG) were cut with BamHI transcribed using T7 RNA polymerase. RNA was labeled at the 5′ end with [γ-32P] ATP. Binding of the randomized RNA pool was performed in buffer X (10 mM HEPES at pH 7.9, 100 mM KCl, 10% glycerol, 0.025% NP40, 400 μg/mL tRNA). We washed the beads with increasing KCl concentrations until ∼10% of the RNA remained bound. The final material was collected by phenol-chloroform extraction and precipitated with ethanol. RT-PCR was then conducted using SELREV and SELT7. After six rounds of amplification/selection, PCR products were cloned as EcoRI–HindIII fragments in pBluescript K+ and sequenced.
Transcription and splicing assays
pSP-Ad and derivatives were linearized with HincII and transcribed with SP6 RNA polymerase (Amersham Biosciences). Plasmid pC3′ −/− and derivatives were linearized with ScaI, and transcription was carried out using T3 RNA polymerase (Amersham Biosciences) in the presence of 32P-UTP. Transcription and RNA purification were as described previously (Chabot 1994). Two femtomoles of the gel-purified pre-mRNA were incubated in HeLa nuclear extracts (Dignam et al. 1983) under standard splicing conditions at 30°C (Krainer et al. 1984). The RNA material was phenol extracted, ethanol precipitated, and fractionated on denaturing acrylamide gels. To investigate the effect of an excess of RNA competitors on splicing, pre-mRNA molecules were mixed with the competing RNA prior to splicing. When recombinant proteins were added to splicing mixtures, a preincubation of 8 min at 30°C was performed before adding the labeled pre-mRNA.
The SRp30c-depleted extract was prepared by transfection a siRNA (80 nM) against SRp30c (sequence of passenger strand: 5′-AGAGGAUGCUAUUUAUGGATT-3′) using lipofectamine in HeLa cells. Seventy-two hours later, cells were collected and a nuclear extract was prepared. Mock-treated cells were used to prepare a control extract.
Gel shift and cross-linking assays
3xCE9 RNA was uniformly labeled with 32P-UTP and gel-purified. RNA oligos were 5′-end labeled using [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs), and purified using a G-25 column (Amersham Biosciences). Gel shift assays were carried out as described previously (Simard and Chabot 2002) in the presence of 1 μg/μL of heparin. Complexes were resolved on nondenaturing 5% polyacrylamide gels. Cross-linking assays were performed on the Stratalinker (Stratagene). Essentially, 6 fmol of uniformly labeled RNA incubated in 12.5 μL of a splicing mixture were subjected to UV radiation (1.5 min at 500 mJ). The mixture was then treated with RNAse A and proteins were fractionated on a 12% acrylamide/SDS gel. Cross-linking assays were also carried out with 5′-end-labeled RNA oligos using the same protocol except that no RNase A treatment was performed.
Transfection, RT-PCR, and Western analyses
Transfection of mini-genes was carried out using lipofectamine, as described previously (Simard and Chabot 2002). The detection of endogenous PTB was performed using antibodies kindly provided by Douglas Black. His-PTB was detected using the anti-His antibody. DUP51 alternative splicing was monitored by RT-PCR using oligos Ge1 and Ge2.
ACKNOWLEDGMENTS
We thank Doug Black for the bacterial PTB expression vector and PTB antibodies and Chris Smith for the eukaryotic PTB expression vector. This work was supported by a grant from the Canadian Institute of Health Research to B.C. B.C. holds a Canada Research Chair in Functional Genomics.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.403607.
REFERENCES
- Ashiya, M., Grabowski, P.J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: Evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Caputi, M., Zahler, A.M. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens, R.P., Wagner, E.J., Garcia-Blanco, M.A. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7388–7400. doi: 10.1128/mcb.20.19.7388-7400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot, B. Synthesis and purification of RNA substrates. In: Higgins S.J., Hames B.D., editors. RNA processing, A practical approach. IRL Press; Oxford: 1994. pp. 1–29. [Google Scholar]
- Chan, R.C., Black, D.L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet, B.N., Logan, P., Singh, G., Cooper, T.A. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- Chou, M.Y., Underwood, J.G., Nikolic, J., Luu, M.H., Black, D.L. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., Roeder, R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estmer Nilsson, C., Petersen-Mahrt, S., Durot, C., Shtrichman, R., Krainer, A.R., Kleinberger, T., Akusjarvi, G. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 2001;20:864–871. doi: 10.1093/emboj/20.4.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, M.E., Gattoni, R., Stevenin, J., Marie, J., Expert-Bezancon, A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the β-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau, D., Revil, T., Fisette, J.F., Chabot, B. hnRNP F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- Gooding, C., Roberts, G.C., Smith, C.W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak, N., Rideau, A., Southby, J., Scadden, A.D., Gooding, C., Huttelmaier, S., Singer, R.H., Smith, C.W. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman, J.S., Meyer, M.I., Wang, Y.C., Mulligan, G.J., Kobayashi, R., Helfman, D.M. The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA. 1998;4:613–625. doi: 10.1017/s1355838298971448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, E.C., Schaal, T.D., Hertel, K.J., Reed, R., Maniatis, T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl. Acad. Sci. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo, J.M., Majos, N., Bonnal, S., Martinez, C., Castelo, R., Guigo, R., Bilbao, D., Valcarcel, J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Jiang, Z., Tang, H., Havlioglu, N., Zhang, X., Stamm, S., Yan, R., Wu, J.Y. Mutations in tau gene exon 10 associated with FTDP-17 alter the activity of an exonic splicing enhancer to interact with Tra2 β. J. Biol. Chem. 2003;278:18997–19007. doi: 10.1074/jbc.M301800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka, A., Mühlemann, O., Aküsjarvi, G. Inhibition by SR proteins of splicing of a regulated adenovirus pre- mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- Kondo, S., Yamamoto, N., Murakami, T., Okumura, M., Mayeda, A., Imaizumi, K. Tra2 β, SF2/ASF and SRp30c modulate the function of an exonic splicing enhancer in exon 10 of τ pre-mRNA. Genes Cells. 2004;9:121–130. doi: 10.1111/j.1356-9597.2004.00709.x. [DOI] [PubMed] [Google Scholar]
- Krainer, A.R., Maniatis, T., Ruskin, B., Green, M.R. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Lavigueur, A., La Branche, H., Kornblihtt, A.R., Chabot, B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes & Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- Lin, C.H., Patton, J.G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- Lou, H., Helfman, D.M., Gagel, R.F., Berget, S.M. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov, V., Nikolic, J.M., Goldman, J.A., Turck, C.W., Chou, M.Y., Black, D.L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras, R., Fisette, J.F., Nasim, F.U., Madden, R., Cordeau, M., Chabot, B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin, A.J., Clark, F., Smith, C.W. Understanding alternative splicing: Toward a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Mulligan, G.J., Guo, W., Wormsley, S., Helfman, D.M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of β-tropomyosin pre-mRNA. J. Biol. Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- Nasim, M.T., Chernova, T.K., Chowdhury, H.M., Yue, B.G., Eperon, I.C. HnRNP G and Tra2β: opposite effects on splicing matched by antagonism in RNA binding. Hum. Mol. Genet. 2003;12:1337–1348. doi: 10.1093/hmg/ddg136. [DOI] [PubMed] [Google Scholar]
- Norton, P.A. Polypyrimidine tract sequences direct selection of alternative branch sites and influence protein binding. Nucleic Acids Res. 1994;22:3854–3860. doi: 10.1093/nar/22.19.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani, F., Buratti, E., Stuani, C., Romano, M., Zuccato, E., Niksic, M., Giglio, L., Faraguna, D., Baralle, F.E. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 2000;275:21041–21047. doi: 10.1074/jbc.M910165199. [DOI] [PubMed] [Google Scholar]
- Park, E., Han, J., Son, G.H., Lee, M.S., Chung, S., Park, S.H., Park, K., Lee, K.H., Choi, S., Seong, J.Y., et al. Cooperative actions of Tra2α with 9G8 and SRp30c in the RNA splicing of the gonadotropin-releasing hormone gene transcript. J. Biol. Chem. 2006;281:401–409. doi: 10.1074/jbc.M505814200. [DOI] [PubMed] [Google Scholar]
- Perez, I., McAfee, J.G., Patton, J.G. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt, S.K., Estmer, C., Ohrmalm, C., Matthews, D.A., Russell, W.C., Akusjarvi, G. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 1999;18:1014–1024. doi: 10.1093/emboj/18.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffetseder, U., Frye, B., Rauen, T., Jurchott, K., Royer, H.D., Jansen, P.L., Mertens, P.R. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J. Biol. Chem. 2003;278:18241–18248. doi: 10.1074/jbc.M212518200. [DOI] [PubMed] [Google Scholar]
- Romano, M., Marcucci, R., Buratti, E., Ayala, Y.M., Sebastio, G., Baralle, F.E. Regulation of 3′ splice site selection in the 844ins68 polymorphism of the cystathionine β-synthase gene. J. Biol. Chem. 2002;277:43821–43829. doi: 10.1074/jbc.M208107200. [DOI] [PubMed] [Google Scholar]
- Screaton, G.R., Cáceres, J.F., Mayeda, A., Bell, M.V., Plebanski, M., Jackson, D.G., Bell, J.I., Krainer, A.R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S., Falick, A.M., Black, D.L. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Kan, J.L., Ghigna, C., Biamonti, G., Green, M.R. A single polypyrimidine tract binding protein (PTB) binding site mediates splicing inhibition at mouse IgM exons M1 and M2. RNA. 2004;10:787–794. doi: 10.1261/rna.5229704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, C., Manley, J.L. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- Simard, M.J., Chabot, B. Control of hnRNP A1 alternative splicing: An intron element represses use of the common 3′ splice site. Mol. Cell. Biol. 2000;20:7353–7362. doi: 10.1128/mcb.20.19.7353-7362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard, M.J., Chabot, B. SRp30c is a repressor of 3′ splice site utilization. Mol. Cell. Biol. 2002;22:4001–4010. doi: 10.1128/MCB.22.12.4001-4010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, P.J., Monie, T.P., Szendroi, A., Davydova, N., Tyzack, J.K., Conte, M.R., Read, C.M., Cary, P.D., Svergun, D.I., Konarev, P.V., et al. Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure. 2004;12:1631–1643. doi: 10.1016/j.str.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Singh, R., Valcárcel, J., Green, M.R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Smith, C.W., Valcarcel, J. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Southby, J., Gooding, C., Smith, C.W. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutally exclusive exons. Mol. Cell. Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoss, O., Schwaiger, F.W., Cooper, T.A., Stamm, S. Alternative splicing determines the intracellular localization of the novel nuclear protein Nop30 and its interaction with the splicing factor SRp30c. J. Biol. Chem. 1999;274:10951–10962. doi: 10.1074/jbc.274.16.10951. [DOI] [PubMed] [Google Scholar]
- Stoss, O., Novoyatleva, T., Gencheva, M., Olbrich, M., Benderska, N., Stamm, S. p59(fyn)-mediated phosphorylation regulates the activity of the tissue-specific splicing factor rSLM-1. Mol. Cell. Neurosci. 2004;27:8–21. doi: 10.1016/j.mcn.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Tacke, R., Manley, J.L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.J., Garcia-Blanco, M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Wang, J., Gao, L., Lafyatis, R., Stamm, S., Andreadis, A. τ exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c.SRp55 complex that either recruits or antagonizes htra2β1. J. Biol. Chem. 2005;280:14230–14239. doi: 10.1074/jbc.M413846200. [DOI] [PubMed] [Google Scholar]
- Wollerton, M.C., Gooding, C., Wagner, E.J., Garcia-Blanco, M.A., Smith, C.W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Leung, D.Y., Kisich, K.O. Serine-arginine-rich protein p30 directs alternative splicing of glucocorticoid receptor pre-mRNA to glucocorticoid receptor β in neutrophils. J. Biol. Chem. 2003;278:27112–27118. doi: 10.1074/jbc.M300824200. [DOI] [PubMed] [Google Scholar]
- Young, P.J., DiDonato, C.J., Hu, D., Kothary, R., Androphy, E.J., Lorson, C.L. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 β 1. Hum. Mol. Genet. 2002;11:577–587. doi: 10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- Zheng, Z.M., Huynen, M., Baker, C.C. A pyrimidine-rich exonic splicing suppressor binds multiple RNA splicing factors and inhibits spliceosome assembly. Proc. Natl. Acad. Sci. 1998;95:14088–14093. doi: 10.1073/pnas.95.24.14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Mayeda, A., Krainer, A.R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]