Abstract
Variants of a previously described small self-aminoacylating ribozyme are tested in order to uncover the potentialities of a 3′ extension responsible for the esterification. The base-composition and the length of this specific part of the ribozyme are investigated. Very short extensions can still reach the active site, reflecting the small persistence length of RNA. The yield of aminoacylation is particularly high for ribozymes with extensions made up of a poly-U, for which a maximum of efficiency is observed for a total length of about 10 nucleotides. A simple model describing the behavior of this region of the ribozyme can account for the data.
Keywords: RNA World, ribozyme, tRNA, polymer, poly-U
INTRODUCTION
Following the idea that the former self-organized biological system was shaped by an RNA chemistry, SELEX experiments (selection-amplification of RNA sequences from a random pool) were conducted by many laboratories in order to find ribozymes with different capabilities (Wilson and Szostak 1999). Yarus and coworkers demonstrated that some RNAs have the ability to self-aminoacylate their 3′ end with aminoacyl adenylate (aa-AMP) as a substrate (Illangasekare et al. 1995), which suggested that primitive tRNAs may not have required any enzyme to load an amino acid. In order to characterize the catalytic site of this type of ribozyme, regions of one of the original 95-mer found in the SELEX experiment were deleted, and the variants were tested for self-aminoacylation. The identified region of the ribozyme responsible for the binding of the aminoacyl adenylate consisted of a short hairpin structure with a characteristic AG/AG bulge in the stem. The end of the single-stranded 3′ extension is thought to reach this active site where aa-AMP is bound, thereby enabling the esterification reaction (Illangasekare et al. 1997). Further deletions showed that an RNA as small as 29 bases was still able to undergo self-aminoacylation at a rate similar to that of the parental molecule (Illangasekare and Yarus 1999).
An optimization of this ribozyme (Fig. 1) could help to understand the critical parameters of the aminoacylation reaction. In this paper, we investigate the respective effects of the length and base composition of the 3′ extension on the rate of self-aminoacylation.
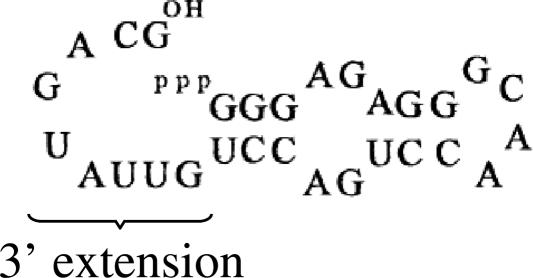
FIGURE 1.
Small self-aminoacylating ribozyme (Illangasekare and Yarus 1999).
RESULTS
Efficiency and base composition of the 3′ extension
The effects of the length and the base composition of the 3′ extension on the rate of self-aminoacylation are tested on several variants of the 29-mer. The experimental conditions are similar to those described in the original work (Illangasekare and Yarus 1999). In all experiments, the buffer is 100 mM NaCl, 80 mM MgCl2, 40 mM CaCl2, 5 mM MnCl2, and 100 mM HEPES (pH 6.8), and the solution kept at 0°C in order to slow down substrate degradation (see below). This RNA requires calcium to display activity, which is noteworthy among ribozymes (Illangasekare et al. 1997). Val-AMP is chosen as a substrate. Alternate amino acids can be used since this type of ribozyme does not display a strong selectivity with regard to the side chain of the amino acid, the binding pocket interacting probably almost exclusively with the AMP moiety (Illangasekare and Yarus 1997). The reaction corresponds to the second step of the aminoacylation reaction on actual tRNAs (the first step being the activation with ATP):
In order to test this reaction, Val-AMP is chemically synthesized following the Berg procedure (Berg 1958). This substrate is highly unstable under the reaction conditions (Berg et al. 1961), and undergoes a parallel first-order exponential decay:
At pH 6.8, we found that the half-life time of Val-AMP is 36 min (first-order decay constant k Val-AMP = 0.019 min−1), very close to the approximate value of 28 min extrapolated from the data for Phe-AMP (Illangasekare and Yarus 1997).
In order to determine the rate constant of a ribozyme, it is thus necessary to take this decay into account. A rate equation that can be satisfactorily fitted to the data is:
where A 0 is the initial Val-AMP concentration, t is time, and k 2nd is the second-order reaction rate constant of reaction 1. The solution to this equation combined with Equation (1) gives (Illangasekare and Yarus 1997):
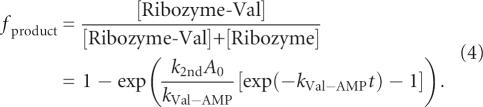 |
According to equation (4), it is possible to evaluate the reaction rate of a ribozyme (k 2nd) while measuring f product at only a single time point. Given the Val-AMP concentration used in the experiments (2–8 mM), as well as the range of variation of k 2nd, 30 min is a convenient time to establish f product for the different variants (see Fig. 2). In order to have A 0 identical in all experiments where a series of variants are tested, the crude mixture of Val-AMP is purified with HPLC and the collected solution is equally split into a series of 0.5 mL microfuge tubes, lyophilized under the same conditions and stored at –80°C prior to use. However, because this procedure does not allow us to know the value of A 0 precisely (in part because Val-AMP undergoes partial degradation during the lyophilization process, which can be different from one series to another), it turns out that only the product [k 2nd A 0] could be determined from equation (4). However, our main purpose is to establish the relative efficiency of each ribozyme, with respect to another. From equation (4), the ratio of two reaction rates k 2nd(a) and k 2nd(b) is calculated as
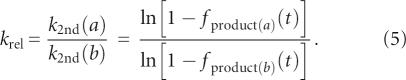 |
FIGURE 2.
Time course of the aminoacylation of three variants of the ribozyme. The 3′extensions (from 5′ to 3′) are: GUUACG (squares), GUUUUACG (triangles), and GUUUUUUACG (circles). The curves show the nonlinear least-square fits of the equation f product = 1−exp{(k 2nd A 0/k Val-AMP)[exp(−k Val-AMP t)−1]}. The bar at 30 min corresponds to the time point where f product was established for all variants, which serves as the basis for comparison in this paper.
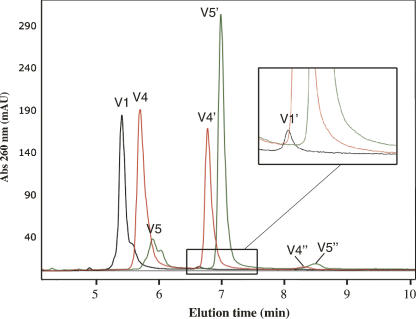
Our measurements with HPLC (Fig. 3) show that the relative reaction rates (k rel) span over more than two orders of magnitude (Table 1). All variants terminate with CG, which had not been randomized during the selection process, and was later found to be a requirement for activity (Illangasekare et al. 1997).
FIGURE 3.
HPLC traces of self-aminoacylation reaction solutions analyzed after an incubation time of 30 min. Each number refers to a particular variant of the ribozyme (V1, V4, and V5; see Table 1). A prime (extension V1′) indicates a peak corresponding to a ribozyme aminoacylated with valine. For the most efficient ribozymes reported here (V4 and V5), a peak corresponding to a peptidyl-RNA (val-val-RNA) is visible (V4″ and V5″). Impurities (usually ribozyme + 1 nt) are present on the sides of the peaks of unacylated ribozymes (V1, V4, V5).
TABLE 1.
Self-aminoacylation efficiency of variants of the 29-mer
Some facts are immediately apparent from Table 1. For a given size of the extension, the aminoacylation efficiency can be dramatically affected by the base composition. For instance, the comparison of V4 with V5 shows that, as a result of two substitutions only (A → U and G → U), k rel is changed by a factor of 3.5. One also notices that extensions with identical base composition but with different base arrangement can change the aminoacylation efficiency (e.g., V7, V8, and V9). Overall, it appears that the presence of purines in the extension tends to slow down the rate of aminoacylation, except when they are located near the 3′ end.
It is well known that purines form stacking interactions whenever possible (e.g., Chen et al. 2005). When localized in the middle of the extension, they might increase the propensity of this region to bind to the core of the ribozyme, which would prevent the terminal nucleotides from reaching the active center. Pyrimidines plausibly minimize this effect, and at the same time confer a higher flexibility to the extension (Goddard et al. 2000). In this connection, our measurements show that an ACG termination provides a better efficiency to the ribozyme compared with UCG (data not shown), confirming that the folding of the extension is mainly driven by purine-mediated interactions.
Ribozymes with very short extensions can still retain some activity
The data of Table 1 show that a variant with an extension as short as 4 nucleotides (nt) can still reach the active site. Although the efficiency is very low (about 1.7% aminoacylation at t = 30 min), it corresponds to a well-defined HPLC peak (see Fig. 3, inset). This provides a clue to infer the path followed by the 3′ extension to the active site. Assuming that the aminoacyl-adenylate binds on the AG/AG bulge, the extension should undergo a very sharp turn at the base of the stem in order for the end to reach this bulge. The crystal structure of a viral RNA pseudoknot (1L3D, Protein Data Bank) shows that such a turn can be achieved with only 1 nt, and that three additional bases would enable the terminal G to reach the bulge, the extension being parallel to the minor groove of the stem. Another interesting fact shown by the crystal structure of 1L3D concerns one role played by Mg2+. Three Mg2+ are localized at the sharp turn present on this pseudoknot (see Egli et al. 2002), evidently to neutralize the negative charges carried by the phosphate group. A high local concentration of divalents is thus necessary to enable a strong torsion of the strand.
The poly-U: Aminoacylation efficiency as a function of the length of the extension
The result obtained with variant V5 suggests that a high k rel is likely to be attributed to the flexibility and the low propensity of stacking of the poly-U compared with A and G. Cytosine was not tested because folding simulations with Mfold (Mathews et al. 1999; Zuker 2003) revealed that the extension would likely interfere with the folding of the core of the ribozyme.
Considering extensions like the one characterizing V5, a low level of interference between the poly-U and other parts of the ribozyme implies that the behavior of the extension should mainly be entropically driven (acknowledging that the binding of the terminal ACG triplet on the site nearby the AG/AG bulge is reversible). One would expect the efficiency of aminoacylation as a function of the number of U to vary monotonically and to decrease as the extension becomes longer. Since short extensions penalize the association due to bending/stretching constraints (enthalpic contribution), a maximum of efficiency for a particular size of the extension, longer than four bases, is expected.
Figure 4 shows f product at t = 30 min for 11 ribozymes under the same experimental conditions. Except for the one with the shortest extension (GUCGOH), all extensions are of the form G(U)xACGOH, with x = 1, 2, 3, 4, 5, 6, 7, 8, 11, 16.
FIGURE 4.
Ribozymes with poly-U in the extension: f product (t = 30 min) versus size of the extension. The smallest extension is GUCG, while all the other ones are of the form G(U)xACG, with x = 1, 2, 3, 4, 5, 6, 7, 8, 11, 16. A0 is ∼ 8 mM, and is identical for all reactions (see Materials and Methods). The errors bars (± 0.05) are set identical for all extensions, and correspond to the maximal value of the error observed. The solid line corresponds to equation (10) with the fitting parameters adjusted to λ = 4.3 and δ = 65. The values of the other parameters are a = 0.43 nm and lp = 1 nm (Caliskan et al. 2005). A ribozyme with another type of extension (V4) is shown together with V5 (a member of the poly-U family) to illustrate the effect of the presence of purines in the poly-U.
A maximum is present for a total length of the extension of 10 bases (x = 6). Remarkably, the efficiency is still comparatively high for the longest extension examined (x = 16), with k rel = 0.42 compared with the maximum.
Among the factors determining the distribution of the values in Figure 4 are A 0 (the initial concentration of Val-AMP) and the concentration of AMP. AMP alone displays a comparable affinity for the binding pocket and is therefore an inhibitor to the reaction (Illangasekare and Yarus 1997). A low initial concentration of Val-AMP simply flattens the distribution but does not change the position of the maximum (data not shown).
We show now that a simple model describing the behavior of the extension can account for the main features of this distribution.
Our hypothesis is that the efficiency of the aminoacylation reaction is proportional to the probability ξ that the end of the extension lies on its binding site within a small interval of time Δt, and that the interaction itself with this site is identical for all ribozymes. We assume that the affinity of the binding pocket for Val-AMP is also invariant. Two main factors account for the value taken by ξ, bending energy and entropic dilution:
Geometric constraint and bending rigidity
Let us define (n + 3) as the number of bases in the extension (“3” referring to the terminal triplet that binds nearby the AG/AG bulge, and which is supposed to add no contribution to the bending energy), a the size of one monomer and R 0 the minimal length of the extension below which the terminal nucleotide is unable to reach the active site. A condition for the reaction to happen is thus (n + 3) > R 0 /a.
For a polymer chain with a persistence length lp, the free energy F associated with the bending of the extension is
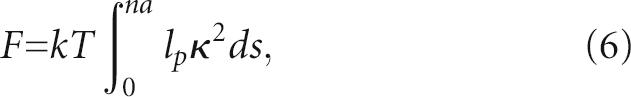 |
(Marko and Siggia 1995; Houchmandzadeh et al. 1997), where s is the curvilinear coordinate along the chain and κ the local curvature. The persistence length depends on the type and concentration of cations. Values reported for poly-U in high concentration of monovalents or divalents are ∼ 0.8–1.0 nm (Seol et al. 2004; Caliskan et al. 2005), i.e., about two to three bases. Since the extension is not significantly longer than the persistence length, we assume that the curvature κ is constant. With κ = 1/R and R ∝ na, this leads to
 |
where λ is a constant of the order of unity that takes into account the geometrical properties of the ribozyme. The probability of observing this bending is simply given by
 |
Entropic dilution
The typical size for a Gaussian chain with a persistence length lp is 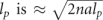 (Cantor and Schimmel 1980). The volume occupied by the chain increases as V ≈ (2nal
p)3/2 and the corresponding concentration of the reaction site decreases like V−1. The entropic dependence of the probability that the 3′ end is on the catalytic site is thus simply given by
(Cantor and Schimmel 1980). The volume occupied by the chain increases as V ≈ (2nal
p)3/2 and the corresponding concentration of the reaction site decreases like V−1. The entropic dependence of the probability that the 3′ end is on the catalytic site is thus simply given by
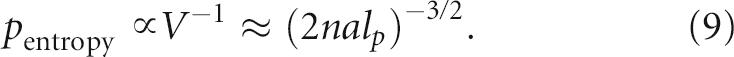 |
Combining the bending and the entropic contributions, we obtain
 |
Figure 4 shows ξ(n + 3) with the parameters λ and δ adjusted to 4.3 and 65, respectively (this adjustment being dependent on the time at which f product is measured), and with a = 0.43 nm and lp = 1 nm. The plot shows that the functional form of ξ can well account for the overall distribution of the experimental values, but that a more detailed description would be required to fit the model to the data. Since the size of the extension is of the order of a few bases only, significant deviations from our statistical description are expected.
DISCUSSION
The unique feature of the small self-aminoacylating ribozyme examined here lies in the versatility offered by its 3′ extension. A core of this extension made up with a poly-U ensures a low level of interaction with the other parts of the ribozyme, and enables the specific ACG termination to reach its binding site. It is already known that poly-U displays the least ordered state in solution compared with the three other homopolymers of RNA (Akeson et al. 1999; Newby Lambert et al. 2006). Our results show that this property can be used to modulate the activity of this ribozyme, and possibly other types of RNA functions. For instance, in systems in which a single-strand RNA is used as a linker (see, e.g., Murakami et al. 2002) a poly-U instead of a poly-A could diminish the perturbation introduced by the linker.
Since this ribozyme constitutes a step toward the primitive tRNA, it is worth noting that short extensions, comparable to the acceptor arm of present-day tRNAs (i.e., four to five bases), can still undergo aminoacylation.
However, in its present form, this ribozyme displays properties that are significantly different from that of tRNA/Synthetase couples, a main difference being that it requires aminoacyl-adenylate as a substrate. Because of the high propensity to hydrolysis of this molecule, the stability of any genetic system built up with such a ribozyme would be compromised. This problem was avoided by Suga and coworkers in another evolved ribozyme (Saito et al. 2001), where the amino acid was activated with cyanomethyl ester (CME), that group being less prone to hydrolysis. It could not, however, be used here because it would not bind to the AG/AG bulge.
These considerations raise the problem of the activation of the amino acid. As a future development, it would be desirable to find a ribozyme that would have the ability to activate the amino acid in situ prior to aminoacylation, preferably with ATP, since this group is already present in a system made up of RNA. It is, however, not clear whether the ribozyme investigated here, the 3′ extension of which already confers a remarkable potential of adaptation, could be derived to perform these two consecutive chemical reactions.
MATERIALS AND METHODS
Ribozyme synthesis and purification
The ribozymes were transcribed from double-strand DNA templates using a T7 RNA polymerase (Megashortscript, Ambion) and purified by HPLC using a C18 analytical column (XTerra C18, Waters) with a mobile phase composed of TEAA 0.1M (pH 6.7) and acetonitrile. The purified samples were desalted using centrifugal filters (Microcon 3000 NMWL, Millipore) and the concentration measured by absorption at 259 nm. The identity and the purity of each ribozyme were checked by mass spectroscopy. The ribozymes could not be chemically synthesized since the 5′ triphosphate, which was found to be required for activity (Illangasekare et al. 1997), could not be added enzymatically. We did not try ribozymes with poly-U extensions larger than 20 nt since long segments of U are poor templates for the T7 polymerase, and makes it difficult to obtain consistent yields of pure ribozymes.
Substrate preparation
Val-AMP was chemically synthesized following the Berg procedure (Berg 1958). The crude mixture was purified by HPLC to remove the excess of AMP. We used a C18 semipreparative column (Vydac); the gradient of the mobile phase was established with ammonium acetate 0.01 M (pH 4.8) and acetonitrile. Once collected, the purified solution of Val-AMP was equally split into a series of 0.5 mL microfuge tubes and immediately frozen into liquid nitrogen. The samples were then lyophilized under high vacuum over several hours to remove the solvent (we used a low concentration of ammonium acetate in the purification step in order to prevent the sample from thawing during this process). The tubes were then stored at – 80°C until use. One tube was used to determine the quality of the stock by HPLC (usually 90% was still Val-AMP and 10% were degradation products). This method made it possible to test series of variants of the ribozyme under exactly the same Val-AMP concentration.
Aminoacylation reactions
The RNA was heated up to 65°C for 10 min and cooled down to room temperature. The buffer was then added to a final concentration of 100 mM NaCl, 80 mM MgCl2, 40 mM CaCl2, 5 mM MnCl2, and 100 mM HEPES (pH 6.8). The solution (usually 50 μL) was cooled down to and maintained at 0°C (on Thermostat Plus, Eppendorf). The final concentration of RNA was usually 2–10 μM. The reaction was started by transferring the solution into a cold tube containing lyophilized Val-AMP (just taken out from – 80°C freezer), vortexed shortly, and placed at 0°C for 30 min. The final concentration of Val-AMP was usually 4–8 mM. At the end of the incubation time, 10% vol of glycogen (20 mg/mL) was added and the reaction was quenched by adding 10% vol of sodium acetate 3.8 M (pH 5.2). The RNA was then precipitated with 2.5 vol of cold ethanol 100% and placed at – 80°C for at least 40 min. For time course experiments, aliquots (6–8 μL, usually taken at time t = 5, 15, 30, 60, and 120 min) were quenched in 200 μL of cold ethanol 75% containing 2.5% vol of glycogen (20 mg/mL) and 2.5% vol of sodium acetate 3.8 M (pH 5.2), and the solution was placed at – 80°C for 40 min.
Products analysis
After centrifugation and recentrifugation with ethanol 75%, the pellets containing the products of the reaction were dried up in vacuum and redissolved in 20 μL of TEAA 0.1 M (pH 6.7) and the solution was injected into the HPLC. We used an analytical column (XTerra C18, Waters) heated up to about 40°C. The gradient of acetonitrile in TEAA 0.1 M, pH 6.7 was 7.5–12.5% in 20 min (1 mL/min). Under these conditions, the ribozyme elution time was usually 5–10 min (depending on the variant). A first peak corresponding to the unacylated ribozyme was followed 2.5 min later by another peak corresponding to the ribozyme acylated with valine. The identity of these peaks was further confirmed by mass spectroscopy. Under such HPLC conditions, no measurable level of deacylation was observed during the time the RNA was in the column, as an earlier work suggested (McCutchan et al. 1975).
The efficiency of aminoacylation was determined as the ratio of the height of the peaks of the acylated versus the acylated plus unacylated ribozymes. The height was preferred to the area because its measure would not include the possible contribution of small impurities, usually present on both sides of the peak signal.
Mass spectroscopy
The RNA transcribed from DNA and the products of the aminoacylation reactions were characterized by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI), using the Voyager-DE STR system (Perspective Biosystems) of the Proteomic Resource Center at the Rockefeller University. For each series of measurements, a fresh matrix made up of 21 mg 3-Hydroxypicolinic acid, 2 mg Hydroxypicolinic acid, and 2 mg diammonium citrate (Sigma-Aldrich), dissolved in 250 μL of RNAse-free water/acetonitrile (50/50), was prepared (Petersson et al. 2002). The RNA was dissolved in 10 μL TEAA 0.1 M (pH 6.7), which was then used to load a Ziptip C4 (Millipore), following the protocol established by the company. The RNA was eluted in 2 μL of the matrix on the well of a gold plate. After evaporation, the gold plate was placed under vacuum for about half an hour to dry the sample, which improves the signal. The samples were analyzed using the linear mode, with an accelerating voltage of 20 kV. The grid voltage was set to 94% and the extraction delay time to 250 nsec. The laser intensity was usually 2300 (A.U.), below which the signal/noise ratio was generally not good enough.
ACKNOWLEDGMENTS
We thank Shixin Ye for invaluable help in the establishment of the experimental procedures. We are also grateful to Haiteng Deng, Suzanne R. Powell, and Henry A. Zebrosky of the Proteomic Resource Center of the Rockefeller University for providing technical support. This work is supported by a fellowship to J.L. from the Swiss National Science Foundation (Grant No. 105059), and in part by the Marie-Josée and Henry Kravis Fellowship at the Rockefeller University to J.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.500907.
REFERENCES
- Akeson, M., Branton, D., Kasianowicz, J.J., Brandin, E., Deamer, D.W. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, P. The chemical synthesis of amino acyl adenylates. J. Biol. Chem. 1958;233:608–611. [PubMed] [Google Scholar]
- Berg, P., Bergmann, F.H., Ofengand, E.J., Dieckman, M. The enzymic synthesis of amino acyl derivatives of ribonucleic acids. J. Biol. Chem. 1961;236:1726–1734. [PubMed] [Google Scholar]
- Caliskan, G., Hyeon, C., Perez-Salas, U., Briber, R.M., Woodson, S.A., Thirumalai, D. Persistence length changes dramatically as RNA folds. Phys. Rev. Lett. 2005;95:268303–268306. doi: 10.1103/PhysRevLett.95.268303. [DOI] [PubMed] [Google Scholar]
- Cantor, C.R., Schimmel, P.R. W.H. Freeman and Company; New York: 1980. Biophysical chemistry. Part III: The behavior of biological molecules. [Google Scholar]
- Chen, G., Znosko, B.M., Kennedy, S.D., Krugh, T.R., Turner, D.H. Solution structure of an RNA internal loop with three consecutive shared GA pair. Biochemistry. 2005;44:2845–2856. doi: 10.1021/bi048079y. [DOI] [PubMed] [Google Scholar]
- Egli, M., Minasov, G., Su, L., Rich, A. Metal ion coordination and conformational flexibility in a viral RNA pseudoknot at atomic resolution. Proc. Natl. Acad. Sci. 2002;99:4302–4307. doi: 10.1073/pnas.062055599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, N.L., Bonnet, G., Krichevsky, O., Libchaber, A. Sequence dependent rigidity of single stranded DNA. Phys. Rev. Lett. 2000;85:2400–2403. doi: 10.1103/PhysRevLett.85.2400. [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh, B., Marko, J.F., Chatenay, D., Libchaber, A. Elasticity and structure of Eukaryote chromosome studied by micromanipulation and micropipette aspiration. J. Cell Biol. 1997;138:1–12. doi: 10.1083/jcb.139.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illangasekare, M., Sanchez, G., Nickles, T., Yarus, M. Aminoacyl–RNA synthesis catalyzed by an RNA. Science. 1995;267:643–647. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]
- Illangasekare, M., Yarus, M. Small-molecule–substrate interaction with a self-aminoacylating ribozyme. J. Mol. Biol. 1997;268:631–639. doi: 10.1006/jmbi.1997.0988. [DOI] [PubMed] [Google Scholar]
- Illangasekare, M., Yarus, M. A tiny RNA that catalyses both aminoacyl-RNA and peptidyl–RNA synthesis. RNA. 1999;5:1482–1489. doi: 10.1017/s1355838299991264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illangasekare, M., Kovalchuke, O., Yarus, M. Essential structures of a self-aminoacylating RNA. J. Mol. Biol. 1997;274:519–529. doi: 10.1006/jmbi.1997.1414. [DOI] [PubMed] [Google Scholar]
- Marko, J.F., Siggia, E.D. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- McCutchan, T.F., Gilham, P.T., Söll, D. An improved method for the purification of tRNA by chromatography on dihydroxyboryl substituted cellulose. Nucleic Acids Res. 1975;2:853–864. doi: 10.1093/nar/2.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, H., Bonzagni, N.J., Suga, H. Aminoacyl-tRNA synthesis by resin-immobilized ribozyme. J. Am. Chem. Soc. 2002;124:6834–6835. doi: 10.1021/ja025872a. [DOI] [PubMed] [Google Scholar]
- Newby Lambert, M., Vöcker, E., Blumberg, S., Redemann, S., Gajraj, A., Meiners, J.-C., Walter, N.G. Mg2+-induced compaction of single RNA molecules monitored by tethered particle microscopy. Biophys. J. 2006;90:3672–3685. doi: 10.1529/biophysj.105.067793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson, E.J., Shahgholi, M., Lester, H.A., Dougherty, D.A. MADLI-TOF mass spectrometry methods for evaluation of in vitro aminoacyl tRNA production. RNA. 2002;8:542–547. doi: 10.1017/s1355838202026146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, H., Kourouklis, D., Suga, H. An in vitro evolved precursor tRNA with aminoacylation activity. EMBO J. 2001;20:1797–1806. doi: 10.1093/emboj/20.7.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol, Y., Skinner, G.M., Visscher, K. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys. Rev. Lett. 2004;93:118102–118105. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- Wilson, D.S., Szostak, J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]