Abstract
We are now aware that RNA-based regulatory mechanisms are commonly used to control gene expression in many organisms. These mechanisms offer the opportunity to exploit relatively short, unique RNA sequences, in altering transcription, translation, and/or mRNA stability, in response to the presence of a small or large signal molecule. The ability of an RNA segment to fold and form alternative hairpin secondary structures—each dedicated to a different regulatory function—permits selection of specific sequences that can affect transcription and/or translation. In the present paper I will focus on our current understanding of the RNA-based regulatory mechanisms used by Escherichia coli and Bacillus subtilis in controlling expression of the tryptophan biosynthetic operon. The regulatory mechanisms they use for this purpose differ, suggesting that these organisms, or their ancestors, adopted different strategies during their evolution. I will also describe the RNA-based mechanism used by E. coli in regulating expression of its operon responsible for tryptophan degradation, the tryptophanase operon.
Keywords: trp operon, trp suboperon, aro supraoperon, tna operon, transcription attenuation, T box regulation, tryptophan as a regulatory signal, tRNATrp as a regulatory signal, peptidyl-tRNA, ribosome mediated regulation
INTRODUCTION
Studies over the past 50+ years have revealed that optimization of gene expression has been a major evolutionary objective for most species. Virtually every event involved in gene expression—and gene product function—in each organism appears to have been subjected to some form of optimization or regulation. The goal must have been to achieve a balanced performance of all life-dependent processes under the variety of environmental conditions each organism experiences. It was once thought that dedicating genetic material to regulatory purposes would be costly, and therefore a single regulatory protein might suffice for each group of functionally related genes. We now know that this view was an oversimplification, for regulated, balanced gene expression was clearly a much more important objective for each organism than we had anticipated. In fact, a significant fraction of the genetic material of each organism is dedicated to regulatory functions.
A second regulatory lesson learned over the years is that information within mRNAs, or other RNAs, as well as small metabolites and other molecules—in addition to DNA and proteins—often provides specific regulatory signals, or performs or modulates the regulatory processes that control gene expression. Discovery of these mechanisms continues. In this article I will focus on the RNA-based mechanisms we have been studying that bacteria use in regulating expression of the genes of tryptophan biosynthesis and the genes of tryptophan degradation. Our findings illustrate the variety of RNA-based regulatory mechanisms that have developed. These include formation of alternative RNA hairpin structures, cross-talk between transcription and translation, regulated RNA–protein interactions, and nascent peptide influence on ribosome translation.
FINDINGS
The tryptophan biosynthetic pathway
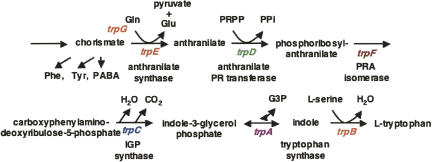
The genes, enzymes, and reactions of the tryptophan biosynthetic pathway appear to have evolved just once, since virtually all organisms with this synthetic capacity have related genes that specify homologous enzymes that catalyze the same reactions (Fig. 1). However, tryptophan biosynthesis is a biologically expensive, complicated process. In fact, the products of four other pathways are essential contributors of carbon or nitrogen during tryptophan formation (Yanofsky and Crawford 1987; Yanofsky et al. 1999). Thus, the principal pathway precursor, chorismate, is also the precursor of the other aromatic amino acids, phenylalanine and tyrosine, as well as serving as the precursor of p-aminobenzoic acid and several other metabolites. In addition, glutamine, phosphoribosylpyrophosphate, and L-serine contribute nitrogen and/or carbon during tryptophan formation. Thus, each organism with tryptophan-synthesizing capacity must have adopted appropriate regulatory strategies to ensure that sufficient levels of chorismate and these other three compounds are produced. In many organisms tryptophan serves as the precursor of other biologically essential compounds, i.e., niacin in most eukaryotes, indoleacetic acid in most plants, and indole in many bacteria. Thus the regulatory strategies designed for the genes of tryptophan biosynthesis of each organism have had to be compatible with other metabolic objectives. An interesting feature of tryptophan biosynthesis is that this capability was dispensed with when organisms evolved that were capable of obtaining tryptophan by feeding on other organisms. Nevertheless, as mentioned, products of tryptophan degradation/metabolism are essential in these organisms.
FIGURE 1.
The genes, enzymes, and reactions of the tryptophan biosynthetic pathway. The seven genes, or genetic segments, seven enzymes, or enzyme domains, and seven reactions, involved in tryptophan formation are shown (Yanofsky and Crawford 1987). Only one of the reactions is reversible. The products of four other pathways contribute carbon and/or nitrogen during tryptophan formation. Two of the tryptophan pathway enzymes often function as polypeptide complexes: anthranilate synthase, consisting of the TrpG and TrpE polypeptides, and tryptophan synthase, consisting of the TrpB and TrpA polypeptides.
Regulatory mechanisms controlling transcription of the trp operon of E. coli
The genes required for tryptophan biosynthesis in Escherichia coli are organized as a single transcriptional unit, the trp operon (Fig. 2; Yanofsky and Crawford 1987). This operon has a single major promoter at which transcription initiation is regulated by a DNA-binding protein, the L-tryptophan-activated trp repressor (Yanofsky and Crawford 1987). This repressor acts by binding at one or more of three operator sites located in the trp operon's promoter region (Fig. 2; Lawson et al. 2004). The structures of the inactive trp aporepressor, the tryptophan-activated trp repressor, and the trp repressor–operator complex have all been determined, and this repressor's mechanism of action is well understood (Fig. 3; Otwinowski et al. 1988; Joachimiak and Zhang 1989; Shakked et al. 1994; Gryk et al. 1996). The aporepressor and repressor are dimers composed of identical helix-turn-helix monomers (Joachimiak and Zhang 1989; Shakked et al. 1994; Gryk et al. 1996). Each repressor half-molecule has a bound tryptophan, and it recognizes an identical nucleotide sequence in each operator (Otwinowski et al. 1988; Joachimiak and Zhang 1989; Shakked et al. 1994). Bound tryptophan alters the spacing between the DNA binding sites in each repressor molecule, positioning them for precise interaction with their operator sequences (Fig. 2). Interactions between separate repressor molecules bound at adjacent trp operator sequences in the trp promoter increase the likelihood that each repressor molecule will remain bound at an operator, thereby increasing the degree of repression (Lawson et al. 2004). The trp repressor also autoregulates its own synthesis, though weakly. It binds at an operator site located in its promoter, inhibiting initiation of transcription (Yanofsky and Crawford 1987). Repressor autoregulation permits E. coli to synthesize increased levels of the aporepressor in preparation for a potential subsequent event—response to the appearance of excess tryptophan. Thus, it prepares itself for more efficient shutdown of tryptophan synthesis whenever the cellular tryptophan concentration becomes elevated following a period of tryptophan starvation.
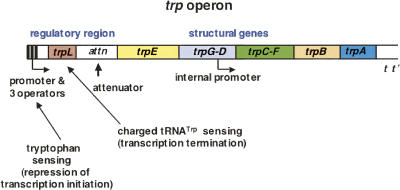
FIGURE 2.
Organization of the trp operon of E. coli. The genes of E. coli required for tryptophan biosynthesis from chorismate are organized in a single operon, or transcriptional unit (Yanofsky and Crawford 1987; Yanofsky 2004). Two pairs of genes are fused: trpG and trpD, and trpC and trpF. The structures of these two bifunctional polypeptides are known, and separate polypeptide segments are concerned with catalysis of each reaction. The relative order of the seven genetic segments, trpEGDFCBA, corresponds roughly to the relative order of the respective enzymatic reactions. The trp operon's regulatory region, located at the beginning of the operon, is designed to sense two signals, L-tryptophan, and charged vs. uncharged tRNATrp (Landick and Yanofsky 1987; Yanofsky 2004). Tryptophan, when in excess, activates the trp aporepressor, while charged and uncharged tRNATrp determine whether transcription will or will not be terminated in the operon's leader region. A poorly expressed internal promoter provides transcripts producing low levels of the last few enzymes of the pathway. This promoter is useful when the principal promoter is turned off. (p = promoter; t = terminator). (Modified from Figs. 1 and 2 in Yanofsky 2004 and reprinted with permission from Elsevier © 2004.)
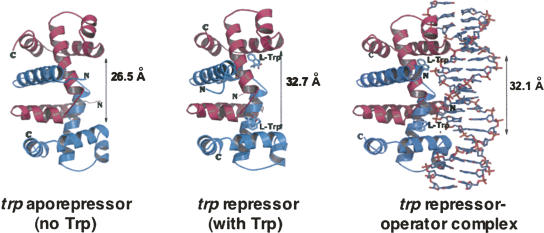
FIGURE 3.
The basic structures of the trp aporepressor, trp repressor, and trp repressor-operator complex. The aporepressor is a dimer of identical polypeptide chains; it has two tryptophan binding sites (Joachimiak and Zhang 1989; Shakked et al. 1994; Gryk et al. 1996). When tryptophan is bound, the repressor's two helix-turn-helix domains assume conformations that prepare them for binding at specific operator sequences. Binding of the trp repressor at a trp operator site—located within the trp promoter—interferes with RNA polymerase binding, hence binding prevents transcription initiation (Otwinowski et al. 1988). Interactions between trp repressor proteins bound at multiple adjacent operators in the trp promoter-operator region increase the stability of repressor-operator complexes, improving the effectiveness of repression (Fig. 2; Lawson et al. 2004). This figure was kindly drawn, and provided, by Dr. Andrzej Joachimiak.
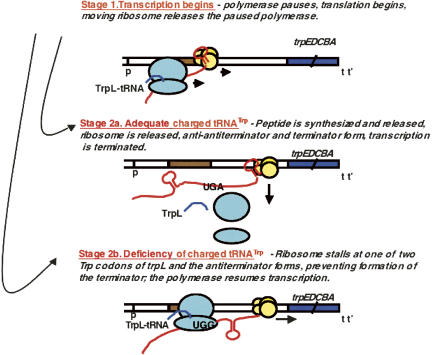
The availability of tryptophan-charged tRNATrp is also sensed as a regulatory signal in controlling trp operon transcription—by a transcription attenuation mechanism. The relevant features of the ∼160 nucleotide (nt) trp operon leader transcript responsible for charged tRNATrp sensing, and for regulation by transcription attenuation, are shown in Figure 4 (Landick and Yanofsky 1987; Yanofsky 2000). Transcription attenuation mechanisms generally involve several sequential stages or events. The stages used in attenuation regulation of the trp operon of E. coli are described in Figure 5 (Landick and Yanofsky 1987; Yanofsky 2000). An essential feature of this attenuation mechanism is the synchronization of translation of a 14-residue leader peptide coding region, trpL, with transcription of the operon's leader region. Synchronization is achieved by exploiting features of the initial segment of the leader transcript, the segment overlapping trpL. This segment can form an RNA hairpin structure, designated hairpin 1–2, called the anti-antiterminator. Hairpin 1:2 also serves as a transcription pause signal (see Figs. 4, 5). Transcriptional pausing is relieved when a ribosome binds at the trpL mRNA start codon and initiates synthesis of the TrpL leader peptide. The moving ribosome appears to disrupt the RNA pause hairpin, releasing the paused RNA polymerase (Fig. 5, Stage 1). Subsequently, either of two events occurs, depending on the availability of uncharged versus charged tRNATrp. When most of the cellular tRNATrp is uncharged, difficulty in translating the two Trp codons of trpL mRNA results in ribosome stalling at one of these Trp codons (Figs. 4, 5, Stage 2b). This allows the antiterminator structure, hairpin 2–3, to form (Fig. 4), which prevents formation of the terminator structure. Prevention of terminator formation allows transcription to continue into the structural genes of the operon (Fig. 5, Stage 2b). When charged tRNATrp is plentiful, however, translation of trpL is completed, and the translating ribosome dissociates at the trpL stop codon. This permits the leader transcript to fold and form the anti-antiterminator and terminator structures, 1:2 and 3:4 (Fig. 4), promoting transcription termination (Fig. 5, Stage 2a). Thus, depending on the availability of charged tRNATrp during translation of trpL, transcription of the structural gene region of the trp operon will or will not proceed. Many amino acid biosynthetic operons of Gram-negative bacterial species are regulated by similar ribosome-mediated transcription attenuation mechanisms (Yanofsky 1981). The unique distinguishing feature of each is the inclusion of codons for the respective amino acid in the corresponding leader peptide coding region.
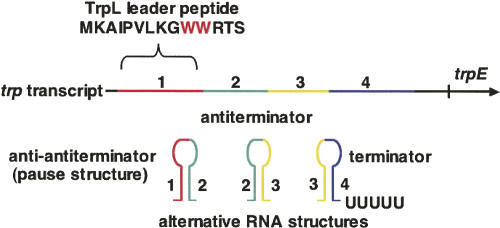
FIGURE 4.
The trp operon leader transcript and its functions. The initial 141 nt of the trp operon transcript can fold and form three alternative RNA hairpin structures: an anti-antiterminator (1:2), an antiterminator (2:3), and a terminator (3:4) (Landick and Yanofsky 1987; Yanofsky 2004). The anti-antiterminator structure (1:2) also serves as a transcription pause structure. The terminator structure is a typical intrinsic terminator which, when formed, directs the transcribing RNA polymerase to terminate transcription. However, whenever the preceding antiterminator structure has formed and persists, it prevents formation of the terminator. Hence transcription is not terminated. The initial leader RNA sequence has an additional role: It encodes a 14 nt leader peptide, TrpL. Translation of the two Trp codons of trpL is used to sense the availability of charged tRNATrp. Whenever the charged tRNATrp level is adequate for rapid translation of the two Trp codons of trpL, translation of trpL is completed, the translating ribosome dissociates, and the anti-antiterminator (1:2) and terminator (3:4) structures form. This results in transcription termination in the leader region of the operon. When the tRNATrp in the cell is largely uncharged, the translating ribosome stalls at one of the trpL Trp codons. The antiterminator RNA structure (2:3) then forms, preventing terminator formation and transcription termination. (See Fig. 5 for more details). Pausing following formation of the anti-antiterminator structure is essential for the coupling of translation of trpL mRNA with transcription of the leader region. The anti-antiterminator, if allowed to form and persist, would prevent formation of the antiterminator, thus the transcription terminator would form. (Modified from Fig. 2 in Yanofsky 2004 and reprinted with permission from Elsevier © 2004.)
FIGURE 5.
The sequential, alternative events regulating transcription termination in the leader region of the trp operon of E. coli. Stage 1: The RNA polymerase molecule that initiates transcription of the trp operon pauses after synthesizing the initial segment of the transcript—the segment that forms the anti-antiterminator pause structure (Landick and Yanofsky 1987; Yanofsky 2004). While the polymerase is paused, a ribosome binds at the trpL mRNA start codon and initiates synthesis of the leader peptide. This translating ribosome then disrupts the anti-antiterminator pause structure, releasing the paused polymerase and allowing it to resume transcription. Stage 2a: When there is sufficient charged tRNATrp in the cell to allow rapid completion of synthesis of the leader peptide, the translating ribosome is released. The anti-antiterminator and terminator structures then form, promoting transcription termination. Stage 2b: When there is a deficiency of charged tRNATrp, the ribosome translating trpL mRNA stalls at one of its two Trp codons. This permits the RNA antiterminator structure to form, which prevents formation of the terminator. Transcription then continues into the operon's structural genes. (Modified from Fig. 2 in Yanofsky 2004 and reprinted with permission from Elsevier © 2004.)
Regulatory subtleties abound, however, therefore each event—at each stage—is not absolute. Other regulatory processes influence trp mRNA synthesis and survival, trp coding region translation, trp enzyme function and turnover, and trp enzyme activity (Landick and Yanofsky 1987; Yanofsky and Crawford 1987). For example, and most importantly, the activity of the first enzyme of the tryptophan biosynthetic pathway, anthranilate synthase, is subject to feedback inhibition by L-tryptophan. This is typical of the first enzyme of many biosynthetic pathways. Feedback inhibition is a physiologically advantageous process because it allows an instantaneous—and readily reversible—reduction in the flow of carbon and nitrogen into a pathway.
trp operon organization and regulation in B. subtilis
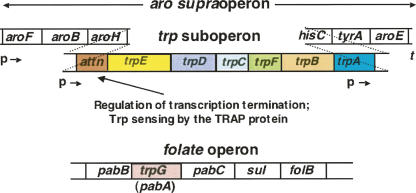
The genes of the trp operon of this organism are organized differently than in E. coli (Fig. 6; Henner and Yanofsky 1993; Yanofsky 2004; Gollnick et al. 2005). Six of the seven trp genes exist as a trp suboperon within an aromatic (aro) supraoperon. This aro supraoperon has three genes preceding and three genes following the trp suboperon (Fig. 6). These upstream and downstream genes encode proteins that act in the chorismate—common precursor—pathway, or in reactions involved in the synthesis of phenylalanine, tyrosine, or histidine. Two promoters are used to initiate transcription of the trp suboperon, one located at the very beginning of the aro supraoperon and the other, a few hundred base pairs before trpE (Fig. 6). Transcription continuation from either promoter into the structural gene region of the trp suboperon is regulated by transcription attenuation, in the region immediately preceding trpE. Regulation is achieved by the action of the tryptophan-activated RNA-binding attenuation protein, TRAP (Gollnick et al. 2005). The seventh trp gene, trpG/pabA, is located in the folate operon (Fig. 6; Gollnick et al. 2005). This gene specifies a glutamine amidotransferase polypeptide that serves as a component of two enzyme complexes, one catalyzing o-aminobenzoate synthesis from chorismate, in tryptophan formation, and the second catalyzing p-aminobenzoate synthesis from chorismate, in folate biosynthesis—hence the designation trpG/pabA (Yakhnin et al. 2007). As will be described, recent studies have shown that trpG/pabA of the folate operon is transcribed from two promoters, and that synthesis of the TrpG/PabA polypeptide is translationally regulated by the action of tryptophan-activated TRAP (Yakhnin et al. 2007).
FIGURE 6.
Organization of the trp suboperon within the aro supraoperon, and the location of trpG in the folate operon, in B. subtilis. Six of the seven trp genes of B. subtilis are organized as a trp suboperon within the aro supraoperon (Gollnick et al. 2005). The three upstream and three downstream genes of this supraoperon encode proteins that participate in the common aromatic pathway or in phenylalanine, tyrosine, or histidine biosynthesis. One of the trp genes, trpG, resides within the folate operon. There is logical, since trpG, or more correctly trpG/pabA, specifies a polypeptide (TrpG/PabA) that serves as a component of two enzyme complexes. This polypeptide is the amino-group donor during the biosynthesis of both o-aminobenzoic acid (anthranilic acid), in tryptophan formation, and p-aminobenzoic acid, in folate formation. Two promoters within the aro supraoperon are used to transcribe the trp suboperon. Transcripts initiated at either promoter can form the alternative RNA structures described in Fig. 7. There is a third promoter—within trpA—that provides a transcript of the terminal three genes of the operon. This transcript is used to provide the polypeptides encoded by these three genes whenever transcription from the two upstream promoters is terminated in the trp suboperon leader region. Transcription, and regulation, of the folate operon is described in Fig. 10 (p = promoter; t = terminator). (Modified from Figs. 1 and 2 in Yanofsky 2004 and reprinted with permission from Elsevier © 2004.)
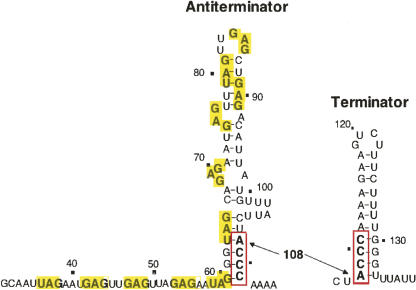
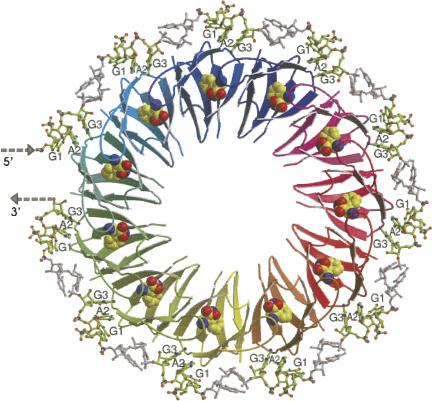
The transcript segment immediately preceding trpE can fold and form either of two hairpin structures, an antiterminator or a terminator (Fig. 7; Babitzke 2004; Gollnick et al. 2005). Choosing between these structures is the role of the RNA-binding protein, TRAP. This protein, when tryptophan-activated, binds to the RNA segment that can form the RNA antiterminator structure and prevents its formation. This permits the alternative transcription terminator structure to form, which terminates transcription (Fig. 7). TRAP is a circular doughnut-shaped molecule consisting of eleven identical subunits (Fig. 8; Antson et al. 1999). A tryptophan-binding site is formed between each pair of TRAP subunits. Each bound tryptophan stabilizes a conformation on the surface of the TRAP protein (McElroy et al. 2002), allowing TRAP to recognize—and bind strongly to—repeat RNA nucleotide sequences (XAG triplets, with the preferred X nucleotide = G > U > A > C) (Gollnick et al. 2005). There are 11 XAG repeats in the transcript sequence preceding and within the antiterminator structure (Fig. 7; each triplet in bold, in a yellow rectangle). A 2–3 nt spacing between adjacent XAGs in RNA has been shown to be optimal for TRAP binding (Gollnick et al. 2005). Eleven KKR motifs in tryptophan-activated TRAP participate in recognition of the 11 XAG triplet repeats in the trp leader sequence. Tryptophan-activated TRAP wraps the transcript around its periphery, freeing the ACCC sequence (Fig. 7, boxed sequence) essential for stabilization of the antiterminator. This allows these four nucleotides to participate in formation of the terminator structure (Fig. 7; Gollnick et al. 2005).
FIGURE 7.
The alternative antiterminator and terminator structures that can form in the transcript of the leader region of the trp suboperon of B. subtilis. The antiterminator and termination hairpin structures that can form in the transcript segment immediately preceding trpE share an ACCC (bold letters, red box) nucleotide segment (Gollnick et al. 2005). Whenever the antiterminator structure forms, and remains stable, it prevents formation of the terminator structure. Therefore transcription termination does not occur in the leader region. A series of (X)AG trinucleotide repeats (bold letters, yellow rectangles) are located in the sequence immediately preceding, and within, the antiterminator sequence. These constitute the binding site for the tryptophan-activated RNA-binding regulatory protein, TRAP. When tryptophan-activated, TRAP binding to the RNA prevents formation of the antiterminator structure—or disrupts it, if it had formed previously. Disruption of the antiterminator structure frees its ACCC-containing 3′ strand, allowing the terminator to form and terminate transcription. Thus TRAP, when tryptophan-activated, binds to leader RNA and promotes transcription termination in the trp suboperon's leader region. Activated TRAP has a second beneficial effect on trp operon expression. When bound to non-terminated trp mRNA, it promotes formation of an RNA secondary structure in trp leader RNA that reduces initiation of translation of trpE. This also reduces trp operon expression (Merino et al. 1995; Du and Babitzke 1998).
FIGURE 8.
Structure of the TRAP–RNA complex. TRAP of B. subtilis is a doughnut-shaped molecule consisting of 11 identical polypeptide subunits (Antson et al. 1999; Gollnick et al. 2005). A tryptophan-binding site is formed between each pair of subunits. In this figure, the atoms of the bound tryptophan molecules are represented as spheres. Tryptophan binding alters TRAP's surface, preparing it for recognition of, and binding to, RNA segments containing XAG repeats. The spacing between the XAG repeats, 2–3 nt, also is crucial (Gollnick et al. 2005). When TRAP binds to trp suboperon leader RNA it wraps the RNA around its periphery, disrupting the antiterminator structure. This allows the alternate, terminator structure to form and terminate transcription. In B. subtilis TRAP also binds to XAG trinucleotide repeats in the transcript segments preceding the start codons for trpG, trpP, and ycbK and inhibits their translation. (Modified from Fig. 3 in Gollnick et al. 2005 with permission from Annual Reviews © 2005; www.annualreviews.org.)
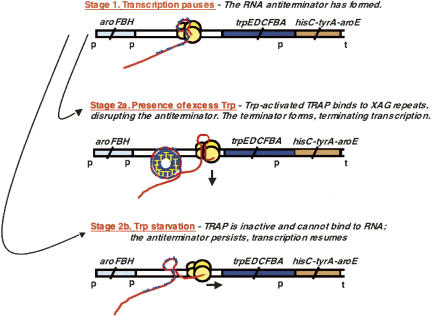
The stages in tryptophan and TRAP regulation of expression of the trp suboperon are summarized in Figure 9. Transcription pausing (Fig. 9, Stage 1) and TRAP's ability to disrupt or prevent formation of the antiterminator (Fig. 9, Stage 2a) are the crucial events that permit this tryptophan-activated protein to regulate transcription of the trp suboperon of the aro supraoperon (Gollnick et al. 2005). But other regulatory options exist here as well. It is important to the organism to be able to shut down translation of trpE mRNA whenever tryptophan is present in excess. Thus, TRAP-induced formation of the terminator structure is only sufficient to cause about 90%–95% transcription termination. In the 5%–10% of TRAP-bound transcripts that have not terminated, a hairpin structure forms that prevents initiation of translation of trpE. This structure reduces synthesis of anthranilate synthase, the enzyme catalyzing the first reaction of the tryptophan biosynthetic pathway. This inhibition spares chorismate for other purposes. Transcriptional pausing has been shown to be instrumental in facilitating this translation inhibition (Yakhnin et al. 2006a,b). This second mechanism further reduces the overall rate of tryptophan synthesis (Merino et al. 1995). As we shall see, TRAP's ability to function is also influenced by a second protein, AT. The level of AT formed by cells is dependent on their levels of uncharged versus charged tRNATrp (Gollnick et al. 2005).
FIGURE 9.
Stages in tryptophan regulation of transcription termination in the leader region of the trp suboperon of B. subtilis. Stage 1: Transcription initiated at either of the two promoters that precede the trp gene cluster pauses in the trp suboperon leader region upon synthesis of the antiterminator structure. Stage 2a: In the presence of excess tryptophan, the TRAP protein is activated and it binds to the mRNA XAG repeats, preventing formation of, or disrupting, the antiterminator structure. This allows the terminator structure to form and terminate transcription. Stage 2b: In the absence of sufficient tryptophan to activate TRAP, TRAP does not bind to RNA, and the polymerase stalled at Stage 1 resumes transcription. The antiterminator then forms, preventing formation of the terminator. Transcription continues into the structural gene region of the trp suboperon (Gollnick et al. 2005). (Modified from Fig. 3 in Yanofsky 2004 and reprinted with permission from Elsevier © 2004.)
TRAP regulates translation of trpG/pabA of the folate operon
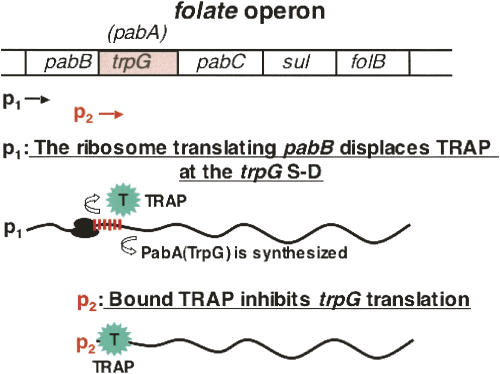
Synthesis of the polypeptide product of the single trp gene not in the trp suboperon, trpG/pabA, is also regulated by tryptophan-activated TRAP. However its regulation is principally translational (Fig. 10; Babitzke et al. 1994; Yakhnin et al. 2007). A series of XAG repeats overlap the Shine–Dalgarno sequence for trpG/pabA. TRAP binding at these XAG's repeats can inhibit initiation of translation of trpG/pabA. Since, as mentioned, the TrpG/PabA polypeptide serves as a component of two enzyme complexes, one required for p-aminobenzoate synthesis and the second for o-aminobenzoate synthesis, special regulatory features are needed to provide sufficient levels of the TrpG/PabA polypeptide to achieve these two objectives. Two promoters are in fact used to transcribe trpG/pabA. One promoter, p1, precedes pabB, the gene located before trpG/pabA in the folate operon; the second promoter, p2, immediately precedes trpG/pabA (Fig. 10; Yakhnin et al. 2007). Tryptophan-activated TRAP regulates translation of trpG/pabA, but principally when the transcript is produced from the p2 promoter (Fig. 10; Yakhnin et al. 2007). Transcripts initiated at the upstream promoter, p1, preceding pabB, allow ribosomes translating pabB to displace TRAP bound at the series of XAG repeats in the trpG/pabA Shine–Dalgarno region. This displacement reduces TRAP inhibition of trpG/pabA mRNA translation of these transcripts (Fig. 10; Yakhnin et al. 2007). On transcripts initiated at the internal promoter, p2, TRAP can bind and remain at the trpG Shine–Dalgarno sequence, effectively blocking trpG/pabA translation (Yakhnin et al. 2007). Thus, synthesis of the TrpG/PabA polypeptide is regulated differentially, depending on the organism's needs for tryptophan and/or folate.
FIGURE 10.
TRAP inhibition of initiation of translation of the trpG/pabA coding region. A series of XAG repeats—a TRAP-binding site—is located immediately preceding the trpG/pabA start codon (Gollnick et al. 2005). Two promoters, p1 and p2, are used to transcribe trpG/pabA, one located before pabB and the second before trpG/pabA (Yakhnin et al. 2007). Transcription from p1 provides a transcript on which ribosomes translating pabB can displace bound TRAP or prevent TRAP from binding, thereby allowing trpG/pabA to be translated. On transcripts initiated at promoter p2, bound tryptophan-activated TRAP masks the trpG/pabA Shine–Dalgarno sequence. This prevents ribosome binding and translation of this coding region. When TRAP is tryptophan-free, and inactive, translation of the trpG/pabA coding region on p2 transcripts can proceed (Yakhnin et al. 2007). The dual promoters allow the TrpG/PabA polypeptide to be synthesized whenever it is needed for either o-aminobenzoic acid or p-aminobenzoic acid synthesis.
How B. subtilis senses unavailability of charged tRNATrp as a regulatory signal
In E. coli charged tRNATrp availability is sensed during attempted translation of the two Trp codons of trpL, the 14-residue leader peptide coding region of the trp operon (Fig. 5). In Bacillus subtilis charged tRNATrp availability is sensed by the at operon, an operon distinct from the mtrB (TRAP) and trp operons (Sarsero et al. 2000). The product of the at operon (Fig. 11), protein AT (Anti-TRAP), is specifically designed to bind to tryptophan-activated TRAP. When AT is bound it prevents TRAP from binding to its target RNAs. This reduces transcription termination in the leader region of the trp suboperon (Valbuzzi and Yanofsky 2001).
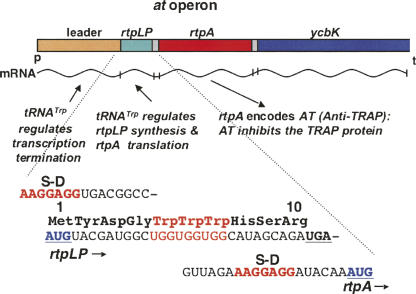
FIGURE 11.
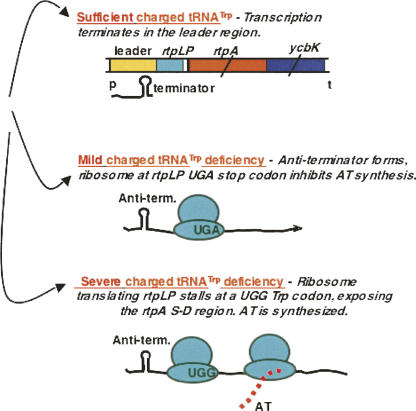
Organization and features of the at operon of B. subtilis. The at operon consists of a leader regulatory region, a 10-residue leader peptide coding region, rtpLP, and two structural genes, rtpA, encoding the AT protein, and ycbK, encoding what it is believed to be an efflux transport protein (Sarsero et al. 2000; Chen and Yanofsky 2004; Gollnick et al. 2005; Yakhnin et al. 2006a,b). Transcription of the at operon is regulated by the T box mechanism, in response to the accumulation of uncharged tRNATrp. Initiation of translation of rtpA, the structural gene for the AT protein, is also regulated by charged and uncharged tRNATrp. The 10-codon leader peptide coding region, rtpLP, immediately preceding the at coding region, contains three Trp codons. This transcript segment is used to regulate AT synthesis translationally in response to the accumulation of uncharged tRNATrp. Thus AT synthesis is regulated both transcriptionally and translationally, using different mechanisms, upon sensing the levels of charged and uncharged tRNATrp.
The at operon's leader region is designed to sense the accumulation of uncharged tRNATrp both transcriptionally and translationally, in regulating synthesis of the AT protein (Figs. 11, 12, 13; Chen and Yanofsky 2004). Transcriptional regulation is based on the T box RNA-based regulatory mechanism, a mechanism used in controlling expression of many genes of Gram-positive bacteria—particularly aminoacyl-tRNA synthetase genes (Henkin 2000; Grundy and Henkin 2004). The transcript of the at operon's leader region forms a typical T box structure, with this T box RNA designed to bind and respond to uncharged tRNATrp (Fig. 12; Sarsero et al. 2000). Whenever uncharged tRNATrp accumulates, it binds to the leader transcript of the at operon, stabilizing an antiterminator (Chen and Yanofsky 2004). This antiterminator prevents formation of the alternative, transcription terminator, hence transcription proceeds into the at operon's structural genes, including rtpA, the structural gene for the AT protein. Charged tRNATrp availability is also sensed translationally, by other features of the at operon's leader transcript. This sensing is performed during attempted translation of three consecutive tryptophan codons in a 10-residue leader peptide coding region, rtpLP, located in the RNA sequence immediately following the T box sequence of the at operon (Figs. 11, 13; Chen and Yanofsky 2004). The three tryptophan codons of rtpLP are located appropriately so that, whenever uncharged tRNATrp-mediated ribosome stalling occurs at any one of these codons, it exposes the AT Shine–Dalgarno sequence, allowing efficient initiation of AT synthesis (Fig. 13). However, when there is sufficient charged tRNATrp for the translating ribosome to complete synthesis of the at operon's leader peptide, this ribosome would reach the rtpLP stop codon, where it would block the rtpA Shine–Dalgarno sequence. This would inhibit initiation of AT synthesis, until the ribosome is released (Fig. 11). We suspect that ribosome release at this leader peptide stop codon is slow, since synthesis of AT is poor whenever cells have moderate levels of charged tRNATrp. The alternative events involved in expression of the at operon are summarized in Figure 13.
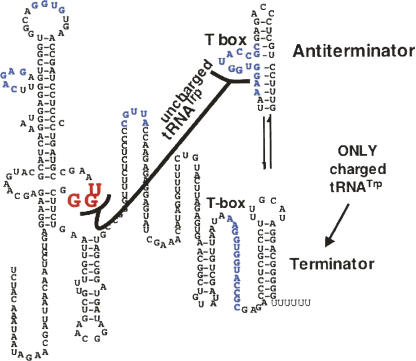
FIGURE 12.
Sequence and presumed alternative structures formed in the T box leader RNA segment of the transcript of the at operon of B. subtilis. The transcript of the at operon leader region has segments analogous to those of many T box sequences that recognize an uncharged tRNA as a regulatory signal (Grundy and Henkin 2004; Sarsero et al. 2000: Gutierrez-Preciado et al. 2005). When uncharged tRNATrp binds to the at operon T box leader RNA segment—by codon–anticodon pairing and by binding of its acceptor end to the T box sequence—the antiterminator structure is stabilized, preventing formation of the terminator. When the cellular tRNATrp is largely charged, it cannot bind to and stabilize the T box antiterminator, therefore the terminator forms, and transcription is terminated. Some critical nucleotides in the leader RNA are colored. (Modified from Fig. 3 in Sarsero et al. 2000 and reprinted with permission from the National Academy of Sciences, USA © 2000.)
FIGURE 13.
The alternative regulatory events involved in the synthesis of the AT protein. In cells with sufficient levels of charged tRNATrp, transcription of the at operon is terminated within its leader region (Gollnick et al. 2005). When there is a mild charged tRNATrp deficiency, uncharged tRNA will occasionally pair with the leader transcript T box sequence and permit transcription to continue into the operon. However, under these conditions, the ribosome translating rtpLP will reach its stop codon and inhibit initiation of translation of rtpA. Thus little or no AT protein will be produced. When there is a severe charged tRNATrp deficiency, the ribosome translating rtpLP will stall at one of its three Trp codons. This stalling will expose the rtpA Shine-Dalgarno region for efficient translation initiation, and the AT protein will be synthesized. (Modified from Fig. 4 in Yanofsky 2004 and reprinted with permission from Elsevier © 2004.)
The structure of the AT protein has been determined (Shevtsov et al. 2005), however the structure of the AT–TRAP complex is not yet known. Since bound AT prevents TRAP from binding to RNA, it is likely that AT recognizes the features of tryptophan-activated TRAP that are essential for RNA binding (Valbuzzi et al. 2002). Consistent with this conclusion, mutant TRAP proteins with single amino acid changes that render them incapable of binding trp leader RNA also prevent these proteins from binding the AT protein (Valbuzzi et al. 2002).
Other features of the DNA, RNA, and proteins of B. subtilis are known to contribute to the regulatory events that fine tune tryptophan biosynthesis. Thus, although E. coli and B. subtilis both recognize tryptophan and uncharged tRNATrp as signals in regulating their respective trp operon, the mechanisms they use to sense these signals are quite different (Yanofsky 2004). This realization supports the presumption stated at the beginning of this article—namely, that during evolution, numerous regulatory mechanisms were undoubtedly developed and tested, and some retained, depending on their effectiveness in allowing each organism to respond to its specific metabolic needs.
trp operon regulation in other Gram-positive bacteria
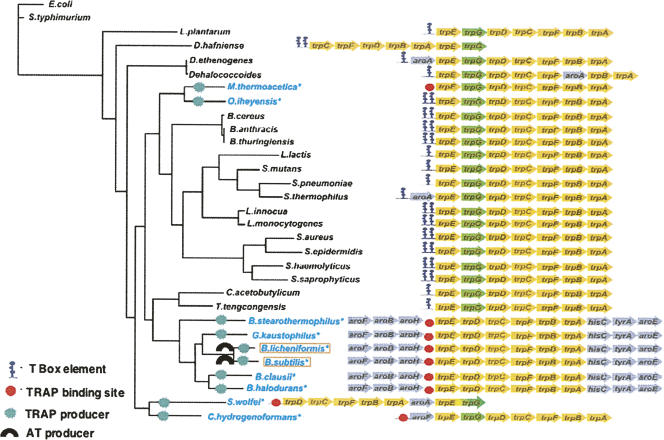
Genomic searches comparing the organization of the trp genes in the trp operon of other Gram-positive species have revealed that, although several have the same trp operon organization as B. subtilis, most do not (Fig. 14; Gutierrez-Preciado et al. 2005, 2007). In fact most of these species have separate trpG and pabA genes, with trpG located within their trp operon, which is separate, and not within an aro supraoperon (Gutierrez-Preciado et al. 2007). Many of these trp operons appear to be transcriptionally regulated by the T box mechanism, presumably responding to uncharged tRNATrp. In several species tandem T box elements precede the first gene of the operon. This arrangement would allow a multiplicative effect of regulation at each of these T box sites. Presumably each T box responds to uncharged tRNATrp, therefore it is not evident if these species do sense tryptophan as a transcription regulatory signal. Of the species that have an aro supraoperon organized identically to the aro supraoperon of B. subtilis, only B. licheniformis appears to produce an AT protein. How these other species sense uncharged tRNATrp, if they do, remains to be discovered.
FIGURE 14.
Organization of the trp operon of other Gram-positive bacteria that have the same or different trp gene arrangements and regulation as in B. subtilis. Several Gram-positive bacterial species have a trp suboperon which is organized similarly to the trp suboperon of B. subtilis (Gutierrez-Preciado et al. 2007). These species have a TRAP protein and there is a TRAP binding site immediately preceding trpE in the trp suboperon. Only one of these species, B. lichiniformis, is also known to form an AT protein (Gutierrez-Preciado et al. 2007). In many other Gram-positive species the trp operon is organized differently, with a trpG within the trp operon. Most of these trp operons do not contain genes of the other aromatic amino acid pathways. In many of these species, all seven trp genes are present in the same operon, and the T box mechanism appears to be the predominant form of regulation of trp operon transcription. (Modified from Fig. 2 in Gutiérrez-Preciado et al. 2007 and reprinted with permission from Elsevier © 2007.)
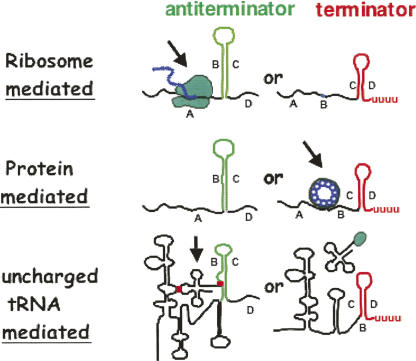
Thus three different transcription attenuation mechanisms are known to regulate trp operon transcription in bacteria (Fig. 15; Henkin and Yanofsky 2002; Merino and Yanofsky 2005). Stabilization of either of two alternative RNA structures is the principal objective of each mechanism. As illustrated in Figure 15, ribosome-, protein-, or tRNA-mediated selection between these alternative RNA hairpin structures is the strategy allowing transcription regulation. These mechanisms are also used in regulating transcription of operons concerned with other functions. There are many translational attenuation mechanisms that are employed in regulating translation of specific coding regions (Lovett and Rogers 1996).
FIGURE 15.
Some common transcription attenuation regulatory mechanisms used in regulating trp operon expression in different bacterial species. Typical leader transcript segments are illustrated that can form alternative RNA hairpin structures, one of which is an intrinsic transcription terminator. The examples shown involve ribosome-mediated, protein-mediated, and tRNA-mediated regulation of formation of either of two alternative RNA hairpin structures. Each of these mechanisms is common in bacteria (Henkin and Yanofsky 2002; Merino and Yanofsky 2005). (Modified from Fig. 1 in Merino and Yanofsky 2005 and reprinted with permission from Elsevier © 2005.)
Regulation of the degradative tryptophanase (tna) operon of E. coli
Many organisms have the ability to degrade tryptophan and use the degradation products for important purposes. Thus, in most eukaryotes tryptophan is the principal precursor of niacin and its derivatives, and in plants tryptophan is a precursor of auxin. Most bacteria do not possess either of these degradative pathways. But many bacteria can degrade tryptophan and use the products for various purposes. In E. coli, for example, indole generated from tryptophan has been shown to serve as a signaling compound in quorum sensing, biofilm formation, and expression of multidrug exporter genes (Wang et al. 2001; Winzer et al. 2002; Ren et al. 2004; Hirakawa et al. 2005). In addition, the tryptophanase operon—and presumably some product of tryptophan degradation—has been shown to be required for pathogenesis by certain strains of E. coli (Anyanful et al. 2005).
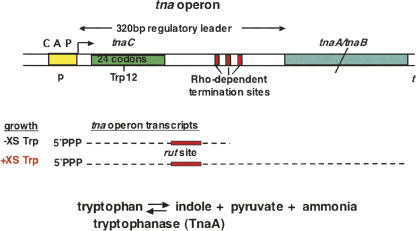
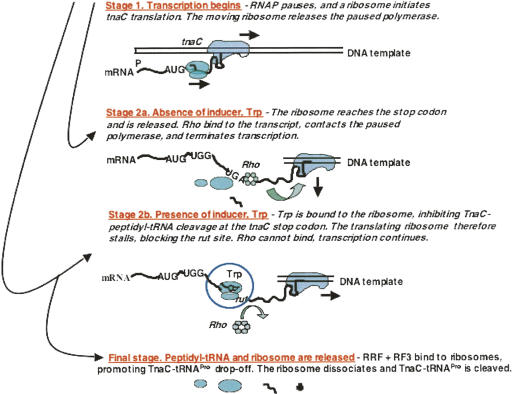
Our studies have been focused on determining the mechanism of tryptophan induction of expression of the tryptophanase (tna) degradative operon of E. coli. We have shown that the leader transcript of this operon contains three segments essential for regulation of tna operon expression: a coding sequence for a 24-residue leader peptide, TnaC; a RNA rut binding site for the Rho termination factor; and RNA pause sites required for Rho-dependent transcription termination (Fig. 16; Deeley and Yanofsky 1981; Stewart and Yanofsky 1985; Gong et al. 2001; Gong and Yanofsky 2002; Cruz-Vera et al. 2005, 2006, 2007). These sites all precede the two structural genes of the operon, tnaA, encoding tryptophanase, the degradative enzyme, and tnaB, encoding a tryptophan permease. The tna operon leader peptide, TnaC, has been shown to regulate Rho action by instructing the translating ribosome to bind tryptophan, inhibiting TnaC–tRNAPro cleavage (Fig. 17; Cruz-Vera et al. 2005, 2007). Inhibition of peptidyl-tRNA cleavage causes the ribosome translating tnaC to stall at the tnaC stop codon. This stalled ribosome blocks the Rho Factor binding site—the rut site—preventing the Rho factor from binding to the transcript and terminating transcription (Fig. 17). When Rho cannot bind and act, the polymerase stalled downstream in the leader region resumes transcription, proceeding into the structural genes of the operon (Gong and Yanofsky 2002). If cells do not contain inducing levels of tryptophan, the ribosome translating tnaC completes TnaC synthesis, the TnaC–peptidyl–tRNA is cleaved, and the translating ribosome is released. The rut site in leader RNA is then free to bind the Rho factor, allowing Rho to bind and contact the polymerase paused in the operon's leader region, terminating transcription. E. coli's tna operon regulatory region resembles the tna operon regulatory region of several other bacterial species.
FIGURE 16.
Features of the leader regulatory region of the tna operon of E. coli. Transcription of the structural gene region of the tna operon is regulated by two mechanisms: catabolite repression, at the tna operon's promoter, and Rho factor-dependent transcription termination, in the operon's leader region (Cruz-Vera et al. 2006). The CAP-dependent promoter is similar to other promoters subject to catabolite repression. Tryptophan induction of tna operon expression is based on relief from Rho-dependent transcription termination in the operon's leader region. The leader transcript segment contains a coding region for a 24-residue peptide, TnaC, that has a crucial Trp residue at position 12. Synthesis of this peptide in the presence of inducer, tryptophan, prevents cleavage of TnaC–peptidyl–tRNA at the tnaC stop codon. The resulting stalled ribosome blocks Rho factor from binding at the rut site, preventing transcription termination. Transcription then continues into the two structural genes of the operon.
FIGURE 17.
Stages in tryptophan induction of tna operon expression in E. coli. When E. coli is grown in a medium lacking a catabolite-repressing carbon source, the CAP protein is activated, and transcription of the tna operon is initiated (Gong et al. 2001; Gong and Yanofsky 2002). Stage 1: An RNA polymerase complex transcribes the initial region of the operon and pauses following synthesis of an RNA segment that forms a pause structure. While the polymerase is paused, a ribosome binds to the nascent tna transcript and initiates synthesis of TnaC. This translating ribosome then releases the paused polymerase, hence both transcription and translation then proceed. Depending on the cellular concentration of free tryptophan, either Stage 2a or Stage 2b occurs. Stage 2a: When free tryptophan is absent, or present at a low concentration, the translating ribosome completes synthesis of TnaC and dissociates from the transcript. Rho factor then binds to the leader RNA. Meanwhile the transcribing RNA polymerase that had transcribed much of the leader region has paused at one of several pause sites located near the end of the leader region. When Rho factor can bind, and act, it contacts this paused polymerase and terminates transcription. Stage 2b: When free tryptophan is plentiful, it binds to ribosomes in the process of TnaC–tRNAPro synthesis and inhibits TnaC–tRNAPro cleavage. These ribosomes remain stalled at the tnaC stop codon, where they mask the mRNA rut site. Rho factor therefore cannot bind, and RNA polymerase molecules paused near the end of leader segment of the tna operon transcript resume transcription into the operon's two structural genes. TnaC–tRNAPro and the stalled ribosome are then released by RRF and RF3 action, the peptidyl–tRNA is hydrolyzed, and the ribosomal subunits are reused for additional protein synthesis (Gong et al. 2007).
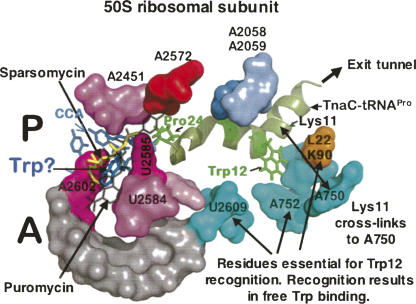
Our recent studies have been concerned with determining how a crucial amino acid in the nascent TnaC–peptidyl–tRNA, Trp12, is recognized, and how this recognition creates a free-tryptophan binding/inhibition site in the translating ribosome (Fig. 18). The approximate location of the crucial Trp12 of TnaC–tRNAPro in the 50S ribosomal subunit was determined by showing that Lys11 of TnaC–tRNAPro can be cross-linked to nucleotide A750 of 23S rRNA, and that Trp12 of TnaC–tRNAPro protects 23S rRNA nucleotide A788 from methylation (Fig. 18; Cruz-Vera et al. 2005). In addition, several 23S rRNA nucleotides and one residue of ribosomal protein L22 have been identified that are essential for tryptophan induction; these reside near the presumed site occupied by Trp12 of TnaC–tRNA in the ribosome exit tunnel (Fig. 18). The likely locations of these residues, U2609 and A752 of 23S rRNA, and K90 of protein L22, are shown in Figure 18. We have also observed that these ribosomal residues appear to act by creating a free-tryptophan binding site—presumably in the A site of the ribosome—at which bound tryptophan inhibits peptidyl transferase cleavage of TnaC–tRNAPro. Thus, in experiments with wild-type ribosomes bearing TnaC–tRNAPro, it was found that free tryptophan inhibits puromycin and sparsomycin binding at the ribosomal A site (Cruz-Vera et al. 2007). However, with mutant ribosomes bearing any of the three residue changes mentioned that prevent induction, free tryptophan no longer protects the ribosome from puromycin or sparsomycin action (Fig. 18; Cruz-Vera et al. 2007). Presumably residues in the ribosomal exit tunnel recognize features of wild-type TnaC–tRNAPro, particularly Trp12, and this creates a free-tryptophan binding site in the wild-type translating ribosome at which bound tryptophan prevents TnaC–tRNAPro cleavage. Tryptophan binding was not detected in ribosomes with any of the three changes mentioned (Cruz-Vera et al. 2007).
FIGURE 18.
Some peptide-ribosomal interactions believed to be responsible for tryptophan induction of tna operon expression. A region of the 50S ribosomal subunit is shown containing bound sparsomycin, puromycin, and presumably tryptophan, in the A site of the translating ribosome (Porse et al. 1999; Hansen et al. 2003; Cruz-Vera et al. 2005, 2007). Also shown is the probable location of TnaC–tRNAPro in the ribosome exit tunnel; identified are likely locations of residues Trp12 and Pro24, residues essential for induction. Residues of 23RNA and of ribosomal protein L22 in the vicinity of Trp12 of TnaC–tRNAPro, some of which have been shown to be required for tryptophan binding and induction, are also labeled. The approximate location of Trp12 of TnaC–tRNAPro has been determined by showing that Lys11 of TnaC–tRNAPro can be cross-linked to nucleotide A750 (Cruz-Vera et al. 2005). This figure is based on the structure of the E. coli ribosome, determined by Schuwirth et al. (2005). (Modified from Fig. 1 in Cruz-Vera et al. 2007 and reprinted with permission from the American Society for Microbiology © 2007.)
We also wondered whether cells have some means of recovering from tryptophan induction of tna operon expression. Are they capable of rescuing ribosomes from stalled TnaC–tRNAPro–ribosome complexes so these ribosomes may participate in new rounds of protein synthesis? Tryptophan induction in wild-type E. coli does not inhibit cell growth. However, when tnaC is overexpressed from a multicopy plasmid, growth is inhibited. This inhibition was shown to be due to the depletion of free tRNA2 Pro, the tRNA attached to TnaC in stalled ribosome complexes (Gong et al. 2006). Further investigation of this phenomenon revealed that the normal half-life of TnaC–tRNAPro under tryptophan induction conditions is about 8 min versus < 1 min in the absence of added tryptophan (Gong et al. 2007). Since TnaC–tRNAPro has a half-life of 8 min, there must be some mechanism of ribosome recycling and TnaC–peptidyl–tRNA cleavage under these extraordinary tryptophan-induction conditions. This mechanism was explored and it was shown that ribosome recycling factor, RRF, plus ribosome release factor 3, RF3, bind to stalled ribosomes containing TnaC–tRNAPro and free tryptophan, and promote TnaC–tRNAPro drop-off from the ribosomes (Fig. 17; Gong et al. 2007). The drop-off TnaC–tRNAPro is then cleaved by peptidyl–tRNA hydrolase. Once the peptidyl–tRNA is released from the ribosome, the ribosome is released and its subunits become available for new rounds of protein synthesis. Thus the system is designed so that tryptophan induction need not have a negative impact on the organism's capacity for new protein synthesis.
CONCLUSIONS
RNA-based regulation was—and is—an obvious option during the course of evolution. We now appreciate that these mechanisms were widely exploited, using a variety of strategies. In addition to the transcription attenuation mechanisms described in this article, there are other regulatory processes in which RNAi or other RNAs play crucial regulatory roles. The tna operon example illustrates an additional regulatory mechanism whereby the RNA encoding a peptide is used to instruct a translating ribosome to resist translation termination. These examples illustrate the diversity of the RNA-based regulatory strategies that organisms use in controlling gene expression. They reveal how essential regulation of gene expression must have been to each organism during the course of its evolution.
ACKNOWLEDGMENTS
I thank Marvin Wickens, Paul Babitzke, Paul Gollnick, Enrique Merino, Ana Gutiérrez-Preciado, and Luis R. Cruz-Vera for their helpful comments on this article. The following individuals either worked in my laboratory or collaborated with us in performing the studies I have described: P. Babitzke, R. Bauerle, D. Bear, G. Bennett, R. Berka, K. Bertrand, K. Brown, J. Carey, G.-N. Chen, K. Cho, I. Crawford, L.R. Cruz-Vera, X. Cui, A. Das, M. Deeley, R. Fisher, K. Gish, P. Gollnick, F. Gong, M. Gong, A. Gutiérrez-Preciado, T. Henkin, D. Henner, V. Horn, B. Hurlburt, K. Ito, E. Jackson, O. Jardetzky, R. Jensen, A. Joachimiak, A. Kamath, R. Kelley, A. Khodursky, K. Kirschner, L. Klig, R. Kolter, V. Konan, L. Korn, M. Kuroda, A. Lee, F. Lee, B. Nichols, T. Platt, R. Landick, E. Merino, E. Miles, G. Miozzari, Y. Nakamura, A. New, D. Oxender, S. Rajagopal, J. Roesser, J. Sarsero, P. Sigler, C. Squires, C.L. Squires, G. Stauffer, V. Stewart, I. Stroynowski, A. Valbuzzi, M. van Cleemput, M. Winkler, W.J. Yang, M.C. Yee, H. Zalkin, and G. Zurawski. I apologize to anyone I have overlooked. Support for the research conducted in my laboratory was provided by the National Institutes of Health, National Science Foundation, American Cancer Society, and the American Heart Association.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.620507.
REFERENCES
- Antson, A.A., Dodson, E.J., Greaves, R.B., Chen, X.-P., Gollnick, P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- Anyanful, A., Dolan-Livengood, J.M., Lewis, T., Sheth, S., DeZalia, M.N., Sherman, M.A., Kalman, L.V., Benian, G.M., Kalman, D. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase. Mol. Microbiol. 2005;57:988–1007. doi: 10.1111/j.1365-2958.2005.04739.x. [DOI] [PubMed] [Google Scholar]
- Babitzke, P. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: The Bacillus subtilis TRAP protein. Curr. Opin. Microbiol. 2004;7:132–139. doi: 10.1016/j.mib.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Babitzke, P., Stults, J.T., Shire, S.J., Yanofsky, C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in trpEDCFBA and trpG transcripts. J. Biol. Chem. 1994;269:16597–16604. [PubMed] [Google Scholar]
- Chen, G.N., Yanofsky, C. Features of a leader peptide coding region that regulate translation initiation for the anti-TRAP protein of Bacillus subtilis . Mol. Cell. 2004;13:703–711. doi: 10.1016/s1097-2765(04)00085-1. [DOI] [PubMed] [Google Scholar]
- Cruz-Vera, L.R., Rajogopal, S., Squires, C., Yanofsky, C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Cruz-Vera, L.R., Gong, M., Yanofsky, C. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. 2006;103:3598–3603. doi: 10.1073/pnas.0600082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Vera, L.R., New, A., Squires, C., Yanofsky, C. Ribosomal features essential for tna operon induction: Tryptophan binding at the peptidyl transferase center. J. Bacteriol. 2007;189:3140–3146. doi: 10.1128/JB.01869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley, M.C., Yanofsky, C. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J. Bacteriol. 1981;147:787–796. doi: 10.1128/jb.147.3.787-796.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H., Babitzke, P. trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis . J. Biol. Chem. 1998;273:20494–20503. doi: 10.1074/jbc.273.32.20494. [DOI] [PubMed] [Google Scholar]
- Gollnick, P., Babitzke, P., Antson, A., Yanofsky, C. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis . Annu. Rev. Genet. 2005;39:47–68. doi: 10.1146/annurev.genet.39.073003.093745. [DOI] [PubMed] [Google Scholar]
- Gong, F., Yanofsky, C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- Gong, F., Ito, K., Nakamura, Y., Yanofsky, C. The mechanism of tryptophan induction of tryptophanase operon expression: Tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNAPro . Proc. Natl. Acad. Sci. 2001;98:8997–9001. doi: 10.1073/pnas.171299298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, M., Gong, F., Yanofsky, C. Overexpression of tnaC of Escherichia coli inhibits growth by depleting tRNA2 Pro availability. J. Bacteriol. 2006;188:1892–1898. doi: 10.1128/JB.188.5.1892-1898.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, M., Cruz-Vera, L.R., Yanofsky, C. RRF and RF3 action promotes TnaC-peptidyl-tRNA dropoff and relieves ribosome stalling during tryptophan induction of tna operon expression in Escherichia coli . J. Bacteriol. 2007;189:3147–3155. doi: 10.1128/JB.01868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, F.J., Henkin, T.M. Regulation of gene expression by effectors that bind to RNA. Curr. Opin. Microbiol. 2004;7:126–131. doi: 10.1016/j.mib.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Gryk, M.R., Jardetzky, O., Klig, L.S., Yanofsky, C. Flexibility of DNA binding domain of trp repressor required for recognition of different operator sequences. Protein Sci. 1996;5:1195–1197. doi: 10.1002/pro.5560050624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Preciado, A., Jensen, R.A., Yanofsky, C., Merino, E. New Insights into regulation of the tryptophan biosynthetic operon in gram positive bacteria. Trends Genet. 2005;21:432–436. doi: 10.1016/j.tig.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Preciado, A., Yanofsky, C., Merino, E. Comparison of tryptophan biosynthetic operon regulation in different gram-positive bacterial species. Trends Genet. 2007 doi: 10.1016/j.tig.2007.05.005. (in press). [DOI] [PubMed] [Google Scholar]
- Hansen, J.L., Moore, P.B., Steitz, T.A. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- Henkin, T.M. Transcription termination control in bacteria. Curr. Opin. Microbiol. 2000;3:149–153. doi: 10.1016/s1369-5274(00)00067-9. [DOI] [PubMed] [Google Scholar]
- Henkin, T.M., Yanofsky, C. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- Henner, D., Yanofsky, C. Biosynthesis of aromatic amino acids. In Bacillus subtilis and other gram-positive bacteria: Biochemistry, physiology, and molecular genetics . In: Sonenshein A.L., et al., editors. American Society for Microbiology; Washington D.C: 1993. pp. 269–280. [Google Scholar]
- Hirakawa, H., Inazumi, Y., Masaki, T., Hirata, T., Yamaguchi, A. Indole induces the expression of multidrug exporter genes in Escherichia coli . Mol. Microbiol. 2005;55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- Joachimiak, A., Zhang, R.G. Crystallographic studies on the E. coli trp aporepressor. Acta Biochim. Pol. 1989;36:195–203. [PubMed] [Google Scholar]
- Landick, R., Yanofsky, C. Transcription attenuation. In Escherichia coli and Salmonella typhimurium: Cellular and molecular biology . In: Neidhardt F.C., et al., editors. American Society for Microbiology; Washington D.C: 1987. pp. 1276–1301. [Google Scholar]
- Lawson, C.L., Benoff, B., Berger, T., Berman, H.M., Carey, J. E. coli trp repressor forms a domain-swapped array in aqueous alcohol. Structure. 2004;12:1099–1108. doi: 10.1016/j.str.2004.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett, P.S., Rogers, E.J. Ribosome regulation by the nascent peptide. Microbiol. Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy, C., Manfredo, A., Wendt, A., Gollnick, P., Foster, M. TROSY-NMR studies of the 91 kDa TRAP protein reveal allosteric control of a gene regulatory protein by ligand-altered flexibility. J. Mol. Biol. 2002;323:463–473. doi: 10.1016/s0022-2836(02)00940-3. [DOI] [PubMed] [Google Scholar]
- Merino, E., Yanofsky, C. Transcription attenuation: A highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Merino, E., Babitzke, P., Yanofsky, C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational and well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 1995;177:6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., Schevitz, R.W., Zhang, R.G., Lawson, C.L., Joachimiak, A., Marmorstein, R.O., Luisi, B.F., Sigler, P.B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Porse, B.T., Kirillov, S.V., Awayez, M.J., Ottenheijm, H.C., Garrett, R.A. Direct crosslinking of the antitumor antibiotic sparsomycin, and its derivatives, to A2602 in the peptidyl transferase center of 23S-like rRNA within ribosome-tRNA complexes. Proc. Natl. Acad. Sci. 1999;96:9003–9008. doi: 10.1073/pnas.96.16.9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D., Bedzyk, L.A., Ye, R.W., Thomas, S.M., Wood, T.K. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli . Appl. Environ. Microbiol. 2004;70:2038–2043. doi: 10.1128/AEM.70.4.2038-2043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarsero, J.P., Merino, E., Yanofsky, C. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. Proc. Natl. Acad. Sci. 2000;97:2656–2661. doi: 10.1073/pnas.050578997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila-Sanjurjo, A., Holton, J.M., Cate, J.H. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Shakked, Z., Guzikevich-Guerstein, G., Frolow, F., Rabinovich, D., Joachimiak, A., Sigler, P.B. Determinants of repressor/operator recognition from the structure of the trp operator binding site. Nature. 1994;368:469–473. doi: 10.1038/368469a0. [DOI] [PubMed] [Google Scholar]
- Shevtsov, M.B., Chen, Y., Gollnick, P., Antson, A.A. Crystal structure of Bacillus subtilis anti-TRAP protein, an antagonist of TRAP/RNA interaction. Proc. Natl. Acad. Sci. 2005;102:17600–17605. doi: 10.1073/pnas.0508728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, V., Yanofsky, C. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 1985;164:731–740. doi: 10.1128/jb.164.2.731-740.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valbuzzi, A., Yanofsky, C. Inhibition of the B. subtilis regulatory protein TRAP by the TRAP-inhibitory protein, AT. Science. 2001;293:2057–2059. doi: 10.1126/science.1062187. [DOI] [PubMed] [Google Scholar]
- Valbuzzi, A., Gollnick, P., Babitzke, P., Yanofsky, C. The anti-trp RNA-binding attenuation protein (anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J. Biol. Chem. 2002;277:10608–10613. doi: 10.1074/jbc.M111813200. [DOI] [PubMed] [Google Scholar]
- Wang, D., Ding, X., Rather, P.N. Indole can act as an extracellular signal in Escherichia coli . J. Bacteriol. 2001;183:4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer, K., Hardle, K.R., Williams, P. Bacterial cell-to-cell communication: Sorry, can't talk now—Gone to lunch! Curr. Opin. Microbiol. 2002;5:216–222. doi: 10.1016/s1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- Yakhnin, H., Yakhnin, A.V., Babitzke, P. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding. Mol. Microbiol. 2006a;61:1252–1266. doi: 10.1111/j.1365-2958.2006.05278.x. [DOI] [PubMed] [Google Scholar]
- Yakhnin, A.V., Yakhnin, H., Babitzke, P. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction. Mol. Cell. 2006b;24:547–557. doi: 10.1016/j.molcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Yakhnin, H., Yakhnin, A.V., Babitzke, P. Translational control of trpG from transcripts originating from the folate operon promoter of Bacillus subtilis is influenced by translation-mediated displacement of bound TRAP, while translational control of transcripts originating from a newly identified trpG promoter is not. J. Bacteriol. 2007;189:872–879. doi: 10.1128/JB.01398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, C. Attenuation in the control of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Yanofsky, C. Transcription attenuation: Once viewed as a novel regulatory strategy. J. Bacteriol. 2000;182:1–8. doi: 10.1128/jb.182.1.1-8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, C. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli vs. B. subtilis. Trends Genet. 2004;20:367–374. doi: 10.1016/j.tig.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Yanofsky, C., Crawford, I.P. The tryptophan operon. In Escherichia coli and Salmonella typhimurium: Cellular and molecular biology . In: Neidhardt F.C., et al., editors. American Society for Microbiology; Washington D.C: 1987. pp. 1452–1472. [Google Scholar]
- Yanofsky, C., Miles, E.W., Kirschner, K., Bauerle, R. TRP operon. In The encyclopedia of molecular biology . In: Creighton T.C., editor. Vol. 4. John Wiley & Sons; New York: 1999. pp. 2676–2689. [Google Scholar]