Abstract
Polypyrimidine tract binding protein (PTB) acts as a regulatory repressor of a large number of alternatively spliced exons, often requiring multiple binding sites in order to repress splicing. In one case, cooperative binding of PTB has been shown to accompany repression. The SM exon of the α-actinin pre-mRNA is also repressed by PTB, leading to inclusion of the alternative upstream NM exon. The SM exon has a distant branch point located 386 nt upstream of the exon with an adjacent 26 nucleotide pyrimidine tract. Here we have analyzed PTB binding to the NM and SM exon region of the α-actinin pre-mRNA. We find that three regions of the intron bind PTB, including the 3′ end of the polypyrimidine tract (PPT) and two additional regions between the PPT and the SM exon. The downstream PTB binding sites are essential for full repression and promote binding of PTB to the PPT with a consequent reduction in U2AF65 binding. Our results are consistent with a repressive mechanism in which cooperative binding of PTB to the PPT competes with binding of U2AF65, thereby specifically blocking splicing of the SM exon.
Keywords: PTB, alternative splicing, hnRNP, pre-mRNA splicing
INTRODUCTION
PTB is an RNA binding protein that acts as a repressive regulator of alternative splicing (for review, see Wagner and Garcia-Blanco 2001; Spellman and Smith 2006), as well as plays roles in pre-mRNA 3′-end processing (Castelo-Branco et al. 2004), mRNA translation (Kaminski et al. 1995; Mitchell et al. 2005), stability (Hamilton et al. 2003), and localization (Cote et al. 1999). Structurally, PTB consists of four RNA recognition motif (RRM) domains, and its optimal binding site determined by SELEX comprises motifs such as UCUU in a generally pyrimidine-rich context (Perez et al. 1997a). Motifs identical or similar to this have been found to act as splicing silencer elements in a wide range of pre-mRNA substrates and are often located adjacent to exons that are included with neuronal or muscle specificity (Mulligan et al. 1992; Ashiya and Grabowski 1997; Chan and Black 1997; Perez et al. 1997a; Gooding et al. 1998; Southby et al. 1999; Zhang et al. 1999; Wagner and Garcia-Blanco 2002; Shen et al. 2004; Izquierdo et al. 2005; Sauliere et al. 2006). Structure–function analysis initially indicated that RRMs 3 and 4 are principally responsible for RNA binding, while the N-terminal half of the protein might be responsible for PTB dimerization (Perez et al. 1997b; Oh et al. 1998). However, this simple division of labor has been challenged in two ways. First, pure recombinant PTB is monomeric in solution (Simpson et al. 2004; Amir-Ahmady et al. 2005; Monie et al. 2005). Second, all four of the RRMs can bind specifically to RNAs containing optimal CUCUCU motifs (Simpson et al. 2004; Oberstrass et al. 2005).
Early models for PTB splicing repressor activity suggested that it bound to the 3′ splice site polypyrimidine tract (PPT) in competition with splicing factor U2AF65 (Lin and Patton 1995; Singh et al. 1995). Binding of the two factors upstream of rat β-tropomyosin exon 6B was subsequently found not to be mutually exclusive (Grossman et al. 1998), although more recent analysis of chicken β-tropomyosin indicated competitive binding of PTB and U2AF65 (Sauliere et al. 2006). Repression of a GABA(A) receptor γ2 exon was shown to occur via prevention of U2 snRNP binding (Ashiya and Grabowski 1997). There are two reported cases of PTB-regulated exons with a single PTB binding silencer element located in the exon (Shen et al. 2004; Izquierdo et al. 2005). However, the vast majority of PTB-repressed exons have multiple associated PTB-binding elements (Mulligan et al. 1992; Ashiya and Grabowski 1997; Chan and Black 1997; Perez et al. 1997a; Gooding et al. 1998; Southby et al. 1999; Zhang et al. 1999; Wagner and Garcia-Blanco 2002; Sauliere et al. 2006). PTB can bind cooperatively to RNAs with multiple binding sites (Chou et al. 2000), and structural analysis of the RRM domains of PTB in complex with RNA has suggested how cooperative binding could be achieved without the necessity for direct PTB–PTB interactions (Oberstrass et al. 2005). Two general models have been proposed for PTB silencing via multiple binding sites. “Zones of repression” are proposed to be induced by cooperative binding of PTB, either along the RNA or by looping of RNA between high affinity sites (Wagner and Garcia-Blanco 2001). In many cases, one or more of the PTB binding sites are in the PPT, and direct binding competition with U2AF65 could occur within a zone of repression. However, in some cases PTB represses exons without any high affinity specific sites in the PPT (Shen et al. 2004; Amir-Ahmady et al. 2005; Izquierdo et al. 2005). PTB-mediated repression has been conferred by artificial tethering of PTB-MS2 coat protein fusions using MS2 binding sites in place of natural PTB binding sites (Wagner and Garcia-Blanco 2002; Robinson and Smith 2006). Deletion analysis showed that the RRM2 domain in combination with the following inter-RRM linker is sufficient to act as a repressor domain when recruited via MS2 downstream from α-TM exon 3. This repression was still dependent upon PTB binding sites upstream of the exon, leaving open the possibility that the tethered PTB repressor domain might interact with PTB bound upstream of the exon (Robinson and Smith 2006).
To date, the best demonstration of PTB-mediated repression involving cooperative binding is provided by the N1 exon of the c-src gene. N1 exon splicing is repressed in nonneuronal cells by PTB binding to sites flanking the exon (Chan and Black 1997; Chou et al. 2000). Mutations in the upstream sites that impair PTB-mediated repression also reduce binding to the downstream sites and vice versa (Chou et al. 2000). While the upstream PTB binding sites are embedded within the PPT, PTB repression can be maintained when they are removed further 5′ of the branch point (Amir-Ahmady et al. 2005). Strikingly, PTB appears to repress the enhancing effect of U1 snRNP bound at the N1 5′ splice site upon U2AF65 binding at the PPT of the downstream constitutive exon 4 (Sharma et al. 2005). Similar findings in the Fas system indicate that PTB bound at an exon splicing silencer inhibits cross-exon interactions between U1 snRNP and U2AF65 (Izquierdo et al. 2005).
We have sought to address the possibility of repression by cooperative PTB binding in a second model system with a different arrangement of PTB binding sites. The SM exon of the α-actinin gene is skipped in most cell types in favor of the usually mutually exclusive NM exon (Waites et al. 1992). In smooth muscle cells, the SM exon is used in place of NM, while in adult brain the predominant isoform contains both the NM and SM exons (Kremerskothen et al. 2002). The SM exon has a distant branch point, 386 nt upstream of the exon, with an adjacent 26 nucleotide (nt) pyrimidine tract (Southby et al. 1999). Transcripts containing the SM exon and associated splice site elements do not splice in HeLa nuclear extract, but splicing is activated by removal of repressor sequences between the exon and its distant branch point, or by depletion of PTB (Southby et al. 1999). This suggests that PTB might repress SM splicing by binding to sites between the exon and its upstream branch point. The nine UCUU motifs within this region represent potential PTB binding sites, although the pyrimidine content of the whole region is >70% and there are multiple other potential PTB binding motifs. In contrast to c-src and other systems, the PTB sites do not flank the repressed exon. Here we describe experiments aimed at delineating the intronic regions upstream of SM to which PTB binds, and analyzing how PTB binding to these sites might affect splicing. In addition to the PPT, we identify two additional regions between the PPT and SM exon that bind PTB. We show that these downstream sites promote binding of PTB at the PPT sites and that this correlates with a decrease in cross-linking of U2AF65 and repression of splicing.
RESULTS
PTB binds to discrete intronic regions
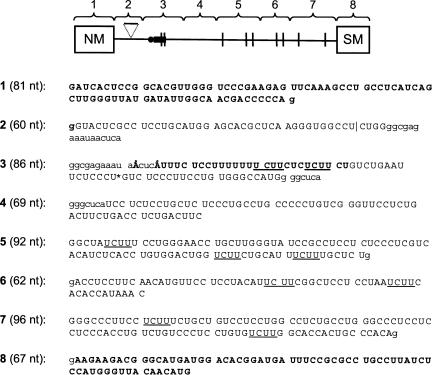
In order to carry out a detailed analysis of PTB interactions with the SM exon regulatory region (Southby et al. 1999), we synthesized eight short (60–96 nt), contiguous RNA fragments encompassing the NM and SM exons and the intron between them (Fig. 1). Each exon constituted an individual fragment (1 and 8, respectively), and a further fragment (3) contained the branch point and PPT. Boundaries were selected so as not to disrupt UCUU or other pyrimidine motifs and such that candidate PTB binding sequences were not located at the extreme termini of a fragment. In some cases a degree of overlap between fragments was necessary, although this was restricted to a minimum and did not duplicate any putative regulatory elements.
FIGURE 1.
Organization and nucleotide sequences of the α-actinin NM-SM PCR fragments. Schematic representation of NM-SM fragments. The branch point is represented by the black circle and the polypyrimidine tract by the black rectangle. UCUU motifs are indicated by vertical lines; the 30-nt spacer element in fragment 2, by the triangle. Regions of overlap between adjacent fragments are identified by lowercase letters. The NM and SM exons (fragments 1 and 8, respectively) and the polypyrimidine tract are indicated in bold. The two branch points are indicated as Å. UCUU motifs are underlined. Fragment lengths are indicated in parentheses. The boundary between the 5′ and 3′ portions of the ligated patch labeled NM-∇-SM transcript (Fig. 6) is indicated by the asterisk within fragment 3.
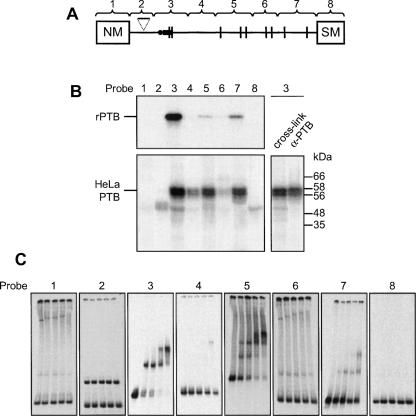
Interactions of recombinant PTB1 and endogenous HeLa nuclear extract proteins with NM-SM fragments 1–8 were first analyzed in UV cross-linking assays (Fig. 2B). The identity of the characteristic HeLa PTB doublet was verified by immunoprecipitation of proteins cross-linking to fragment 3. The cross-linking profiles of the recombinant and endogenous HeLa PTBs were similar, and no distinction was apparent between the behavior of the HeLa PTB1 and PTB4 isoforms, consistent with previous in vivo studies (Wollerton et al. 2001). PTB cross-linked selectively to a subset of RNA fragments. The most intense cross-linking signal occurred consistently with fragment 3, containing the PPT and branch points. Fragments 5 and 7 also showed a high degree of cross-linking, while fragment 4 cross-linked only weakly. Fragments 1, 2, 6, and 8 all cross-linked very weakly or not at all. In order to rule out the possibility that the relative intensity of PTB cross-linking with the RNA fragments was a function merely of transcript length or radiolabeled U content, the shortest fragment (2) was expanded to 90 nt by the insertion of the 30-nt random sequence spacer required for activation of splicing (Fig. 1; Southby et al. 1999). The expanded probe, containing five additional U residues, showed no increase in PTB cross-linking relative to the original (data not shown). Further evidence for the sequence specificity of PTB cross-linking is the observation that point mutations within probe 3 that disrupt the UCUU elements but increase the radiolabeled U content caused a reduction in the PTB signal (Fig. 5B).
FIGURE 2.
PTB binds selectively to specific NM-SM transcript fragments. (A) Schematic representation of RNA probes 1–8. (B) Cross-linking of recombinant 6xHis tagged PTB (upper panel) or endogenous HeLa nuclear extract proteins (lower panel) to NM-SM fragments 1–8. [32P]-labeled NM-SM transcripts were incubated with recombinant PTB or HeLa nuclear extract for 25 min. Following UV cross-linking and RNase digestion, proteins were analyzed by SDS-PAGE and autoradiography. Endogenous nuclear extract proteins cross-linked to NM-SM fragment 3 were immunoprecipitated with α-PTB antibody and analyzed alongside a total cross-linking sample (lower right panel). The two PTB isoforms are visible. (C) Electrophoretic mobility shift assay with [32P]-labeled probes 1–8 and recombinant PTB. For each probe, PTB was titrated (0, 0.25, 0.5, 1, and 2 μM; left to right).
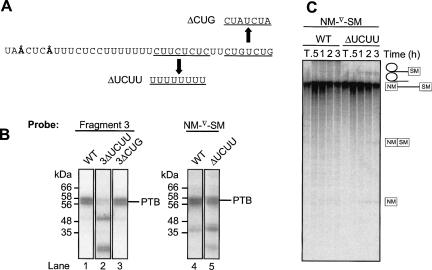
FIGURE 5.
UCUU elements in the polypyrimidine tract are important for PTB binding and splicing repression. (A) Sequence of NM-SM PCR fragment 3. Nucleotide substitutions in the ΔUCUU and ΔCUG mutants are indicated. (B) UV cross-linking of HeLa nuclear extract proteins to [32P]-labeled WT and mutant transcripts after incubation in nuclear extract for 25 min. Lane 1, fragment 3; lane 2, fragment 3ΔUCUU; lane 3, fragment 3ΔCUG; lane 4, NM-∇-SM WT; and lane 5, NM-∇-SMΔUCUU. All five lanes are derived from the same gel. (C) NM-∇-SM transcripts were spliced in HeLa nuclear extract for 0.5, 1, 2, or 3 h. RNA species were analyzed by 12% PAGE. T indicates transcript alone. Transcripts were WT NM-∇-SM or had the ΔUCUU point mutations in fragment 3. Quantitation of splicing of these two transcripts is shown in Figure 6D.
Electrophoretic mobility shift assays (EMSAs) were then carried out using recombinant PTB and fragments 1–8 (Fig. 2C). These confirmed that PTB bound with highest affinity to the PPT-containing fragment 3, followed by fragments 5 and 7. All three of these RNAs showed evidence of binding to more than one PTB molecule. Fragment 4 showed a small amount of binding only at the highest PTB concentrations, while fragments 1, 2, 6, and 8 did not bind at all. The cross-linking and EMSA data therefore indicate that fragments 3, 5, and 7 contain the main PTB-binding regions associated with the SM exon.
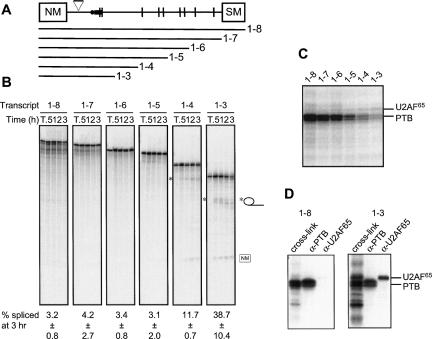
Interactions of PTB were then analyzed with fragments of the full-length NM-∇-SM transcript, which contain the 30 nt spacer to relieve steric interference between the SM branch point and NM 5′ splice site. Previous work has shown that splicing of NM-∇-SM can be activated by truncation of the transcript at restriction sites immediately downstream from the PPT, which removes all but two of the nine UCUU motifs that constitute potential PTB binding sites (Southby et al. 1999). This suggests that PTB exerts a part of its repressive activity via interaction with some or all of these downstream elements. The relative contributions of these sites to repression of the SM exon were investigated by in vitro splicing of NM-∇-SM transcripts progressively truncated from the 3′ terminus (Fig. 3A). Transcripts other than 1–8 (which is equivalent to the full-length NM-∇-SM substrate) are potentially competent for step 1 of splicing, although they cannot undergo step 2 due to the lack of a 3′ exon. Activation of splicing was apparent only when all sequences downstream from fragment 4 were absent (Fig. 3B), corresponding to the removal of all seven UCUU motifs downstream from the PPT. Interactions of HeLa PTB with the truncated NM-∇-SM transcripts were investigated by UV cross-linking (Fig. 3C,D). The intensity of the PTB signal was diminished with each progressive 3′ deletion. In contrast, U2AF65 cross-linking increased between 1–8 and 1–3 (Fig. 3D). This suggests that sites downstream of the PPT appear to be important both for PTB binding and for repression of SM exon splicing.
FIGURE 3.
Progressive removal of PTB binding sites downstream of the polypyrimidine tract activates SM exon splicing. (A) Organization of the α-actinin NM-∇-SM transcript. The triangle represents the 30-nt spacer sequence. Note that NM-∇-SM is identical to 1–8 in Figure 2. The various truncated transcripts are indicated below. (B) [32P]-labeled probes were spliced in 30% HeLa nuclear extract for 0.5, 1, 2, or 3 h and the RNA species resolved by 6% PAGE. T indicates transcript alone. Intron lariats produced by 1–3 and 1–4 are indicated by the asterisks. Values below each set of lanes represent the percentage of splicing at 3 h (mean ± SD, n = 3). (C) Splicing reactions were incubated for 25 min prior to UV cross-linking and RNase treatment; proteins were resolved by SDS-PAGE. The positions of PTB and U2AF65 are indicated to the right. (D) Immunoprecipitation of cross-linked PTB and U2AF65 with the 1–8 (left) and 1–3 (right) RNAs. Following UV cross-linking and RNase treatment, proteins were immunoprecipitated with α-PTB or α-U2AF65 and analyzed by SDS-PAGE alongside a total cross-linking sample.
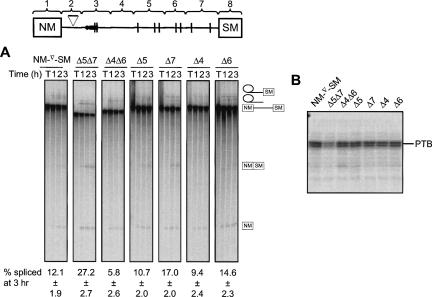
In order to determine more precisely which of the downstream regions contribute to the effects of PTB, internally deleted versions of the NM-∇-SM transcript were created, in which fragments 4, 5, 6, and 7 were removed individually or in particular combinations. Removal of fragment 7 alone resulted in a 1.4-fold enhancement of splicing relative to the wild-type (WT) NM-∇-SM RNA; the Δ5Δ7 combination produced further activation of the SM exon (2.2-fold increase over WT levels) (Fig. 4A), although Δ5 alone did not enhance splicing significantly. In contrast, deletions Δ4 and Δ6 had no effect on splicing, while the combined deletion Δ4Δ6 reduced splicing efficiency, possibly by bringing the PTB-binding fragments into closer proximity. These observations are consistent with the relative ability of PTB to interact with the individual transcript fragments (Fig. 2). In parallel UV cross-linking assays (Fig. 4B), the Δ5Δ7 deletion reduced the PTB signal to ∼50% of the WT level, whereas the Δ4Δ6 deletion had only a marginal effect. These data indicate that the reduction of PTB cross-linking is due to deletion of specific sequences, rather than due to the length of the transcript. Removal of any individual fragment did not cause a substantial reduction in PTB cross-linking.
FIGURE 4.
Internal deletions that remove PTB binding sites activate splicing. (A) [32P]-labeled probes were spliced in 30% HeLa nuclear extract for 1, 2, or 3 h and the RNA species resolved by 12% PAGE. T indicates transcript alone. Positions of splicing products and intermediates are indicated to the right. (B) Splicing reactions were incubated for 25 min prior to UV cross-linking and RNase treatment; proteins were resolved by SDS-PAGE. Values below each set of lanes represent the percentage of splicing at 3 h (mean ± SD, n = 4).
PTB binding to the PPT
The role of PTB binding by fragment 3 could not be investigated by deletion analysis as it contains the essential branch point and PPT, removal of which would abolish splicing. The two UCUU motifs located at the 3′ end of the PPT were identified as probable PTB binding sites within fragment 3 (Fig. 2A). This was tested by UV cross-linking of HeLa nuclear extract proteins with a mutant form of the transcript (3ΔUCUU), in which the four C residues encompassed by the UCUU motifs were mutated to U (Fig. 5A,B; Gromak et al. 2003). The mutation resulted in a >70% reduction in PTB cross-linking despite the increase in radiolabeled U content of the probe (Fig. 5B, lanes 1,2). In contrast, a control RNA containing mutations in adjacent CUG elements (3ΔCUG) (Gromak et al. 2003) behaved almost identically to the WT (Fig. 5B, lane 3). These data provide further evidence for the specificity of PTB interactions with the NM-SM fragments. Two novel cross-linking species of ∼20 and ∼50 kDa appeared with the ΔUCUU mutant form of fragment 3; these remain to be identified.
The functional effect of the fragment 3 ΔUCUU mutation was tested in the context of the full-length NM-∇-SM substrate. The mutation resulted in a ∼twofold increase in the level of SM exon splicing in vitro (Figs. 5C, 6D). Cross-linking of HeLa PTB to the full-length RNA was reduced by only ∼35% with the mutant relative to the WT transcript (Fig. 5B, lanes 4,5); this was accompanied by an increased interaction of unidentified ∼20 and ∼40 kDa species. The substantial residual cross-link observed with the mutant full-length RNA presumably reflects the interactions of PTB with downstream sequences within fragments 5 and 7. These data indicate that the UCUU motifs within the PPT constitute a binding site of functional importance for PTB-mediated SM exon repression. However, it is important to note that the mutation of four Cs to Us that reduced PTB binding might also have increased U2AF65 binding (Singh et al. 1995). The increased efficiency of SM splicing could therefore arise from a combination of decreased affinity for PTB and increased affinity for U2AF65.
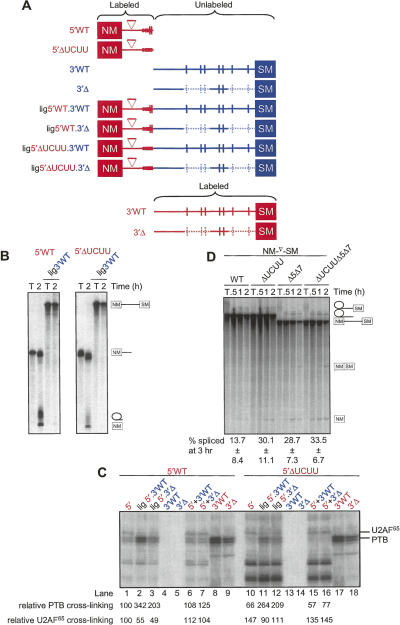
FIGURE 6.
Downstream binding sites cooperatively enhance interaction of PTB with the polypyrimidine tract region. (A) Schematic diagram of transcripts. Radiolabeled RNA is indicated in red and unlabeled in blue. The deletions of fragments 5 and 7 in the 3′Δ RNAs are indicated by the light blue dashed lines. Patch labeled RNAs were prepared by DNA oligonucleotide-splinted ligation. (B) [32P]-labeled 5′ and ligated RNAs were spliced in 30% HeLa nuclear extract for 2 h and the RNA species resolved by 12% PAGE. T indicates transcript alone. (Left) WT 5′ RNA; (right) 5′ΔUCUU RNA. (C) UV cross-linking of ligated RNAs. Transcripts were incubated for 25 min in HeLa nuclear extract. Following cross-linking and RNase treatment, HeLa nuclear extract proteins were resolved by SDS-PAGE. The intensity of PTB and U2AF65 cross-linking is shown below each lane as a percentage relative to the intensity of cross-linking of each protein to the 5′WT probe (lane 1). Note that within each set of lanes (5′WT, lanes 1,2,3,6,7 and 5′ΔUCUU, lanes 10,11,12,15,16) the labeled part of the RNA is derived from the same transcription reaction. Labels above the lanes are color-coded to indicate the parts of the transcript that were radiolabeled (red text) and unlabeled (blue). (D) NM-∇-SM transcripts were spliced in HeLa nuclear extract for 0.5, 1, or 2 h. RNA species were analyzed by 12% PAGE. T indicates transcript alone. Transcripts were WT NM-∇-SM or had the indicated point mutations in fragment 3 (ΔUCUU) or deletions of fragments 5 and 7 (Δ5Δ7). Quantitation of splicing at the 2-h timepoint is shown below each set of lanes (mean ± SD, n = 4 for WT and ΔUCUU, n = 3 for Δ5Δ7 and ΔUCUUΔ5Δ7).
PTB binding at the PPT is enhanced by downstream sites
A patch-labeling approach (Moore and Query 1998) was adopted to investigate the functional relationship between the PTB binding sites within the PPT and those further downstream. Radiolabeled 5′ transcripts, corresponding approximately to probe 1–3 (Fig. 1), were ligated to trace-labeled 3′ transcripts comprising the remaining downstream sequence of NM-∇-SM (Fig. 6A). The 5′ fragments were composed of WT or ΔUCUU mutant sequence, while the 3′ fragment was either WT or Δ5Δ7. Only the PPT region of the 5′ portion interacts detectably with PTB (Fig. 2A; Gromak et al. 2003), so any cross-linking to the patch-labeled RNAs represents PTB interacting with the PPT.
The 5′ fragments alone spliced efficiently, but ligation of the unlabeled 3′ ends led to substantial inhibition of splicing (Fig. 6B, cf. 2-h lanes within each panel). Parallel UV cross-linking assays indicated a 3.4-fold enhancement of PTB cross-linking to the WT PPT upon ligation to the WT 3′ sequences (Fig. 6C, cf. lanes 1 and 2). An equimolar mixture of the unligated 5′ and 3′ fragments showed cross-linking identical to that of the 5′ fragment alone (lane 6), indicating that the 3′ RNA is effective at promoting PTB binding to the PPT only when linked in cis. Ligation of the unlabeled 3′Δ5Δ7 RNA (Fig. 6C, lane 3, lig 5′WT.3′Δ5Δ7) also promoted PTB cross-linking to the PPT region, but to a lesser degree (twofold enhancement) than observed with 3′WT. These data indicate that downstream sequences promote PTB binding to the PPT.
The ΔUCUU mutant 5′ RNA alone showed reduced PTB cross-linking and increased U2AF65 cross-linking relative to its WT counterpart (Fig. 6C, cf. lanes 10 and 1). However, coupling of the ΔUCUU form to unlabeled downstream sequences again promoted interaction of PTB with the mutant PPT (Fig. 6C, lanes 10–12). Once again, ligation of the Δ5Δ7 mutant 3′ end increased PTB cross-linking to a lesser degree than did the WT 3′ end (Fig. 6C, lanes 11, 12). The relative degree of PTB cross-linking to the four ligated products (Fig. 6C, lanes 2, 3, 11, 12) varied inversely with the relative efficiency of splicing of the corresponding body-labeled transcripts (Fig. 6D). Interactions of U2AF65 with the ligated transcripts and their corresponding 5′ constituent RNAs also varied inversely with those of PTB (e.g., Fig. 6C, lanes 1 and 2, 10 and 11). Furthermore, the intensity of the U2AF65 signal was greater with the ΔUCUU 5′ probe than with the WT form. These data are consistent with a mechanism whereby PTB binding to the PPT, promoted by the downstream PTB binding sites, represses the SM exon.
DISCUSSION
Our data indicate the repression of α-actinin SM exon splicing involves PTB binding to sites spread over a region of ∼350 nt encompassing most of the distance between the PPT and the SM exon. PTB binding is not uniform across this region, as indicated by the lack of PTB binding to fragment 6 and the weak binding to fragment 4, despite the fact that both fragments are pyrimidine rich. Fragment 3, containing the PPT, bound PTB most strongly (Fig. 2). The UCUUCUCUCUU motif at the 3′ end of the PPT was crucial for PTB binding to fragment 3 alone (Fig. 5), and mutation of this element partially activated splicing of the full-length substrate (Fig. 5). However, PTB binding by fragment 3 was not sufficient for splicing repression, as indicated by the splicing activity of RNA substrates truncated at the 3′ end of fragment 3 (Figs. 3, 6). PTB binding at the PPT was enhanced by downstream PTB-binding elements, and it was this enhanced level of binding that correlated with repression of splicing. The simplest explanation of the enhanced PTB binding at the PPT is cooperative binding of PTB to RNAs with more than one high affinity site.
Although our approach was initially guided by the distribution of UCUU motifs, these are not able to account adequately for the differential PTB binding by the fragments. The distribution of other pyrimidine motifs, such as UCU, between the different RNA fragments also did not correlate particularly well with PTB binding, although UCUU-like motifs were most highly enriched in fragment 3. In particular, the distribution of CCU motifs, which in double-stranded form have been reported to act as PTB binding IRES elements (Mitchell et al. 2005), showed no correlation with PTB binding. Tellingly, the UCUU motifs in fragment 3 lie adjacent within an unbroken 26-nt pyrimidine tract, while those in fragment 6, which did not cross-link PTB, are both within 5–6 nt pyrimidine stretches. Indeed, PTB binding correlates more strongly with the length of the longest pyrimidine tract in each fragment than with the distribution of any individual motif. Fragments 3, 5, and 7 have pyrimidine tracts of 26, 13, and 14 nt, respectively, while the longest pyrimidine stretch in any other fragment was 9 nt.
Fragments 5 and 7, both of which cross-linked to PTB, contain UCUUs located in pyrimidine stretches of 6–8 nt (fragment 5) and 14 and 4 nt (fragment 7). The functional importance of fragments 5 and 7 was indicated by the activation of splicing upon their deletion, whereas deletion of fragments 4 and 6 had no effect (Fig. 4). Despite the intrinsic ability of fragment 3 to bind PTB, this binding alone is not sufficient for splicing repression. Transcripts terminated at the end of fragment 3 were able to splice efficiently (Figs. 3, 6), but when unlabeled 3′ RNAs were ligated to them, splicing was repressed, PTB cross-linking to the PPT increased, and U2AF65 cross-linking decreased (Fig. 6). Ligation of unlabeled Δ5Δ7 3′ end, containing the PTB binding site deletions, caused less repression of splicing and enhancement of PTB cross-linking to the PPT. The observation that deletion of the major downstream PTB binding regions (Δ5Δ7) did not activate splicing to the same extent as complete deletion of the 3′ end (Fig. 3) could be explained by PTB binding to fragment 4 (Fig. 2C). While fragment 4 alone does not bind PTB strongly, in combination with fragment 3 it might be sufficient to promote a degree of cooperative binding.
Our results provide an interesting comparison with data from other systems. While the requirement for multiple PTB binding sites has been reported in a number of model systems (e.g., Ashiya and Grabowski 1997; Perez et al. 1997a; Wagner and Garcia-Blanco 2002), repression mediated by cooperative binding of PTB has been demonstrated only with the N1 exon of c-src (Chou et al. 2000; Amir-Ahmady et al. 2005). Moreover, PTB-mediated repression of both N1 and exon 6 of Fas involves decreased binding of U2AF65 (Izquierdo et al. 2005; Sharma et al. 2005). However, in both cases the binding sites for PTB were located between binding sites for U2AF65 and U1 snRNP but were distant from both. The effect of PTB was to prevent activation of U2AF65 binding by U1 snRNP bound at a 5′ splice site. In the α-actinin SM system, the primary PTB binding sites are within the PPT adjacent to the U2AF65 binding site, a location that would be consistent with direct binding competition between PTB and U2AF65. However, our data do not rule out the possibility that PTB binding here may prevent the activation of U2AF65 binding by the upstream NM exon 5′ splice site (i.e., an intron definition interaction).
The SM exon is regulated not only by PTB but also by members of the CELF (Suzuki et al. 2002; Gromak et al. 2003) and Fox (Jin et al. 2003) families of RNA binding proteins. Both families antagonize the repressive effect of PTB, and in the case of CUG-BP, the functional antagonism results from direct binding competition at the 3′ end of the PPT (Gromak et al. 2003). CUG-BP binding competes with PTB binding, leading to a consequent increase in U2AF65 binding and splicing. These data are also consistent with PTB-mediated inhibition of SM splicing by direct interference with U2AF65 binding.
We analyzed the interaction of PTB with patch-labeled RNA by UV cross-linking. This has the advantage that it allows analysis of protein binding in extracts under splicing conditions, and to specific regions of the RNAs. It does, however, have the accompanying disadvantage of being less quantitative than approaches such as electrophoretic mobility shift, filter binding, or NMR chemical shift titration (e.g., Simpson et al. 2004; Amir-Ahmady et al. 2005; Clerte and Hall 2006). For example, our data do not distinguish whether the cooperative binding represents increased occupancy of a single PTB-binding site within the PPT, or cooperative enhancement of a second lower affinity interaction. Analysis of PTB binding to the N1 3′ splice, which comprises a direct 9 nt pyrimidine repeat within an overall ∼35 nt pyrimidine tract, indicated tight binding of one PTB monomer followed by a second lower affinity binding event (Amir-Ahmady et al. 2005). Significantly, the second binding event was cooperative if a short low affinity downstream PTB binding site was present, and this correlated with repression of splicing. The SM exon PPT is comparable in length to that of N1, with apparently weaker additional sites downstream, so the two binding event model may also apply to the actinin SM exon.
The two general models for PTB-mediated repression involving multiple PTB monomers involve either RNA looping between high affinity sites, or propagative binding of PTB along the RNA between high affinity sites (Wagner and Garcia-Blanco 2001). Results from the c-src system are more supportive of looping. In particular, U1 snRNP is stably bound at the N1 5′ splice site, even when the RNA is repressed by cooperative PTB binding to flanking sites (Chou et al. 2000; Sharma et al. 2005). Propagative binding requires direct protein–protein interactions between PTB monomers. This was consistent with early suggestions that PTB is dimeric (Perez et al. 1997b; Oh et al. 1998). More recent findings suggest that PTB is monomeric (Simpson et al. 2004; Amir-Ahmady et al. 2005; Monie et al. 2005), although it remains possible that weak interactions between RNA bound monomers could occur (Clerte and Hall 2006). Similar looping and propagation binding models have been proposed to explain splicing repression by hnRNPA1, and evidence for both forms of binding has been obtained in different systems (Zhu et al. 2001; Nasim et al. 2002). Therefore, both RNA looping and propagative binding are plausible possibilities for the cooperative binding of PTB in different model systems. Future experiments will aim to distinguish between these modes of binding upstream of the SM exon.
MATERIALS AND METHODS
Constructs
Constructs for in vitro transcription were based on NM-SM reporters described by Southby et al. (1999) and were prepared by standard cloning techniques (Sambrook et al. 1989). Point mutations and oligonucleotide insertions were created by site directed mutagenesis (Southby et al. 1999).
Transcription templates for individual fragments 1–8 were prepared by PCR; a T7 promoter sequence was incorporated in the forward primer in each case. Primers for fragments 1–4 are described by Gromak et al. (2003). Fragments 5–8 were generated using the following oligonucleotides:
5F: 5′-CGTAATACGACTCACTATAGGCTATCTTTCCTGGGAA-3′;
5R: 5′-CAGAGCAAAGAAATGCAGA-3′;
6F: 5′-CGTAATACGACTCACTATAGGACCTCCTTCAACATCTT-3′;
6R: 5′-GGTTTATGGTGTGAAGATT-3′;
7F: 5′-CGTAATACGACTCACTATAGGGCCCTTCCTCTTTCTG-3′;
7R: 5′-CTGTGGGCAGTGGTGC-3′;
8F: 5′-CGTAATACGACTCACTATAGGAAGAAGACGGGCATGATG-3′; and
8R: 5′-CATGTTCTAACCCATGGAG-3′.
PCR products were treated with T4 DNA polymerase, to remove Taq-generated 3′ overhangs, and gel purified.
In vitro transcription and splicing reactions
Transcription of 32P-labeled or trace-labeled RNAs from linearized plasmid or PCR-generated templates was performed using T7 polymerase as described previously (Southby et al. 1999). Transcripts were capped, except for 3′ RNAs used in ligation reactions, and were gel purified as necessary. Quantitation was carried out by scintillation counting and verified in some cases by PhosphorImager analysis of samples subjected to denaturing PAGE. In vitro splicing reactions (10 μL) typically contained 4–20 fmol of 32P-labeled RNA and 30% HeLa nuclear extract and were carried out as described previously (Southby et al. 1999; Gromak et al. 2003). Incubation at 30°C for time periods indicated in Figures 3–6 was followed by proteinase K digestion, phenol extraction, and ethanol precipitation; splicing intermediates and products were analyzed by 6% or 12% PAGE in the presence of 8 M urea, followed by PhosphorImager analysis. Splicing was quantitated using ImageQuant software by measuring the intensity of the 5′ exon and spliced product and normalizing for their radiolabeled U-content relative to full-length precursor transcript. Lariats were often not readily quantitated as the small size of the lariat loop means that the lariats run quite close to the precursor RNA upon electrophoresis.
Ligation of RNA fragments
PCR templates for RNA ligation experiments were generated using primers described previously (Gromak et al. 2003). For the production of patch-labeled RNAs, trace-labeled 3′ transcripts and high specific activity 5′ transcripts were combined in a 15:1 or 30:1 ratio and ligated following annealing to splint 6 (WT: 5′-CCATGGCCCACAGGAAGGGAGACAGGGAGAATTCAGACAGA-3′) or splint 7 (ΔUCUU: GGACAGACAGGTGGGAGGAGGAGCAGGGAGAATTCAGACAGA), using T4 DNA ligase (Moore and Query 1998; Gromak et al. 2003). After removal of the bridging oligonucleotode by digestion with DNase RQ1, products were gel purified to separate them from unligated substrates.
RNA binding
UV cross-linking was performed using high specific activity transcripts as described previously (Gromak et al. 2003). Splicing reactions were incubated for 25 min at 30°C in the absence of PVA. Individual NM-SM fragments that did not constitute splicing substrates were incubated with HeLa nuclear extract in the presence of rRNA nonspecific competitor. Following RNase treatment, radiolabeled cross-linked proteins were resolved by SDS-PAGE and analyzed by autoradiography. For cross-linking reactions with patch-labeled RNAs (Fig. 6), each 5′ RNA and its corresponding ligated products contained identical radiolabeled transcript derived from a single reaction. This enabled quantitative comparison of cross-linking intensities within each group of ligated and unligated RNAs (i.e., Fig. 6C, lanes 1–7 and 10–16). Moreover, WT and ΔUCUU mutant 5′ transcripts were prepared from a common transcription reaction mix, facilitating comparison across the two groups.
Immunoprecipitation was carried out with α-PTB or α-U2AF65 anti-serum following RNase treatment of cross-linked samples (Southby et al. 1999). Multiple identical reactions were pooled when required for enhanced signal intensity. Cross-linked proteins were incubated with antibody at 4°C for 1 h; incubation for a further hour was carried out following the addition of Protein A–Sepharose beads (Pharmacia). Beads were washed three times in NETS buffer (150 mM NaCl, 50 mM Tris-HCl at pH 7.5, 5 mM EDTA, 0.05% Nonidet P-40) and boiled in SDS loading buffer for 5 min, and then proteins were resolved by electrophoresis.
EMSAs were carried out as previously described (Gooding et al. 1998). Binding reactions (5 μL) contained 5 fmol RNA and a titration of recombinant PTB4 (0, 0.25, 0.5, 1, and 2 μM).
ACKNOWLEDGMENTS
This work was supported by program grants from the Wellcome Trust (059879 and 077877). A.J.M. was supported by a studentship from the MRC.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.219607.
REFERENCES
- Amir-Ahmady, B., Boutz, P.L., Markovtsov, V., Phillips, M.L., Black, D.L. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiya, M., Grabowski, P.J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: Evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco, P., Furger, A., Wollerton, M., Smith, C., Moreira, A., Proudfoot, N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, R.C., Black, D.L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, M.Y., Underwood, J.G., Nikolic, J., Luu, M.H., Black, D.L. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- Clerte, C., Hall, K.B. Characterization of multimeric complexes formed by the human PTB1 protein on RNA. RNA. 2006;12:457–475. doi: 10.1261/rna.2178406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote, C.A., Gautreau, D., Denegre, J.M., Kress, T.L., Terry, N.A., Mowry, K.L. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- Gooding, C., Roberts, G.C., Smith, C.W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- Gromak, N., Matlin, A.J., Cooper, T.A., Smith, C.W. Antagonistic regulation of α-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA. 2003;9:443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman, J.S., Meyer, M.I., Wang, Y.C., Mulligan, G.J., Kobayashi, R., Helfman, D.M. The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA. 1998;4:613–625. doi: 10.1017/s1355838298971448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, B.J., Genin, A., Cron, R.Q., Rigby, W.F. Delineation of a novel pathway that regulates CD154 (CD40 ligand) expression. Mol. Cell. Biol. 2003;23:510–525. doi: 10.1128/MCB.23.2.510-525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo, J.M., Majos, N., Bonnal, S., Martinez, C., Castelo, R., Guigo, R., Bilbao, D., Valcarcel, J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Jin, Y., Suzuki, H., Maegawa, S., Endo, H., Sugano, S., Hashimoto, K., Yasuda, K., Inoue, K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski, A., Hunt, S.L., Patton, J.G., Jackson, R.J. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- Kremerskothen, J., Teber, I., Wendholt, D., Liedtke, T., Böckers, T.M., Bernekow, A. Brain-specific splicing of α-actinin 1 mRNA. Biochem. Biophys. Res. Commun. 2002;295:678–681. doi: 10.1016/s0006-291x(02)00734-9. [DOI] [PubMed] [Google Scholar]
- Lin, C.H., Patton, J.G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Bushell, M., Evans, J.R., Stoneley, M., Le Quesne, J.P., Spriggs, R.V., Willis, A.E. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes & Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monie, T.P., Hernandez, H., Robinson, C.V., Simpson, P., Matthews, S., Curry, S. The polypyrimidine tract binding protein is a monomer. RNA. 2005;11:1803–1808. doi: 10.1261/rna.2214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J., Query, C.C. Uses of site-specifically modified RNAs constructed by RNA ligation. In: Smith C.W.J., editor. RNA protein interactions: A practical approach. Oxford University Press; Oxford, UK: 1998. pp. 75–108. [Google Scholar]
- Mulligan, G.J., Guo, W., Wormsley, S., Helfman, D.M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of β-tropomyosin pre-mRNA. J. Biol. Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- Nasim, F.U., Hutchison, S., Cordeau, M., Chabot, B. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA. 2002;8:1078–1089. doi: 10.1017/s1355838202024056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass, F.C., Auweter, S.D., Erat, M., Hargous, Y., Henning, A., Wenter, P., Reymond, L., Amir-Ahmady, B., Pitsch, S., Black, D.L., et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Oh, Y.L., Hahm, B., Kim, Y.K., Lee, H.K., Lee, J.W., Song, O., Tsukiyama-Kohara, K., Kohara, M., Nomoto, A., Jang, S.K. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem. J. 1998;331:169–175. doi: 10.1042/bj3310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, I., Lin, C.H., McAfee, J.G., Patton, J.G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997a;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- Perez, I., McAfee, J.G., Patton, J.G. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997b;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- Robinson, F., Smith, C.W. A splicing repressor domain in polypyrimidine tract-binding protein. J. Biol. Chem. 2006;281:800–806. doi: 10.1074/jbc.M510578200. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., Maniatis, T. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sauliere, J., Sureau, A., Expert-Bezancon, A., Marie, J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the β-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol. Cell. Biol. 2006;26:8755–8769. doi: 10.1128/MCB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S., Falick, A.M., Black, D.L. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Kan, J.L., Ghigna, C., Biamonti, G., Green, M.R. A single polypyrimidine tract binding protein (PTB) binding site mediates splicing inhibition at mouse IgM exons M1 and M2. RNA. 2004;10:787–794. doi: 10.1261/rna.5229704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, P.J., Monie, T.P., Szendroi, A., Davydova, N., Tyzack, J.K., Conte, M.R., Read, C.M., Cary, P.D., Svergun, D.I., Konarev, P.V., et al. Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure. 2004;12:1631–1643. doi: 10.1016/j.str.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J., Green, M.R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Southby, J., Gooding, C., Smith, C.W. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutally exclusive exons. Mol. Cell. Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman, R., Smith, C.W. Novel modes of splicing repression by PTB. Trends Biochem. Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Suzuki, H., Jin, Y., Otani, H., Yasuda, K., Inoue, K. Regulation of alternative splicing of α-actinin transcript by Bruno-like proteins. Genes Cells. 2002;7:133–141. doi: 10.1046/j.1356-9597.2001.00506.x. [DOI] [PubMed] [Google Scholar]
- Wagner, E.J., Garcia-Blanco, M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.J., Garcia-Blanco, M.A. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- Waites, G.T., Graham, I.R., Jackson, P., Millake, D.B., Patel, B., Blanchard, A.D., Weller, P.A., Eperon, I.C., Critchley, D.R. Mutually exclusive splicing of calcium-binding domain exons in chick α-actinin. J. Biol. Chem. 1992;267:6263–6271. [PubMed] [Google Scholar]
- Wollerton, M.C., Gooding, C., Robinson, F., Brown, E.C., Jackson, R.J., Smith, C.W. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–832. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Liu, W., Grabowski, P.J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Mayeda, A., Krainer, A.R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]