Abstract
Mamit-tRNA (http://mamit-tRNA.u-strasbg.fr), a database for mammalian mitochondrial genomes, has been developed for deciphering structural features of mammalian mitochondrial tRNAs and as a helpful tool in the frame of human diseases linked to point mutations in mitochondrial tRNA genes. To accommodate the rapid growing availability of fully sequenced mammalian mitochondrial genomes, Mamit-tRNA has implemented a relational database, and all annotated tRNA genes have been curated and aligned manually. System administrative tools have been integrated to improve efficiency and to allow real-time update (from GenBank Database at NCBI) of available mammalian mitochondrial genomes. More than 3000 tRNA gene sequences from 150 organisms are classified into 22 families according to the amino acid specificity as defined by the anticodon triplets and organized according to phylogeny. Each sequence is displayed linearly with color codes indicating secondary structural domains and can be converted into a printable two-dimensional (2D) cloverleaf structure. Consensus and typical 2D structures can be extracted for any combination of primary sequences within a given tRNA specificity on the basis of phylogenetic relationships or on the basis of structural peculiarities. Mamit-tRNA further displays static individual 2D structures of human mitochondrial tRNA genes with location of polymorphisms and pathology-related point mutations. The site offers also a table allowing for an easy conversion of human mitochondrial genome nucleotide numbering into conventional tRNA numbering. The database is expected to facilitate exploration of structure/function relationships of mitochondrial tRNAs and to assist clinicians in the frame of pathology-related mutation assignments.
Keywords: mitochondrial genome, tRNA genes, secondary structures, alignments, mammalian
INTRODUCTION
Transfer RNAs (tRNAs) are key actors in translation of the genetic information from mRNA into proteins. Several thousands of tRNA gene sequences are presently known (Sprinzl and Vassilenko 2005), supporting the existence of precise common structural rules responsible for cloverleaf secondary folding and L-shaped tertiary architectures as recognized decades ago when the sequence of yeast tRNAAla (Holley et al. 1965) and the crystallographic structure of yeast tRNAPhe (Robertus et al. 1974; Suddath et al. 1974) were solved (for review, see Rich and Schimmel 1977; Giegé et al. 1993; Dirheimer et al. 1995). Size of structural domains, location of conserved or semi-conserved nucleotides, and tertiary interactions are of textbook knowledge for prokaryotic and archaeal tRNAs as well as for tRNAs from eukaryotic cytosols and from chloroplasts. Analysis of nucleotides not involved in folding within subgroups of tRNAs revealed sets of signals for particular tRNA functions such as recognition by aminoacyl-tRNA synthetases (Giegé et al. 1998; Beuning and Musier-Forsyth 1999) or recognition by post-transcriptional modification enzymes (Grosjean and Benne 1998) or other domain specific features (Marck and Grosjean 2002). Interestingly, the situation for several mitochondrial tRNAs is different. Already at the time of sequencing the first complete mitochondrial genome, that of bovine (Anderson et al. 1981), it appeared that, despite a cloverleaf-like secondary structure, the mitochondrial tRNAs markedly deviate from classical tRNAs. These unexpected features have been confirmed for mitochondrial tRNAs from other organisms (e.g., Wolstenholme et al. 1987) and especially from mammals. Compilation and comparison of tRNAs from 31 fully sequenced mammalian mitochondrial genomes within 22 groups (one per amino acid specificity, two for leucine, and two for serine according to the anticodon triplet) demonstrated that these tRNAs distinguish from classical tRNAs in regard of the size of D- and T-domains (especially loops) and the absence of expected conserved and semi-conserved nucleotides, leaving the understanding of three-dimensional (3D) folding of these tRNAs mainly open (Helm et al. 2000), despite efforts in this direction (Steinberg et al. 1997).
Thus, mammalian mitochondrial tRNAs are interesting molecules in the frame of RNA folding studies, RNA recognition rules, definition of tRNA identity elements for aminoacylation, and more generally toward understanding evolution of RNAs in these different fields, evolution of the translational machineries, and of mitochondria in general.
Further, mammalian mitochondrial tRNAs gained interest since a growing number of human disorders could be correlated to point mutations in mitochondrial tRNA genes. Since the first discovery of mutation A3243G in the gene for tRNALeu(UUR) (Goto et al. 1990), about 130 pathology-related mutations have been reported (Brandon et al. 2005). Genes of tRNA represent only 10% of the mitochondrial genome but contain about 50% of pathology-related mutations, highlighting the importance of these genes. Assignment of mutations as pathological (McFarland et al. 2004) as well as understanding the molecular mechanisms underlying the vast diversity of phenotypic expressions of point mutations are tackled by a variety of approaches (e.g., Florentz et al. 2003; Jacobs 2003; Wittenhagen and Kelley 2003; Shapira 2006). All require basic knowledge on structure/function relationships of human mitochondrial tRNAs and of consensus sequences of mammalian mitochondrial tRNAs or of phylogenetically related subgroups of tRNAs. Thus, specific features of the 22 tRNA genes and their evolution have been addressed in animal mitochondrial genomes (Higgs et al. 2003; Jameson et al. 2003), features of mammalian mitochondrial tRNAs have been highlighted (Helm et al. 2000), differences between pathology-related and polymorphisms in human mitochondrial tRNAs searched (Florentz and Sissler 2001), and the possibility for predicting pathology-related mutations computed (Kondrashov 2005).
The outcomes of our initial compilation (Helm et al. 2000) of 679 genes from 31 mammalian mitochondrial genomes lead to 22 consensus sequences (conserved and semi-conserved nucleotides at defined positions) and 22 typical sequences (most frequent nucleotide at each position), posted in a frozen state on an initial Mamit-tRNA Web site. Due to the exponential availability of new mammalian mitochondrial genomes in genome databases, Mamit-tRNA sequence information has now been transformed and transferred into a database and was reshaped and restructured in a dynamic and interactive way. Mamit-tRNA is a database displaying the alignment of >3000 mammalian mitochondrial tRNA gene sequences in 22 subgroups, which have been retrieved from 150 fully sequenced genomes available at GenBank, NCBI. Individual sequences have been carefully verified and erroneous sequences removed. Mamit-tRNA gives access to individual tRNA sequence information as well as to a Web-based compilation project of multiple sequences (organism, order, or superorder) in any combination. The result appears dynamically on cloverleaf-shaped secondary structures either as typical or consensus sequences with Supplemental statistical data for each position within the tRNA. Mamit-tRNA contains now each of the 22 cloverleaf structures of human mitochondrial tRNAs highlighting positions of all pathogenic and polymorphic mutations reported so far in the literature. In addition, Mamit-tRNA contains a straightforward conversion table of nucleotide numbering within the human mitochondrial genome and within conventional tRNA cloverleaves.
DATA COLLECTION
Mammalian mitochondrial genome sequences have been imported from available fully sequenced genomes (GenBank, NCBI) by specifying the accession number or the locus name with “Lasergene sequence and analysis software” (DNASTAR) using the EditSeq module. In EditSeq, DNA sequences as well as all associated sequence features and references are displayed and allow for corrections if necessary. The MagAlign module then allows the generation of multiple sequence alignments for all mitochondrial tRNA genes of a given specificity. At this level all sequences become framed and aligned manually in respect to the structural domains of a tRNA (acceptor-stem, D-loop and stem, anticodon loop and stem, variable region, T-loop and stem). This is an absolute requisite to assure all further alignments and is inherent to mammalian mitochondrial tRNA genes. Indeed, opposite to classical tRNAs, D- and T-loops not only vary dramatically in size, but also miss the expected milestone nucleotides. Manual alignments are done according to rules established previously (Helm et al. 2000). During this manual step, up to 30% of the annotated tRNA genes revealed annotation errors (erroneous annotation of 5′ and 3′ ends, reverse complement sequence, absence of large terminal domains, or presence of large terminal additional sequences). Most errors have been corrected by individual sequence screening at the level of the flanking sequences in the full-length genome (using EditSeq), new annotation, saving, and new import to the MagAlign alignment project. Those sequences, for which no correct version could be found, were removed from the database. It cannot be excluded that the data set of annotated tRNA sequences still contains internal sequencing errors, which cannot be distinguished from specific evolutionary mutations. Indeed, natural polymorphisms distinguish haplogroups within species (e.g., Ingman and Gyllensten 2006). Further, it should be recalled that the database deals with DNA sequences while mature tRNAs may undergo editing events at the RNA level (e.g., Börner et al. 1996). The alignment project of one or several sequences is finally saved in a text format (PAU) and implemented to the database by a specifically developed administrator interface. Sequences are further organized according to their amino acid specificity of the tRNA and to the phylogeny of the corresponding organism. The handy full process will be used for a regular (bisemestrial) update of the database.
MAMMALIAN MITOCHONDRIAL tRNA GENE SEQUENCES
More than 3000 linear sequences aligned according to secondary structural domains
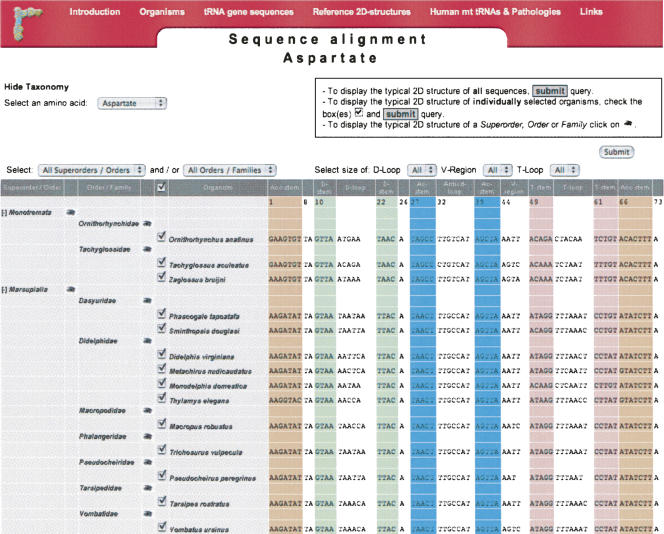
The database contains presently 3064 tRNA gene sequences from 150 organisms, which are classified in 22 families according to the amino acid identity. Linear sequences of individual mammals are displayed according to the molecular phylogeny classification (Murphy et al. 2001; Delsuc et al. 2002), where mammals are divided into Monotremes (Prototheria: Monotremata), Marsupials (Metatheria: Marsupialia), and Placentals (Eutheria: Placentalia). Among Eutherians, four major clades or superorders of placental mammals are identified by phylogenetic analyses of concatenated mitochondrial and nuclear DNA/protein markers. Each clade or order (distinguished by a specific color code) is subdivided into orders/families and further to individual organisms (Fig. 1). All tRNA sequences have been carefully aligned according to the two-dimensional (2D) structural domains, and complementary base-paired stems are highlighted using a color code, according to the reference 2D structure recalled on the Mamit-tRNA Web site.
FIGURE 1.
Screen picture of linear sequence alignments of mitochondrial tRNAs on Mamit-tRNA.
Marsupial mitochondrial genomes are missing the gene for tRNALys (Janke et al. 1994) and have instead a sequence for which no tRNA feature can be found. For other organisms a total of 11 tRNA genes were not annotated on the corresponding mitochondrial genome due to erroneous automatic annotation (Bos indicus tRNAAsp, Caenolestes fuliginosus tRNAVal, Dromiciops gliroides tRNAPro, Macrotis lagotis tRNAPro, Monodelphis domestica tRNAVal, Muntiacus crinifrons tRNAMet, Notoryctes typhlops tRNAPhe, Perameles gunnii tRNAPro, Rhyncholestes raphanurus tRNAVal, Urotrichus talpoides tRNAIle).
On-line sequence analysis tools: Establishment of individual, consensus, and typical 2D structures
Mamit-tRNA implements a dynamic Web interface that allows researchers to browse its collection of tRNA gene sequences. Simple searches may be performed and each sequence can be displayed linearly with color codes indicating secondary structural domains, which further can be converted into a printable 2D cloverleaf structure. Multiple sequences can be selected by organism, order, or superorder in any combination for compilation. tRNA sequences can also be selected according to the size of their D- and/or T-loop and/or variable region. The outcome appears dynamically on cloverleaf-shaped secondary structures either as typical or consensus sequences with Supplemental statistical data for each position within the tRNA and for base pairs in stems (Watson–Crick, G–T, mismatches). Due to large variations in size, D- and T-loop sequence comparisons escape the analysis. Cloverleaves can be exported as jpeg files by simple screen sliding and used in any drawing program (e.g., Fig. 2).
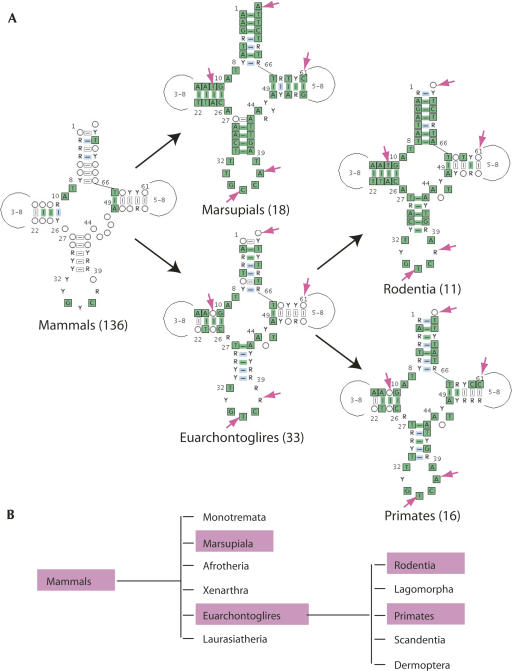
FIGURE 2.
Example of sequence conservation within phylogenetic subgroups: the case of tRNAAsp. (A) Outputs of database searches. Consensus sequences obtained for indicated subgroups. The number of considered sequences is indicated in parenthesis. Y stands for pyrimidine, R for purine. The color code for nucleotides is the following: green stands for 100% conservation, blue for >90% conservation, white for >50% conservation. The color code for base pairing is the following: green for 100% conservation of Watson–Crick (WC) pairs, blue for high frequency of WC pairs and <50% G–T pairs, white for high frequency of WC and G–T pairs but <50% mismatches. Specific nucleotide positions of interest (discussed in the text) are pointed by arrows. (B) Overview of the phylogenetic links of the considered examples.
As first outcome of the fivefold enriched database (as compared to the initial set of sequences; Helm et al. 2000), it is possible to gain a refined view on conserved, semi-conserved, and typical nucleotides at each position within each of the 22 mammalian tRNAs. In the case of tRNAAsp chosen as example (Fig. 2), only 7 nucleotides (nt) are strictly conserved within the 136 considered sequences, while 10 nt were conserved when comparing 31 sequences. More generally, the enlarged set of considered molecules confirms all major structural features determined for the smaller set (31 organisms) and does not reveal major new features in terms of size and nucleotide composition of 2D structural domains. The large database offers, however, the new possibility of sequence comparison in subgroups of organisms which presently contain a sufficient number of members for significant results. As example (Fig. 2), it becomes possible to distinguish specific sequence elements within tRNAAsp from Marsupials and from Euarchontoglires and further distinguish specific features between two subgroups of the Euarchontoglires order such as Rodentia and Primates. Five positions can be compared within these five compilations: (1) the discriminator base 73 is strictly conserved as A in Marsupials, while it is not conserved in Euarchontoglires; (2) the central anticodon nucleotide 34 is C in Marsupial and T in the other sequences, in agreement with editing events (Börner et al. 1996); (3) purine 37 is strictly A in Marsupials and Primates, and A or G in the two other consensus sequences; (4) pair 11–24 in the D-stem is strictly conserved in Marsupials but not in the full group of Euarchontoglires, where it is strictly conserved in Rodentia and not at all in Primates; and (5) base pair 61–53 is strictly conserved in Marsupials but not in Euarchontoglires. Similar distinctions can be made in each of the 22 tRNA families.
HUMAN MITOCHONDRIAL tRNAs AND PATHOLOGIES
Location of mutations on 2D structures of human mitochondrial tRNAs
Human mitochondrial tRNA genes are hot spots for mutations. About 130 mutations have been correlated to diverse pathologies and more than 110 mutations are polymorphisms (harmless point mutations distinguishing individual human sequences). Mamit-tRNA displays each of the 22 human mitochondrial tRNA 2D structures in individual windows, each with the full set of polymorphic and pathology-related mutations described in the literature presently. Each cloverleaf structure is accompanied by a table listing the pathology-related mutations, names of pathologies, and corresponding references.
Conversion table for tRNA nucleotide numbering
Mutations in the human mitochondrial genome are reported and numbered by clinician according to the reference sequence reported by Anderson and colleagues (Anderson et al. 1981). Best known examples are A3243G in tRNALeu(UUR) and A8344G in tRNALys. This numbering system complicates easy localization of the corresponding positions in tRNA secondary structural domains. A straightforward numbering conversion table is offered within the Web site allowing for an easy application of the international tRNA nucleotide numbering system (Sprinzl and Vassilenko 2005). The table is split in two parts, each covering a subset of the 22 tRNAs. A first part is dedicated to the only four human tRNAs displaying all expected classical structural features, including those within the D- and T-loops. The corresponding conversion table is precise for each individual nucleotide. A second part is dedicated to the 18 other tRNAs for which nucleotide numbering conversions are given for all domains of the tRNA except D- and T-loops, for which no internal reference nucleotide is present.
CONCLUSION
The database on mammalian mitochondrial tRNA gene sequences is available via http://mamit-tRNA.u-strasbg.fr. It corresponds to a sound collection of sequences from which errors have been removed and alignments made manually. Due to the large variation in size of two structural domains of mammalian mitochondrial tRNAs, the manual approach guarantees highest possible quality which would escape automatically generated databases and make them not reliable. However, it should be stressed that the possibility of sequencing errors remains, since it is not distinguishable whether deviations from consensus nucleotides at a given position correspond to errors or reveal indeed natural nucleotide changes in some organisms. tRNA secondary structures can be retrieved in jpeg format and used for figure constructions.
The database with its integrated interfaces is expected to become a tool for basic research on structural properties and peculiarities of mammalian mitochondrial tRNAs. It will, for example, allow to distinguish the presence (or absence) of recognition signals for partner macromolecules, to tackle evolutionary considerations in the fields of RNA structures, tRNA aminoacylation identity signals, translation machineries in organelles, evolution of the mitochondrial genome in general, and its coevolution with the nuclear genome. The database is also expected to facilitate advances in the field of tRNA mutations and pathologies. Interestingly the human mitochondrial genomes encodes only for 13 proteins and two rRNAs in addition to the 22 tRNA genes. All 13 proteins are subunits of the respiratory chain complexes (I–IV) and ATP synthase (complex V) (to be complemented by about 70 subunits of nuclear origin) and are thus directly linked to the energy metabolism. More and more pathologies linked to the mitochondrial translation machinery become discovered (Jacobs and Turnbull 2005), including aminoacyl-tRNA synthetases as major targets (Antonellis et al. 2006; Scheper et al. 2007). This is to say the central importance of tRNA genes to mitochondrial activity and the importance of understanding both their diversity and their uniqueness.
ACKNOWLEDGMENTS
We thank the members of ULP Multimedia, in particular Jean Crusot, Matthieu Koch, and Pierre Notter for developing and building up the relational database of Mamit-tRNA and for their long-term assistance. Special thanks to Emmanuel Douzery (University of Montpellier) for molecular phylogeny-related aspects, to Elisabeth Zielonka for careful proofreading of the database, to Dagmar Friede for initial analysis of tRNA subgroups, and to Richard Giegé for suggestions and support. This work was funded by CNRS, Université Louis Pasteur, Association Française contre les Myopathies (AFM), and EC contract FP5 RTD QLG2-CT-00660.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.588407.
REFERENCES
- Anderson, S., Bankier, A.T., Barrel, B.G., de Bruijn, M.H.L., Coulson, A.R., Drouin, J., Eperon, J.C., Nierlich, D.P., Roe, B.A., Sanger, F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Antonellis, A., Lee-Lin, S.Q., Wasterlain, A., Leo, P., Quezado, M., Goldfarb, L.G., Myung, K., Burgess, S., Fischbeck, K.H., Green, E.D. Functional analyses of glycyl-tRNA synthetase mutations suggest a key role for tRNA-charging enzymes in peripheral axons. J. Neurosci. 2006;26:10397–10406. doi: 10.1523/JNEUROSCI.1671-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuning, P.J., Musier-Forsyth, K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999;52:1–28. doi: 10.1002/(SICI)1097-0282(1999)52:1<1::AID-BIP1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Börner, G.V., Mörl, M., Janke, A., Pääbo, S. RNA editing changes the identity of a mitochondrial tRNA in marsupials. EMBO J. 1996;15:5949–5957. [PMC free article] [PubMed] [Google Scholar]
- Brandon, M.C., Lott, M.T., Nguyen, K.C., Spolim, S., Navathe, S.B., Baldi, P., Wallace, D.C. MITOMAP: A human mitochondrial genome database—2004 update. Nucleic Acids Res. 2005;33:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc, F., Scally, M., Madsen, O., Stanhope, M.J., de Jong, W.W., Catzeflis, F.M., Springer, M.S., Douzery, E.J. Molecular phylogeny of living xenarthrans and the impact of character and taxon sampling on the placental tree rooting. Mol. Biol. Evol. 2002;19:1656–1671. doi: 10.1093/oxfordjournals.molbev.a003989. [DOI] [PubMed] [Google Scholar]
- Dirheimer, G., Keith, G., Dumas, P., Westhof, E. Primary, secondary and tertiary structures of tRNAs. In: Söll D., RajBhandary U.L., editors. tRNA: Structure, biosynthesis, and function. American Society for Microbiology; Washington, DC: 1995. pp. 93–126. [Google Scholar]
- Florentz, C., Sissler, M. Disease-related versus polymorphic mutations in human mitochondrial tRNAs: Where is the difference? EMBO Rep. 2001;2:481–486. doi: 10.1093/embo-reports/kve111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentz, C., Sohm, B., Tryoen-Tóth, P., Pütz, J., Sissler, M. Human mitochondrial tRNAs in health and disease. Cell. Mol. Life Sci. 2003;60:1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé, R., Puglisi, J.D., Florentz, C. tRNA structure and aminoacylation efficiency. Prog. Nucleic Acid Res. Mol. Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- Giegé, R., Sissler, M., Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, Y., Nonaka, I., Horai, S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Benne, R. Modification and editing of RNA. ASM Press; Washington, DC: 1998. [Google Scholar]
- Helm, M., Brulé, H., Friede, D., Giegé, R., Pütz, J., Florentz, C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, P.G., Jameson, D., Jow, H., Rattray, M. The evolution of tRNALeu genes in animal mitochondrial genomes. J. Mol. Evol. 2003;57:435–445. doi: 10.1007/s00239-003-2494-6. [DOI] [PubMed] [Google Scholar]
- Holley, R.W., Apgar, J., Everett, G., Madison, J., Marquisee, M., Merrill, S., Penswick, J.R., Zamir, A. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- Ingman, M., Gyllensten, U. mtDB: Human mitochondrial genome database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–D751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, H.T. Disorders of mitochondrial protein synthesis. Hum. Mol. Genet. 2003;12:R293–R301. doi: 10.1093/hmg/ddg285. [DOI] [PubMed] [Google Scholar]
- Jacobs, H.T., Turnbull, D.M. Nuclear genes and mitochondrial translation: A new class of genetic disease. Trends Genet. 2005;21:312–314. doi: 10.1016/j.tig.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Jameson, D., Gibson, A.P., Hudelot, C., Higgs, P.G. OGRe: A relational database for comparative analysis of mitochondrial genomes. Nucleic Acids Res. 2003;31:202–206. doi: 10.1093/nar/gkg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, A., Feldmaier-Fuchs, G., Thomas, W.K., von Haeseler, A., Pääbo, S. The marsupial mitochondrial genome and the evolution of placental mammals. Genetics. 1994;137:243–256. doi: 10.1093/genetics/137.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, F.A. Prediction of pathogenic mutations in mitochondrially encoded human tRNAs. Hum. Mol. Genet. 2005;14:2415–2419. doi: 10.1093/hmg/ddi243. [DOI] [PubMed] [Google Scholar]
- Marck, C., Grosjean, H. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland, R., Elson, J.L., Taylor, R.W., Howell, N., Turnbull, D.M. Assigning pathogenicity to mitochondrial tRNA mutations: When “definitely maybe” is not good enough. Trends Genet. 2004;20:591–596. doi: 10.1016/j.tig.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Eizirik, E., O'Brien, S.J., Madsen, O., Scally, M., Douady, C.J., Teeling, E., Ryder, O.A., Stanhope, M.J., de Jong, W.W., et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Rich, A., Schimmel, P.R. Structural organization of complexes of transfer RNAs with aminoacyl-transfer RNA synthetases. Nucleic Acids Res. 1977;4:1649–1665. doi: 10.1093/nar/4.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus, J.D., Ladner, J.E., Finch, J.T., Rhodes, D., Brown, R.S., Clark, B.F.C., Klug, A. Structure of yeast phenylalanine tRNA at 3Å resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Scheper, G.C., van der Klok, T., van Andel, R.J., van Berkel, C.G., Sissler, M., Smet, J., Muravina, T.I., Serkov, S.V., Uziel, G., Bugiani, M., et al. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat. Genet. 2007;39:534–539. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- Shapira, A.H.V. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- Sprinzl, M., Vassilenko, K.S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, S., Leclerc, F., Cedergren, R. Structural rules and conformational compensations in the tRNA L-form. J. Mol. Biol. 1997;266:269–282. doi: 10.1006/jmbi.1996.0803. [DOI] [PubMed] [Google Scholar]
- Suddath, F.L., Quigley, G.J., McPherson, A., Sneden, D., Kim, J.J., Kim, S.H., Rich, A. Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0 angstroms resolution. Nature. 1974;248:20–24. doi: 10.1038/248020a0. [DOI] [PubMed] [Google Scholar]
- Wittenhagen, L.M., Kelley, S.O. Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem. Sci. 2003;28:605–611. doi: 10.1016/j.tibs.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Wolstenholme, D.R., Macfarlane, J.L., Okimoto, R., Clary, D.O., Wahleithner, J.A. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc. Natl. Acad. Sci. 1987;84:1324–1328. doi: 10.1073/pnas.84.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]