Abstract
The foot-and-mouth disease virus (FMDV) RNA contains two in-frame AUG codons separated by 84 nt that direct translation initiation of the viral polyprotein. The mechanism of initiation at the IRES-proximal AUG codon (AUG1) has been previously analyzed, whereas no data on factor requirements for AUG2 have been reported. Here, using the method of 48S translation initiation complex reconstitution, we show that eIF1 is indispensable in forming the 48S initiation complex at AUG2. In contrast, it reduces the assembly of this complex at AUG1. Stabilization of a stem–loop between the initiation triplets induces a small decrease in the toeprint intensity at AUG2, accompanied by an increase in the AUG1/AUG2 ratio as well as a moderate reduction of protein synthesis initiated at AUG2 in transfected cells. PTB and ITAF45 exerted an additive positive effect on the 48S complex at AUG2, although a substantial reconstitution on both AUGs occurs on omission of either of these proteins. Relative to the β-globin mRNA, the 48S complex formation at AUG1 and AUG2 is slow and occurs with the same kinetics as revealed by the “kinetic” toeprint assay. Mutation of AUG1 to AUA does not abrogate protein synthesis in transfected cells, and has no effect on the rate of the 48S complex formation at AUG2. We conclude that the AUG2 initiation region is selected independently of 48S complex formation at the upstream AUG1. The kinetic toeprint assay also shows that cap-dependent assembly of the 48S complex in vitro occurs faster than the FMDV IRES-mediated complex assembly.
Keywords: translation initiation, AUG selection, IRES-elements, foot-and-mouth disease virus RNA, picornavirus
INTRODUCTION
Initiation of protein synthesis in eukaryotes can occur by two distinct mechanisms, cap-dependent scanning and internal ribosome entry. The second mode of initiation was first described for picornavirus RNAs (for review, see Belsham and Jackson 2000; Martinez-Salas et al. 2001; Pestova et al. 2001), and the mechanisms of action of picornavirus IRES elements still remain a paradigm of internal translation initiation in eukaryotes. According to their secondary structures, picornavirus IRESs can be classified into three groups: type I includes enteroviruses and rhinoviruses; type II includes cardioviruses and aphthoviruses; and type III, hepatoviruses. As has been recently found, the IRESs of some members of the family Picornaviridae (porcine enterovirus 8, simian virus 2, and porcine teschovirus 1) form a separate group showing a striking similarity to the IRES from hepatitis C virus RNA (Pisarev et al. 2004; Chard et al. 2006).
The current view of the mechanism of operation of picornavirus IRES elements derives from studies performed with RNAs from encephalomyocarditis (EMCV) and Theiler murine encephalomyelitis viruses (TMEV) (cardioviruses), foot-and-mouth disease virus (FMDV) (aphthoviruses), polioviruses (enteroviruses), and rhinoviruses (for review, see Belsham and Jackson 2000; Belsham and Martinez-Salas 2004; Martinez-Salas and Fernandez-Miragall 2004). Although the secondary structures of type I and type II IRESs are quite distinct, there is little doubt that the 40S ribosomal subunit enters the respective mRNAs within the 3′ parts of these IRESs, which harbor a conserved pyrimidine tract. However, the AUG triplet that immediately follows this tract is used to initiate translation only in the case of the EMCV and FMDV IRESs, being silent for poliovirus and rhinovirus IRES elements. In the latter case, the functional start codon is positioned downstream and separated from this silent AUG triplet by ∼160 and ∼32 nucleotides (nt), respectively. It has been speculated that after a primary binding of 40S subunits at the silent AUG triplet, the true start codon is attained by a scanning process similar to that used in the standard cap-dependent initiation. However, no direct experimental evidence for such a hypothesis has been presented.
Although, as noted above, the secondary structures of the EMCV and FMDV IRES elements are similar, the respective RNAs differ significantly in organization of the downstream part of their translation initiation regions. The proximal to the IRES AUG 11 in the EMCV RNA is recognized as the only start codon, whereas two in-frame AUG codons separated by 84 nt are used to initiate translation on the FMDV RNA (Belsham and Jackson 2000; Belsham and Martinez-Salas 2004). Both AUG codons as well as their relative distance are conserved in FMDV field isolates. The second start triplet, termed here AUG2, is more often used in infected cells as well as in vitro, irrespective of the AUG1 context (Lopez de Quinto and Martinez-Salas 1999). However, the mechanism of initiation at the preferred AUG2 codon has not been elucidated.
The molecular mechanism of translation initiation on the EMCV, FMDV, and TMEV IRES elements (type II) has been dissected in detail by using the method of reconstitution of 48S translation initiation complexes from purified components combined with toeprinting (Pestova et al. 1996, 1998a; Dmitriev et al. 2003b). This approach has allowed one to determine a set of canonical translation initiation factors required by these IRESs, to identify a minimum set of IRES transacting factors (ITAFs) involved in the initiation process, to determine binding sites of canonical and auxiliary initiation factors within the secondary structures of these IRESs, and finally to understand their functional topography. It has been well documented that the location of the authentic start codon of the EMCV RNA and the AUG1 of FMDV RNA by the 40S ribosomal subunit may occur in the absence of eIF1 and, hence, does not necessarily involve scanning (Pestova et al. 1996, 1998a; Pilipenko et al. 2000). By analogy with the poliovirus RNA (see above), the involvement of scanning in the selection of AUG2 of FMDV RNA has been suggested (Belsham 1992; Belsham and Jackson 2000), but as noted above, not experimentally tested. The set of canonical eukaryotic initiation factors (eIFs) that is required to initiate translation at the FMDV AUG2 has not been determined either.
Individual members of type I and type II IRES elements are clearly distinct in their requirement for auxiliary mRNA-binding proteins, or ITAFs. Whereas all IRESs are strongly dependent on the pyrimidine tract binding protein (PTB) (Borovjagin et al. 1994; Hunt and Jackson 1999; Gosert et al. 2000; Pilipenko et al. 2000), the FMDV IRES additionally requires ITAF45 (Pilipenko et al. 2000), and poliovirus and rhinovirus IRESs require PCBP2 (Poly[rC]- binding protein 2) (Blyn et al. 1997) and unr (Boussadia et al. 2003). The stimulatory effect of La autoantigen on the translation of poliovirus RNA has also been repeatedly reported (Costa-Mattioli et al. 2004, and references therein). As for FMDV, whether or not PTB and ITAF45 are required to initiate translation not only at the AUG1 of FMDV RNA but also at the remote AUG2 has not been elucidated.
This study is mostly devoted to the analysis of the molecular mechanism of translation initiation at the AUG2 of the FMDV RNA. Using the method of reconstitution of 48S initiation complexes combined with toeprinting, we show that the initiation of translation at the downstream AUG codon of FMDV RNA does not occur in the absence of eIF1. Both PTB and ITAF45 strongly stimulate the assembly of the 48S initiation complex at the second AUG codon, although, according to our data, the initiation can proceed with only one of these ITAFs. We also demonstrate that the scanning factor eIF1 works against initiation at AUG1, thereby favoring initiation at the downstream start site. Additionally, we describe for the first time a new method—“kinetic toeprint assay.” This assay has enabled us to show a similar rate of accumulation of 40S subunits at the first and second AUGs and to present compelling evidence that the presence or absence of the proximal AUG (AUG1) does not affect the rate of 48S complex formation at the downstream start codon, in agreement with data in transfected cells. Finally, our results show that the rate of the 48S complex assembly directed by the FMDV IRES is a slow process relative to the 48S complex formation during the cap-dependent translation initiation.
RESULTS
Requirement for initiation factors eIF1 and eIF1A to assemble 48S translation initiation complexes at AUG1 and AUG2 of FMDV-Luc RNA
It has been reported that the reconstitution of 48S complexes proximal to the FMDV IRES initiation triplet requires factors eIF4F, eIF4A, eIF3, eIF2, PTB, and ITAF45, facilitated by eIF4B, but does not need scanning factors eIF1 and eIF1A (Pilipenko et al. 2000; Pestova et al. 1998a; Pestova and Kolupaeva 2002), which are present in the cell anyway. To determine the requirements for eIFs and ITAFs to assemble the 40S ribosomal subunit at the second (AUG2) initiation codon of FMDV RNA and compare them with those for AUG1 (AUG1 and AUG2 of the FMDV RNA are also referred to as the Lab and Lb initiation sites, respectively) (e.g., see Belsham and Jackson 2000), the same method of reconstitution of 48S complexes from purified components combined with toeprinting has been used (Pestova et al. 1996). As a basic model RNA construct, we employed the FMDV IRES (strain C-S8c1) linked to the luciferase (Luc) reporter gene at AUG2. The toeprints from the initiation at AUG1 and AUG2 were obtained using two different primers annealed to the FMDV-Luc sequences (see Materials and Methods). To analyze the 48S complex assembly at AUG1 only, we used the FMDV-Luc construct wherein AUG2 was mutated to AUA (see Materials and Methods). This allowed us to avoid possible interference by the 48S complex reconstitution at AUG2 with annealing the respective primer. Preliminary experiments showed that the native eIF4F could be substituted with the recombinant C-terminal fragment of eIF4G (p100) plus eIF4A (data not shown), and therefore p100 was used throughout all the experiments described in this study.
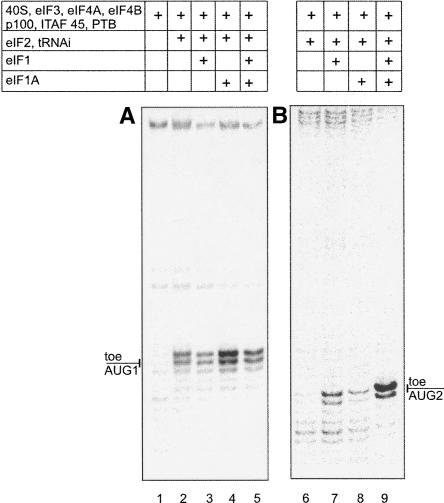
As shown in Figure 1A, the assembly of the 48S complex at AUG1 occurred when eIF1 and eIF1A were excluded from the reconstitution mixture (Fig. 1A, lane 2). When the reconstitution system was supplemented with both eIF1 and eIF1A (Fig. 1A, lane 5), the yield of 48S complexes did not change significantly, suggesting that these factors were dispensable for the assembly of the 48S complex at the AUG codon proximal to the IRES (AUG1), in accordance with observations published before (Pilipenko et al. 2000). However, addition of eIF1 alone led to inhibition of the 48S complex assembly, whereas in contrast addition of eIF1A produced a remarkable positive effect (Fig. 1A, cf. lanes 2, 3, and 4). As noted above, for the system supplemented with both factors, the yield of the 48S complex was similar to that revealed in their absence (Fig. 1A, cf. lanes 2 and 5). These data suggest opposite effects of eIF1 and eIF1A on the formation of 48S complexes at AUG1. A different situation was observed for AUG2. In this case (Fig. 1B), the presence of eIF1 was found to be critical for the assembly at AUG2 (Fig. 1B, lanes 6 and 7), and the yield of the 48S complex was strongly enhanced by a further addition of eIF1A (Fig. 1B, lane 9). However, eIF1A alone was not sufficient to promote complex formation (Fig. 1B, lane 8).
FIGURE 1.
Dependence of the 48S complex formation at (A) AUG1 and (B) AUG2 on the initiation factors eIF1 and eIF1A. Positions of toeprint signals (denoted on the left of the gels) were determined using dideoxynucleotide sequences generated with the same primers (not shown in this figure; see Fig. 4A,B).
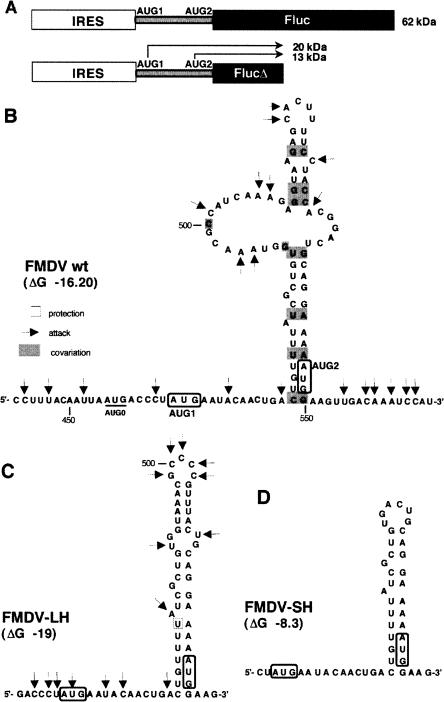
In order to test the influence of a stem–loop structure predicted to form in front of AUG2, which may cause a negative effect on the efficiency of start recognition dependent on the scanning model, the mutant constructs FMDV–LH and FMDV–SH were prepared using a truncated version of the luciferase reporter that unequivocally allowed us to distinguish initiation at AUG1 or AUG2 (Fig. 2A). The secondary structure of the spacers for the wild-type FMDV construct based on the chemical probing with DMS, RNase T2, and A partial digestion and phylogenetic covariation is shown in Figure 2B. It is of interest that several residues around AUG1 in the wild-type FMDV construct appeared accessible to nucleases and chemicals, whereas AUG2 was embedded in a U:A-rich stem–loop structure that was phylogenetically conserved and not accessible to attack by DMS or RNase T2. A large internal bulge was found around nucleotide 500 as deduced from both RNA attack and nucleotide sequence variability. The estimated internal energy of this RNA structure is −16.20 kcal/mol.
FIGURE 2.
(A) Schematic representation of the FMDV constructs used for translation assays in transfected cells to analyze simultaneously initiation at AUG1 and AUG2. (Empty rectangle) The FMDV IRES; (black rectangle) the luciferase gene fused to AUG2. Initiation at triplets AUG1 and AUG2, underlined on the sequence, gives rise to the synthesis of 20- or 13-kDa polypeptides, respectively, in constructs with a deleted version of the luciferase-coding region. (B) The sequence encompassing the first 84 nt of the FMDV polyprotein including the two functional initiation codons, AUG1 and AUG2. The nucleotide sequence of in-frame mutants (C) FMDV–LH and (D) FMDV-SH is also shown. (B–D) Secondary structures of the spacer region for the wild-type FMDV RNA and its mutants FMDV–LH and FMDV–SH, respectively. A transcript encompassing the entire IRES sequence extended to the second AUG was incubated in vitro with DMS or RNase A or T2, and then analyzed by primer extension using a 5′-end-labeled primer. (Arrows) Attacks were used in conjunction with (gray solid rectangles) the RNA phylogenetic conservation to represent the secondary RNA structure of (B) wild-type sequence, and (C) FMDV–LH. The secondary structure of FMDV–SH was built using computer modeling. The functional FMDV start codons AUG1 and AUG2 are boxed. Nucleotide numbering is used according to previous FMDV IRES literature (Fernandez-Miragall and Martinez-Salas 2003). The genetic variability in the region between the AUGs of 31 FMDV sequences deposited in databanks, belonging to serotypes C (12), O (nine), A (seven), and SAT (three), were used to derive the phylogenetic conservation of RNA structure. Compensatory nucleotide substitutions, consistent with base-pair formation, affected 16 bases (gray boxes in B); natural variation tolerated changes that converted G:C pairs to G.U or to A:U (in five positions), and U:A pairs to C:G or to U.G (in two positions). A strong tendency to covariate G492 and C500 to form a G.U pair was noticed. Independent nucleotide substitutions were also noticed in 15 positions, all located in loops in the secondary structure depicted in B.
A more stable stem–loop structure (ΔG, −19 kcal/mol) was observed in FMDV–LH (Fig. 2C). In this modified RNA the AUG1 region was more accessible than in FMDV–wt, and the stem–loop encompassing AUG2 was mostly base-paired. The least stable secondary structure (ΔG, −8.3 kcal/mol) was represented by the construct FMDV–SH, in accordance with RNA prediction models (Fig. 2D). As seen in Figure 3A, lanes 2 and 3, stabilization of the stem–loop structure formed by the FMDV–LH spacer resulted in a small (to 85%) but reproducible decrease of the yield of the 48S complex at AUG2. In contrast, shortening of this stem–loop structure showed some enhancement (to 125% of wild type) of the toeprint signal (Fig. 3A, lanes 2–4). The analysis of protein synthesis initiated from AUG1 and AUG2 in transfected BHK-21 cells (Fig. 3B), as well as in in vitro translation assays (data not shown), indicated that the stabilization of the spacer stem–loop structure resulted in an increase of the initiation at AUG1 (Fig. 3B, cf. lanes 1 and 2). The ratio of AUG1/AUG2 for FMDV RNA was 0.12, whereas that of the FMDV–LH construct was 0.74. This result is in accordance with observations that a stem–loop structure positioned downstream from the initiation codon stimulates the 48S complex formation at this codon (Kozak 1990). Some increase of the AUG1/AUG2 ratio was also observed for the FMDV–SH construct (Fig. 3B, lane 3). However, this increase was significantly smaller than that of the FMDV–LH RNA (0.44 versus 0.74). This effect may be accounted for by an improvement of the structural context of the AUG1 initiation site owing to a less stable downstream stem–loop (ΔG of the stem is −8.3 kcal/mol). Collectively, these data suggested that the selection of AUG2 involved the initiation factor eIF1 acting against AUG1 and in favor of AUG2. The scanning of the spacer is presumably accompanied by some unwinding of the stem–loop structure between the two start codons.
FIGURE 3.
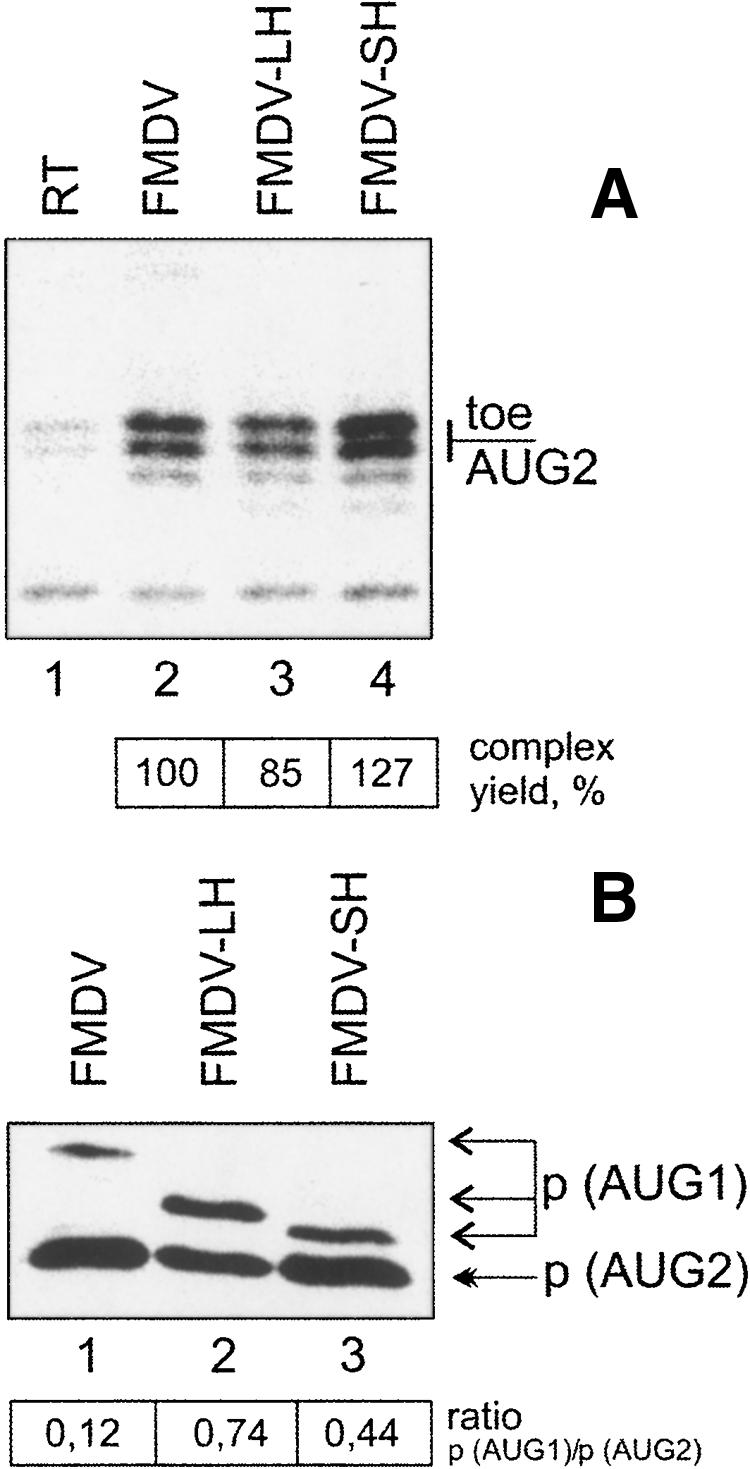
Effect of the secondary structure of the spacer region on the translation initiation at AUG1 and AUG2. (A) The gel showing toeprint assays in the reconstitution system from purified components. (Lane RT) No initiation factors added; (FMDV) control wild-type RNA. (B) Western blot assay of the luciferase truncated proteins schematically shown in Figure 2 synthesized in BHK-21 cells, 20 h post-transfection with the indicated constructs. (Three upper arrows) Point to the peptides (p) initiated at the AUG1 in FMDV wt, FMDV–LH, and FMDV–SH, respectively. (Solid arrow) The protein initiated at AUG2.
It should be noted that impairment of the eIF4G binding site in domain 4 of the FMDV IRES (Lopez de Quinto and Martinez-Salas 2000; Lopez de Quinto et al. 2001) by nucleotide substitution or deletion completely abrogated the assembly of the 48S complex at the downstream codon (data not shown). The same effect was observed when a deletion was introduced into the functionally important apical part of domain 3 of the FMDV IRES (Lopez de Quinto and Martinez-Salas 1997), suggesting that all elements of the FMDV IRES essential to initiate translation at AUG1 also were indispensable for the initiation at AUG2.
Translation initiation at the downstream AUG codon of FMDV RNA is strongly dependent on ITAFs
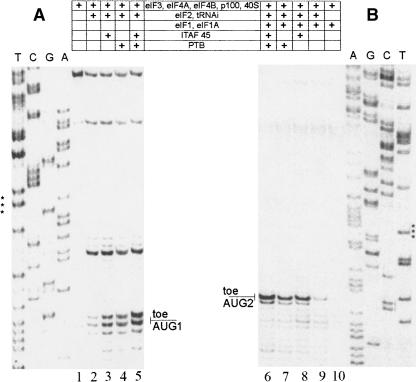
It has been shown that both PTB and ITAF45 are essential for formation of the 48S initiation complex at the AUG initiation triplet proximal to the FMDV IRES (Pilipenko et al. 2000). The ITAFs requirement for the initiation at the downstream start site has not been investigated before. Therefore, one could not rule out that these ITAFs, or one of them, were dispensable in forming the 48S complex at the second AUG separated from the first initiation triplet by 84 nt. As seen in Figure 4, A and B, PTB and ITAF45 caused an additive positive effect on the formation of the 48S complex at both AUGs. However, we observed a remarkable stimulation effect on the assembly of this complex at both start codons even when only one of these ITAFs was present in the incubation system. In contrast, in the earlier study (Pilipenko et al. 2000), the 48S toeprint was hardly seen when one of these factors was excluded from the reconstitution. It should be noted that the requirement of having either of them being present could not be compensated for by increasing the amount of the other ITAF in the reconstitution mixture (data not shown).
FIGURE 4.
Effect of PTB and ITAF45 on the reconstitution of 48S complexes at (A) AUG1 and (B) AUG2. Positions of the toeprint bands were determined using dideoxynucleotide sequences generated with the same respective primers (see Materials and Methods). These sequences are shown on the left and right of the gels, respectively.
Assembly of 48S translation initiation complexes at AUG1 and AUG2 of FMDV-Luc occurs with similar kinetics
The experiments described above present compelling evidence that to initiate translation at the downstream AUG codon of the FMDV RNA, 40S subunits first enter onto a common entry site within the FMDV IRES and then cross the spacer between AUG1 and AUG2 with participation of eIF1. As may be inferred from previously published data (Lopez de Quinto and Martinez-Salas 1999), the efficiency of translation initiation at AUG2 is little affected by the presence or absence of AUG1 in the FMDV RNA in either transfected cells or RRL (data not shown). However, according to the linear mode of the scanning process, AUG1 should be recognized first, followed by the binding of the 40S ribosome at the downstream initiation codon. The question arises as to whether the presence of AUG1 affects the kinetics of the reconstitution of the 48S complex at AUG2. To this end, we elaborated a kinetic toeprint approach. Aliquots were taken from the reconstitution mixture at defined time intervals and quenched by 30 mM Mg2+, followed by analysis of the yield of 48S complexes by primer extension inhibition. Preliminary experiments showed that at this concentration of magnesium a further assembly of 48S complexes was completely blocked, whereas primer extension reaction was not affected.
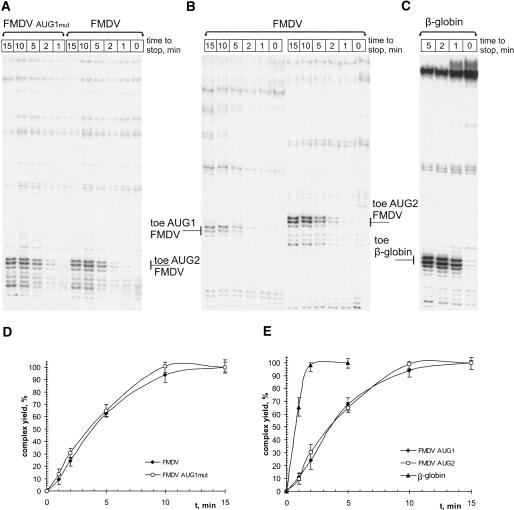
Using this approach, the rate of 48S complex reconstitution at the second AUG codon was compared for two FMDV-Luc constructs, one that contained both AUG codons and the other one with AUG1 mutated to AUA (Fig. 5A). To have balanced concentrations of initiation factors, 40S ribosomes and auxiliary proteins, the assembly of 48S complexes was performed in RRL, which efficiently initiates translation from both AUGs as it does in living cells (data not shown). The yield of 48S complexes determined from the toeprint assays was plotted against time (Fig. 5D). In addition, time courses of the assembly of 48S initiation complexes, separately for AUG1 and AUG2, were also obtained (Fig. 5B,E). No significant difference between the rate of assembly at AUG1 and AUG2 was observed, i.e., the presence of the upstream AUG did not cause any influence on the rate of the 48S complex formation on AUG2.
FIGURE 5.
Kinetic toeprint analysis in RRL of the 48S complex formation on AUG1 and AUG2 of FMDV-Luc and β-globin mRNA. (A) Gel showing toeprint bands corresponding to aliquots withdrawn from the same reconstitution mixture at different time points for the wild-type FMDV-Luc and the FMDV-Luc with the mutated AUG1 (AUG1→AUA). (B) Similar to A but for the FMDV-Luc constructs with either AUG1 or AUG2 mutated. (C) Similar to A and B but for β-globin mRNA. (D) Time course of formation of the 48S complex at AUG 2 of FMDV-Luc mRNAs (filled diamonds) with or (open circles) without AUG1. The plot was generated on the basis of the data presented in A. (E) Comparison of the time courses of the 48S complex accumulation at AUG1 and AUG2 of FMDV-Luc mRNA with that for the initiation codon of β-globin mRNA. The plot was created on the basis of the data presented in B and C. The points in the plots represent the average of several independent experiments. The standard deviation was within 10%.
Rates of the FMDV IRES- and cap-mediated assembly of the 48S translation initiation complex are remarkably different
The intriguing feature about the results presented in Figure 5, D and E, is that the reconstitution of 48S complexes at both AUG codons occurred with unexpectedly slow kinetics. Moreover, the rates of formation of 48S complexes at AUG1 and AUG2 were similar in spite of the quite distinct primary and secondary structure contexts of these start codons. In fact, a moderate increase in the yield of the 48S complex was still observed between 5 and 10 min of incubation. This result prompted us to compare the rate of 48S complex accumulation on the FMDV IRES with that for the cap-dependent β-globin mRNA. As follows from the data presented in Figure 5, C and E, the assembly of 48S complexes with the β-globin mRNA was complete after 2 min of incubation under the same ionic conditions (100 mM KCl, 1 mM Mg2+). It should be noted that the time courses shown in Figure 5 for the FMDV constructs did not change significantly by increasing the KCl concentration from 100 to 140 mM (i.e., more optimal for the translation of FMDV and EMCV RNAs in vitro). These results showed that at least in vitro the cap-dependent initiation is kinetically more advantageous over the IRES-mediated initiation.
DISCUSSION
In this study, we have investigated the mechanisms of FMDV IRES-directed translation initiation at two alternative start codons separated by the natural RNA sequence using the reconstitution of 48S complexes from purified components. In agreement with existing hypothesis (see Belsham and Jackson 2000), we present here compelling evidence that the selection of the downstream start codon involves the scanning factor eIF1. In some disagreement with previous reports, we have shown that both eIF1 and eIF1A affect the recognition of the AUG codon proximal to the FMDV IRES. Whereas eIF1A significantly stimulates the 48S complex formation at AUG1, eIF1 acts in the opposite way. On the other hand, eIF1 is indispensable for 48S assembly at AUG2. PTB and ITAF45 stimulate 48S complex formation at both AUG codons to an approximately similar extent. The positive effects of PTB and ITAF45 appear to be additive. In contrast to the earlier study (Pilipenko et al. 2000), a significant assembly of the 48S complex occurred when only one of these ITAFs was present in the reconstitution mixture. This discrepancy may be accounted for by the improved reconstitution system (Dmitriev et al. 2003b) employed in this study. For instance, instead of a large excess of the total tRNA where only tRNAi Met was aminoacylated, we have used the individual Met-tRNAi Met. As has been shown by Dmitriev et al. (2003b), a large excess of tRNA results in sequestering some canonical initiation factors and, presumably, mRNA-binding ITAFs as well, thereby reducing their active concentrations in the reconstitution mixture.
A striking feature about the assembly of 48S complexes at the FMDV IRES is that, at least in a cell-free system, it occurs with a rather slow rate as compared to the β-globin mRNA. Our preliminary experiments show that the same is true for the EMCV IRES. This may be accounted for by the fact that the formation of the 48S complex on the FMDV and presumably on other picornavirus IRES elements involves the accommodation of multiple mRNA-binding components, e.g., eIF4G, eIF3, eIF4B, PTB, and ITAF45. The binding of these factors may be accompanied by conformational transitions within the FMDV IRES structure. Consequently, the 48S complex assembly both at AUG1 and AUG2 of the FMDV RNA takes a remarkably longer time than on the AUG codon of the standard cap-dependent β-globin mRNA. This finding provides a mechanistic basis of why the translation initiation apparatus is preferentially recruited to cap-dependent rather than to IRES-driven mRNAs in living cells (Svitkin et al. 2005). It may also explain the various mechanisms evolved in picornaviruses that interfere with the function of eIF4E in infected cells.
Similar rates of assembly of 48S complexes at AUG1 and AUG2 separated by 84 nt suggest that scanning of the spacer by the 40S ribosomal subunit is a very fast process, which has no significant contribution to the overall rate of the 48S complex formation at the downstream AUG. Thus, the initiation at AUG2 does not need the 48S complex at AUG1 to be first formed to allow a scanning process to be initiated in search of the more optimal downstream AUG context. We hypothesize, therefore, that the selection of the downstream AUG occurs independently of the recognition of AUG1. We assume that different initiation regions may have distinct intrinsic probabilities to be positioned in the 40S ribosome mRNA-binding cleft, to accomplish the 48S assembly steps, and to be ultimately locked in the complex competent for the subsequent 80S complex formation. These probabilities may be determined not only by such parameters as the proximity of an AUG to the ribosome entry site or the immediate nucleotide context of the initiation triplet, but also by the structural organization of the initiation region. Unfortunately, except for the importance of positions −3 and +4 in an initiation region, we know very little about other features that determine the optimal structural context of the initiation codon. For instance, as we have previously demonstrated for the FMDV AUG1 nucleotide context, these features may be also affected by nucleotides at positions as far from the initiation triplet as −15 and +23 (Lopez de Quinto and Martinez-Salas 1999). Why the structural context of the downstream FMDV initiation codon is more powerful than that of AUG1 remains unclear. A separate investigation involving a detailed analysis of this region by point and deletion mutagenesis is required to clarify this issue. One may note several features of the region surrounding AUG2 that are not present in AUG1: more optimal A and G at positions −3 and +4, respectively; in the case of AUG2, a conserved A-stretch preceding AUG2; the single-stranded nature of the coding sequence downstream from AUG2; and finally, the existence of an unstable stem formed by U- and A-tracts encompassing AUG2.
Thus, our previous model in which initiation on the second AUG codon of the FMDV RNA does not depend on the presence of the first initiation triplet (Lopez de Quinto and Martinez-Salas 1999) has found strong mechanistic support in this study. The experimental strategy described in this study may be applied to other picornavirus IRESs as well, as soon as investigators succeed in assembling on them 48S complexes from purified components. The kinetic toeprint described here may also be useful for such a project.
MATERIALS AND METHODS
Generation of FMDV IRES constructs
All constructs encoded a monocistronic RNA encompassing the IRES of FMDV C-S8 extended to the second initiator AUG fused to the luciferase reporter gene (Lopez de Quinto and Martinez-Salas 1999). The nonfunctional AUG triplet located 8 nt upstream of AUG1 (AUG0) was removed in some constructs with no effect on the efficiency of translation initiation both in vitro and in vivo. Constructs AUG0, AUG1, and AUG2 carrying a single substitution of each AUG to AUA were performed by PCR using sense oligonucleotides 5′-AATTAATAACCCTATGAATACAAC-3′, 5′-CCTATAAATACAACTGACTGTTTTATC-3′, or 5′-CGGACTGCAGGAAAAATAGAAGTTGAC-3′, respectively, in combination with external primers. Constructs AUG0 and AUG1 were generated by insertion of a HindIII–PstI fragment in pFMDVluc; construct AUG2 was generated using PstI and EcoRV restriction sites. Combinations of AUG0 with AUG1 and/or AUG2 were performed by exchanging appropriate HindIII–PstI fragments.
To generate constructs FMDV–LH and FMDV–SH (Fig. 2) containing in-frame nucleotide substitutions in the spacer region between AUGs, the antisense primers 5′-CACTCCTGCAGTAAACGGGGCGTTTACCAC-3′ and 5′-GCGTCCTGCAGTCACAGCGATAAAACAGTCAG-3′, respectively, were used in a double PCR reaction with external primers (Fernandez-Miragall and Martinez-Salas 2003). The modified products were then inserted in pFMDVluc via HindIII and PstI restriction sites.
The nucleotide sequence of the entire length of each cDNA under study was determined, using automatic sequencing (ABI PRISM dye terminator cycle sequencing ready reaction kit; Perkin-Elmer).
For studies aimed at verifying the translation initiation codon, all FMDV mutants were also inserted in a monocistronic plasmid expressing a short version of the luciferase open reading frame (Lopez de Quinto and Martinez-Salas 1999), which allowed the synthesis of peptides of ∼20 and 13 kDa, corresponding to protein synthesis initiation at AUG1 or AUG2, respectively (see Figs. 2A, 3B). For FMDV–LH or FMDV–SH, initiation at AUG1 resulted in polypeptides of ∼16 and 14 kDa, whereas initiation at AUG2 remained unaffected.
In vitro transcription
Prior to RNA synthesis, plasmids were linearized to generate monocistronic transcripts encoding the full-length IRES extended to the second initiator AUG upstream of the complete or a truncated version of luciferase using restriction enzymes BbuI or Hpa1, respectively. Transcription was performed for 1 h at 37°C using the Megashortscript kit (Ambion) as recommended by the manufacturer. The transcripts for toeprint analysis were obtained by T7 transcription from the corresponding plasmids linearized with MfeI.
RNA structure probing
For RNA structure mapping, transcripts encompassing the entire FMDV IRES extended to the second AUG were prepared from monocistronic plasmid DNA linearized with BbuI restriction enzyme. About 1.5 μg of each RNA was treated in buffers N (50 mM sodium cacodylate at pH 7.5, 300 mM KCl, 10 mM MgCl2), N′ (100 mM KCl, 2 mM MgAc2, 20 mM HEPES at pH 7.5, 0.1 mM EDTA), or D (10 mM Tris-HCl at pH 8, 1 mM EDTA) with DMS using 1 μL of a freshly made 1:5 dilution in EtOH for 15 min at 20°C (Brunel and Romby 2000). For enzymatic probing, transcripts were treated with RNase T2 (0.05 U/μL; 20 min at 37°C), RNase A (5 × 10−6 μg/μL, 30 min at 37°C) (Fernandez-Miragall et al. 2006). Untreated RNAs were used as negative controls.
A 5′-end-labeled oligonucleotide (5′-CCCGGGTGTGGGTACC-3′ AUG2as) was used in a primer extension reaction to analyze chemical and enzymatic attacks. Template RNA (1.5 μg) was denatured for 3 min at 95°C and cooled down on ice. Annealing and extension of the labeled antisense primer were carried out in 15 μL of reverse transcriptase (RT) buffer (20 mM Tris-HCl at pH 7.5, 15 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.01% [v/v] NP-40, 50% [v/v] glycerol, 1 mM each dNTP in the presence of 100 U of de SuperScript II RT), during 1 h at 45°C. The RNA template was then hydrolyzed, and the RT-extension products were fractionated in denaturing 6% acrylamide, 7 M urea gels as described (Fernandez-Miragall and Martinez-Salas 2003). To prepare the sequence ladder, the Thermo sequenase cycle sequencing kit (Amersham) was used with the same 5′-labeled antisense oligonucleotide used for primer extension.
In vivo IRES activity
Monocistronic FMDV constructs were used to transfect 80%–90% confluent BHK-21 monolayers, 1 h after infection with the vaccinia virus recombinant vT7F-3 expressing the T7 RNA polymerase. Extracts from 1–2 × 105 cells were prepared 20 h post-transfection in 100 μL of 50 mM Tris-HCl (pH 7.8), 120 mM NaCl, and 0.5% NP-40 as described (Lopez de Quinto and Martinez-Salas 1997). Experiments were performed on duplicate wells, and each experiment was repeated at least three times. IRES activity was quantified in two ways: first, as the expression of firefly luciferase activity measured in a luminometer (Berthold) normalized to the protein content in the extract calibrated by Bradford (Bio-Rad); and second, as the intensity of luciferase protein detected in a Western blot performed with equal amounts of protein loaded in 15% SDS-gels, stained with a specific rabbit polyclonal luciferase antibody (1:2000) (Cortex), followed by the appropriate secondary antibody and ECL staining (Pierce).
Assembly and analysis of 48S translation initiation complexes
The isolation of translation initiation components, the reconstitution of 48S complexes from purified components, and their analysis by toeprinting were performed as described (Pestova et al. 1996, 1998a,b; Dmitriev et al. 2003a,b) with one significant modification: individual initiator tRNAi Met from Escherichia coli was used instead of the total mammalian tRNA in which only tRNAi Met was aminoacylated. The recombinant PTB and ITAF45 were isolated as described by Pestova et al. (1996) and Pilipenko et al. (2000), respectively. To assemble 48S complexes in RRL, the incubation mixture (the final volume was 10 μL), containing 5 μL of RRL (Promega) and 1 unit of RNase inhibitor Ribolock (Fermentas), was adjusted to 20 mM HEPES/KOH (pH 7.5), 1 mM DTT, 0.5 mM spermidine-HCl, 100 mM KAc, 2 mM GMPPNP, and 1 mM MgAc2. The mixture was incubated for 5 min at 30°C, followed by addition of 0.5 pmol of mRNA and further incubation for 10 min at 30°C. The formation of complexes was stopped by addition of MgAc2 to 30 mM. To perform kinetic toeprint assays, a 5–10-fold larger volume of RRL was supplied with the same components except mRNA, and the mixture was incubated for 5 min at 30°C. Then, the proportionally increased amount of mRNA was added, and aliquots (10 μL) were taken out from this mixture at defined time points. The reconstitution reaction was immediately “quenched” by transferring the aliquots to 30 mM of MgAc2. The reverse transcription reaction and analysis of the resulting cDNAs were performed using denaturing 6% PAGE as described before (Dmitriev et al. 2003b). Radioactive bands were visualized with a PhosphorImager (Molecular Dynamics). The yield of 48S and 80S complexes was determined as the ratio of the radioactivity in the toeprint bands compared to the total radioactivity in the corresponding lanes, including the toeprint bands, using Image-Quant 5.0 software.
To obtain toeprints from AUG1 and AUG2, primer 5′-CCTGCAGTCCGTGGTAGG-3′ complementary to the spacer at positions 62–80 nt downstream from AUG1 and primer 5′-TGCAGTTGCTCTCCAGC-3′ complementary to the FLuc sequence positioned 63 nt downstream from AUG2 were used, respectively. The toeprinting of β-globin mRNA was performed with the primer annealed at positions 60–80 downstream from the start codon of this mRNA.
ACKNOWLEDGMENTS
We thank E. Pilipenko for kindly providing us with the ITAF 45 expression plasmid. This work was supported by grants RFBR 05-04-49704 to I.N.S., BFU-2005-00948 to E.M.-S., and by an Institutional grant from Fundación Ramón Areces to CBMSO.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.469707.
REFERENCES
- Belsham, G.J. Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 1992;11:1105–1110. doi: 10.1002/j.1460-2075.1992.tb05150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham, G.J., Jackson, R.J. Translation initiation on picornavirus RNA. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 869–900. [Google Scholar]
- Belsham, G.J., Martinez-Salas, E. Genome organization, translation, and replication of foot-and-mouth disease virus RNA. In: Domingo E., Sobrino F., editors. Foot-and-mouth disease: Current perspectives. Horizon Scientific Press; Norwich, UK: 2004. pp. 19–52. [Google Scholar]
- Blyn, L.B., Towner, J.S., Semler, B.L., Ehrenfeld, E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovjagin, A., Pestova, T., Shatsky, I. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 1994;351:299–302. doi: 10.1016/0014-5793(94)00848-5. [DOI] [PubMed] [Google Scholar]
- Brunel, C., Romby, P. Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol. 2000;318:3–21. doi: 10.1016/s0076-6879(00)18040-1. [DOI] [PubMed] [Google Scholar]
- Boussadia, O., Niepmann, M., Creancier, L., Prats, A.-C., Dautry, F., Jacquemin-Sablon, H. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 2003;77:3353–3359. doi: 10.1128/JVI.77.6.3353-3359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chard, L.S., Bordeleau, M.E., Pelletier, J., Tanaka, J., Belsham, G.J. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J. Gen. Virol. 2006;87:927–936. doi: 10.1099/vir.0.81546-0. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli, M., Svitkin, Y., Sonenberg, N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 2004;24:6861–6870. doi: 10.1128/MCB.24.15.6861-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev, S.E., Pisarev, A.V., Rubtsova, M.P., Dunaevsky, Y.E., Shatsky, I.N. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 2003a;533:99–104. doi: 10.1016/s0014-5793(02)03776-6. [DOI] [PubMed] [Google Scholar]
- Dmitriev, S.E., Terenin, Y.M., Dunaevsky, Y.E., Merrick, W.C., Shatsky, I.N. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol. Cell. Biol. 2003b;23:8925–8933. doi: 10.1128/MCB.23.24.8925-8933.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Miragall, O., Martinez-Salas, E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Miragall, O., Ramos, R., Ramajo, J., Martinez-Salas, E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert, R., Chang, K.H., Rijnbrand, R., Yi, M., Sangar, D.V., Lemon, S.M. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 2000;20:1583–1595. doi: 10.1128/mcb.20.5.1583-1595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, S.L., Jackson, R.J. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Martinez-Salas, E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Martinez-Salas, E. Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology. 1999;255:324–336. doi: 10.1006/viro.1999.9598. [DOI] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Martinez-Salas, E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo . RNA. 2000;6:1380–1392. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Lafuente, E., Martinez-Salas, E. IRES interaction with translation initiation factors: Functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7:1213–1226. doi: 10.1017/s1355838201010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas, E., Fernandez-Miragall, O. Picornavirus IRES: Structure function relationship. Curr. Pharm. Des. 2004;10:3757–3767. doi: 10.2174/1381612043382657. [DOI] [PubMed] [Google Scholar]
- Martinez-Salas, E., Ramos, R., Lafuente, E., Lopez de Quinto, S. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 2001;82:973–984. doi: 10.1099/0022-1317-82-5-973. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U., Shatsky, I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Borukhov, S.I., Hellen, C.U.T. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998a;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J., Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 1998b;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G., Lomakin, I.B., Pilipenko, E.V., Shatsky, I.N., Agol, V.I., Hellen, C.U.T. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Pestova, T.V., Kolupaeva, V.G., Khitrina, E.V., Poperechnaya, A.N., Agol, V.I., Hellen, C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes & Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- Pisarev, A.V., Chard, L.S., Kaku, Y., Johns, H.L., Shatsky, I.N., Belsham, G.J. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y.V., Herdy, B., Costa-Mattioli, M., Gingras, A.C., Raught, B., Sonenberg, N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]