Abstract
Repair of oxidative damage to DNA bases is essential to prevent mutations and cell death. Endonuclease III is the major DNA glycosylase activity in Escherichia coli that catalyzes the excision of pyrimidines damaged by ring opening or ring saturation, and it also possesses an associated lyase activity that incises the DNA backbone adjacent to apurinic/apyrimidinic sites. During analysis of the area adjacent to the human tuberous sclerosis gene (TSC2) in chromosome region 16p13.3, we identified a gene, OCTS3, that encodes a 1-kb transcript. Analysis of OCTS3 cDNA clones revealed an open reading frame encoding a predicted protein of 34.3 kDa that shares extensive sequence similarity with E. coli endonuclease III and a related enzyme from Schizosaccharomyces pombe, including a conserved active site region and an iron/sulfur domain. The product of the OCTS3 gene was therefore designated hNTH1 (human endonuclease III homolog 1). The hNTH1 protein was overexpressed in E. coli and purified to apparent homogeneity. The recombinant protein had spectral properties indicative of the presence of an iron/sulfur cluster, and exhibited DNA glycosylase activity on double-stranded polydeoxyribonucleotides containing urea and thymine glycol residues, as well as an apurinic/apyrimidinic lyase activity. Our data indicate that hNTH1 is a structural and functional homolog of E. coli endonuclease III, and that this class of enzymes, for repair of oxidatively damaged pyrimidines in DNA, is highly conserved in evolution from microorganisms to human cells.
Reactive oxygen species generated either as a by-product of cellular respiration or by ionizing radiation and other oxidizing agents can cause extensive damage to DNA bases and disrupt the phosphodiester backbone. Unrepaired oxygen radical-derived DNA lesions have been implicated as a causative factor in both cancer and aging (1, 2). Repair of most forms of oxidative DNA damage is mediated via the base excision-repair pathway (3, 4), which has been well characterized in Escherichia coli. One of the key components of the base excision-repair pathway in that organism is endonuclease III, the product of the nth gene (5). The crystal structure of this iron/sulfur protein has been determined recently at 1.85 Å resolution (6, 7).
Although originally isolated as an activity that incised at certain sites of DNA damage generated by ultraviolet (UV) irradiation in a Mg2+-independent reaction (8), endonuclease III was subsequently shown to act primarily as a DNA glycosylase that hydrolyses the N-glycosyl bond linking damaged pyrimidines to the DNA backbone. Numerous different substrates for endonuclease III have been identified representing saturated, opened, or contracted pyrimidine rings, such as thymine and cytosine glycol, urea, and N-substituted urea, 5-hydroxy-5-methylhydantoin, and 5,6-dihydrothymine (9–11). The originally observed DNA nicking activity associated with endonuclease III was subsequently shown to result not from hydrolytic cleavage of phosphodiester bonds, but from β-elimination of the 3′ phosphate at the site of the excised damaged base, and this activity has been termed apurinic/apyrimidinic (AP) lyase (12). Endonuclease III can also incise the DNA backbone at AP sites generated chemically or by other DNA glycosylases in vitro, but a physiological relevance for this activity has yet to be established.
Gene products representing functional homologs of E. coli endonuclease III have been identified in several species, including other bacteria and lower eukaryotes (see ref. 13 and Fig. 1 for details), suggesting that this class of enzymes performs an important role in DNA repair. However, mutants of E. coli lacking endonuclease III activity are fully viable and are not hypersensitive to DNA damaging agents that generate oxidized bases (5). The possible explanation for this apparent contradiction is that E. coli shows extensive functional redundancy for many of the enzymatic activities required for base excision-repair. In this case, the action of the related endonuclease VIII and/or other glycosylases might compensate for loss of endonuclease III (14). Evidence that this family of glycosylases/AP lyases is required for repair of oxidative DNA damage in eukaryotes comes from the finding that deletion of the gene encoding an endonuclease III homolog in Saccharomyces cerevisiae, NTG1, confers hypersensitivity to H2O2 and menadione (15).
Figure 1.
A comparison of the sequences (species code and GenBank accession no. in brackets) of endonuclease III-like proteins of human (hNTH1), S. pombe (Nth-Spo; Q09907), Caenorhabditis elegans (Ce; Z50874), Bacillus subtilis (Bs; P39788), and E. coli (Ec; P20625); UV N-glycosylase from Micrococcus luteus (Ml; P46303) and Mut Y of E. coli (Ec; P17802). The cysteine residues (Cys-Xaa6-Cys-Xaa2-Cys-Xaa5-Cys) involved in binding the [4Fe-4S] cluster are marked with asterisks. The S. pombe endonuclease III homolog, Nth-Spo (13), has the last cysteine displaced by two residues. The highly conserved helix–hairpin–helix motif, which is thought to interact with DNA, is indicated. The hairpin consists of GVG, usually flanked by P. The proposed active site lysine residue is indicated by a solid triangle and the conserved aspartic acid is marked with a dot. The potential nuclear targeting sequences in the N-terminal domain of the human protein are underlined. Positions with a majority of identical residues are boxed (black) and shown in the consensus. Amino acids similar to the consensus are shaded grey. The C-terminal regions that extend beyond the termini of the human and E. coli proteins are not shown.
During a search for the tuberous sclerosis gene (TSC2) on human chromosome 16, a number of genes located in regions deleted in one or more affected patients were identified. In addition to the gene subsequently shown to be TSC2 (16) and the polycystic kidney disease gene (PKD1) that lies immediately proximal to TSC2 (17), two genes lying immediately distal to TSC2 were partially characterized (16). We show here that one of these genes, OCTS3 (which lies next to TSC2 in a head-to-head orientation), encodes a human homolog of E. coli endonuclease III. We have overexpressed the human protein (designated hNTH1) in E. coli and purified it to apparent homogeneity. We show that hNTH1 possesses DNA glycosylase activity against a range of oxidized pyrimidines, as well as an associated AP lyase activity. Our results indicate that hNTH1 is a functional homolog of E. coli endonuclease III.
MATERIALS AND METHODS
Enzymes.
Nth protein from E. coli was prepared using an overexpressing strain λN99/pHIT1 (18) and was kindly provided by Drs. L. Vilpo and R. D. Wood (Imperial Cancer Research Fund, Clare Hall Laboratories, London). The homologous Schizosaccharomyces pombe enzyme, Nth-Spo, was produced in an E. coli overexpression system and purified as described previously (13). Human AP endonuclease 1 (HAP1) protein was purified to homogeneity as reported previously (19). Human uracil-DNA glycosylase was a gift from Dr. G. Slupphaug (University of Trondheim, Norway). E. coli Fpg protein was isolated from an overproducer strain (20). Urease was purchased from Boehringer Mannheim.
Isolation and Sequencing of cDNA Clones Encoding hNTH1.
An OCTS3 cDNA in pCDM8 was isolated from a library prepared from HT29 (colon adenocarcinoma) RNA using standard procedures. Anchor-ligated human fetal brain cDNA (Marathon Ready cDNA, CLONTECH) was used for 5′ rapid amplification of cDNA ends (RACE), according to the manufacturer’s instructions using the specific primer OC2 (5′-TCAGCATCCTCGCGCTCAAGGCGGTCAT-3′) and an anchor primer. Products were subcloned into GEM-T (Promega). Reverse transcription–PCR to confirm the 5′ RACE was performed with the primers O3/EL1 (5′-ATGGGCCGCCGGGATGTG-3′) and O3/EL2 (5′-GCCACACGCAGTCTCTGTG-3′) using lymphoblast cDNA and the following PCR conditions: 31 cycles of 94°C for 1 min; 64°C for 1 min; and 72°C for 1 min; and then once at 72°C for 10 min. Products were cloned into pZErO (Invitrogen). Nucleotide sequencing was with an Applied Biosystems model 373A or model 377 sequencer using cycle sequencing with AmpliTaq FS Ready Mix (Perkin–Elmer). For Northern blot analysis, a CLONTECH human multiple tissue blot was used containing 2 μg of tissue mRNA per gel lane.
Overexpression and Purification of hNTH1 Protein.
The cDNA encoded by the OCTS3 gene was amplified from pCDM8/OCTS3 using the following primers that incorporated XhoI sites (underlined) to permit cloning into pET14b (Novagen). PCR conditions were 10 cycles of 92°C, 30 s; 55°C, 45 s; 72°C, 1 min. The primers were as follows: 5′ primer, AGA TTC CTC GAG ACC GCC TTG AGC GCG AGG; 3′ primer, AGA TTC CTC GAG TCA GAG ACC CTG GGC GGC.
The pET-14b/hNTH1 construct was transformed into BL21.pLysS cells, an E. coli strain incorporating a chromosomally integrated T7 polymerase gene under the control of the lac promoter. Expression of hNTH1 protein was induced for 2 hr by the addition of 0.4 mM isopropyl β-d-thiogalactoside (final concentration) to bacterial cultures. Following cell lysis, more than 90% of the recombinant hNTH1 protein was found in the soluble fraction. This fraction was loaded onto a phosphocellulose P11 column and the bound proteins were eluted with a 100–1250 mM NaCl gradient. Fractions containing the hNTH1 protein (eluting at ≈700 mM NaCl) were pooled and dialyzed against binding buffer (5 mM imidazole/500 mM NaCl/20 mM Tris·HCl, pH 7.9). The dialyzed protein was loaded onto a Ni2+-charged His.Bind resin column (Novagen), the column was washed, and bound proteins were eluted with elution buffer (1 M imidazole/500 mM NaCl/20 mM Tris·HCl, pH 7.9). The purified protein was kept at −70°C in storage buffer (50% glycerol/125 mM NaCl/20 mM Tris·HCl, pH 7.9).
Preparation of DNA Substrates.
Double-stranded polydeoxyribonucleotides containing scattered 14C-labeled thymine glycol or urea residues were prepared by treatment of single-stranded polymers with OsO4 or KMnO4 before annealing with a complementary strand as described previously (13). To prepare an oligonucleotide substrate containing a single AP site, a 43 base oligonucleotide containing a single uracil residue was annealed to a complementary 35-mer. The annealed DNA was purified by polyacrylamide gel electrophoresis and end-labeled using T4 polynucleotide kinase. The polymer was incubated with human uracil DNA glycosylase to create an AP site as described (21).
Enzyme Assays.
DNA glycosylase assays with polynucleotides containing 14C-labeled thymine glycol or urea residues were performed as described previously (13). Release of 8-hydroxyguanine from a double-stranded oligonucleotide by DNA glycosylase activity was measured by electrochemical detection. For AP site cleavage assays, the end-labeled oligonucleotide substrate was incubated at 37°C with purified HAP1 (19), E. coli endonuclease III, or hNTH1 protein in a reaction buffer containing 66 mM Tris·HCl (pH 7.5), 5 mM MgCl2, and 1 mM 2-mercaptoethanol. The reaction was stopped by addition of formamide gel loading buffer (80% formamide/1 mM EDTA, pH 8.0/0.1% bromophenol blue/0.1% xylene cyanol). Samples were denatured and run on a 12% denaturing polyacrylamide gel.
RESULTS
Isolation and Characterization of the OCTS3 cDNA.
Previous studies identified a gene, designated OCTS3, lying immediately adjacent to and in a 5′-to-5′ orientation with TSC2 in human chromosome region 16p13.3 (16). As the OCTS3 gene is constitutionally deleted in a number of patients with tuberous sclerosis (refs. 16 and 22; unpublished work), we decided to characterize this gene further. A near full-length cDNA, OCTS3C, of 1028 bp was isolated by hybridizing the genomic clone, CW24, which contains the 5′ ends of both OCTS3 and TSC2 (16), to a cDNA library. RACE experiments indicated that the 5′ end of the OCTS3 gene may lie 33 bp upstream of the 5′ limit of OCTS3C, and this was confirmed by analysis of an reverse transcription–PCR product of 212 bp (nucleotides 7–219 of the full transcript) that extends over an intron/exon boundary. Sequencing of the full-length transcript of 1061 bp revealed that the longest open reading frame from the first potential initiation codon was 936 bp (nucleotides 20–955). Interestingly, two additional in-frame methionines are close to the predicted translation start site (positions 9 and 16; Fig. 1), each associated with a partial, but not perfect, Kozak consensus sequence, raising the possibility of alternative translational starts for the protein.
The OCTS3 Gene Encodes a Homolog of E. coli Endonuclease III.
The predicted product of the OCTS3 gene is 312 amino acids in length with a calculated molecular mass of 34,300 Da and a pI of 10.1. Comparison of this amino acid sequence with protein sequences in the data bases revealed a high level of sequence similarity with prokaryotic and eukaryotic endonuclease III-like proteins, and therefore the OCTS3 gene product was designated hNTH1 (human endonuclease III homolog 1). Fig. 1 shows an alignment of the predicted hNTH1 amino acid sequence with those of other members of the endonuclease III family of repair enzymes, including a recently characterized S. pombe endonuclease III homolog, Nth-Spo (13). Notable features of the hNTH1 sequence and its homologs are the presence of four highly conserved cysteine residues comprising a binding motif for the [4Fe-4S] cluster, and a helix–hairpin–helix motif that has been shown in the case of E. coli endonuclease III to contribute to DNA binding of the enzyme (6). Several other conserved blocks of amino acids are also evident. It should be noted that the amino acid sequence of hNTH1 has an N-terminal domain of 98 amino acids that is not conserved in E. coli endonuclease III and that contains a possible nuclear targeting sequence (Fig. 1).
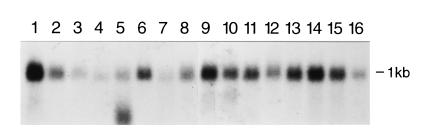
Northern blot analysis showed that the OCTS3 gene encodes a mRNA of approximately 1.0 kb that is expressed in all human tissues analyzed (Fig. 2). The level of expression varied severalfold between different tissues with the highest level present in heart and the lowest in lung and kidney. A possible alternatively spliced form of ≈0.5 kb was detected in liver.
Figure 2.
Northern blot analysis of mRNA (2 μg) from human tissues. Lanes: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas; 9, spleen; 10, thymus; 11, prostate; 12, testis; 13, ovary; 14, small intestine; 15, colon; 16, leukocytes. The ubiquitously expressed OCTS3 mRNA is ≈1.0 kb. The smaller product (≈0.5 kb) present only in liver may be due to alternative splicing or specific degradation (although other probes detected no degradation of the RNA in this sample).
Expression of hNTH1 in E. coli and Purification of the Recombinant Protein.
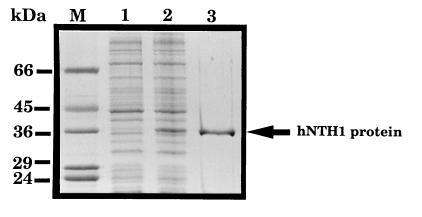
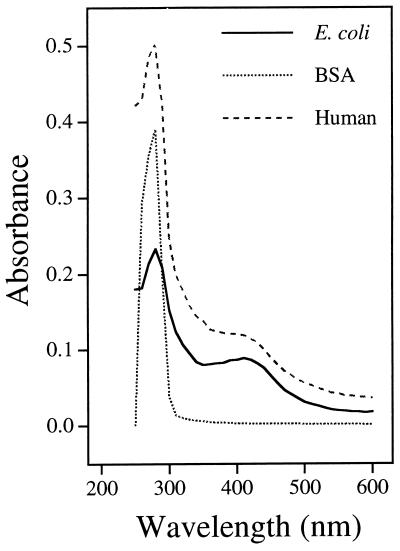
The OCTS3 cDNA (initiating at Met-9) was subcloned into pET14b, and the hexahistidine-tagged hNTH1 protein was overexpressed in E. coli and purified to apparent homogeneity by a combination of phosphocellulose chromatography and affinity chromatography on a nickel chelate column. The purified hNTH1 protein had an apparent Mr of 36,000 on SDS/polyacrylamide gels (Fig. 3), in close agreement to its molecular mass predicted from the cDNA sequence. Consistent with the presence of an iron/sulfur cluster within hNTH1, the purified protein had a brown coloration and an absorption spectrum with a peak at ≈415 nm (Fig. 4). E. coli endonuclease III (Fig. 4) and the Nth-Spo enzyme (data not shown) had similar absorption spectra.
Figure 3.
Purification of recombinant hNTH1 protein. A Coomassie blue-stained SDS/polyacrylamide gel is shown of a lysate of BL21 cells containing the pET14b/hNTH1 plasmid before (lane 1) and after (lane 2) induction of hNTH1 protein expression by isopropyl β-d-thiogalactoside. The hNTH1 protein eluting from the nickel-chelate column in the presence of 1 M imidazole is in lane 3, and is indicated on the right. Lane M contains molecular mass standards (Sigma) as indicated on the left.
Figure 4.
Absorption spectra in the 415 nm range of purified E. coli endonuclease III and hNTH1 proteins. BSA is shown as a control.
DNA Glycosylase Activity of hNTH1 on DNA Substrates Containing Oxidized Pyrimidine Residues.
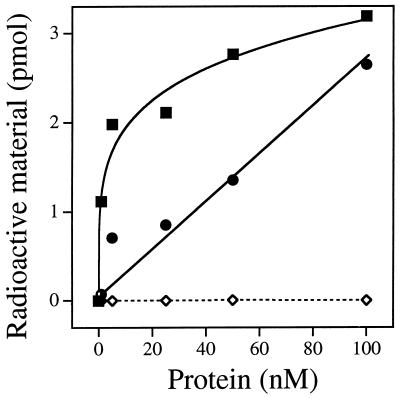
E. coli endonuclease III possesses DNA glycosylase activity on a wide variety of oxidized pyrimidines. The purified hNTH1 protein was tested for its ability to excise thymine glycol and urea residues from double-stranded polydeoxyribonucleotide substrates. Fig. 5 shows that hNTH1 could liberate OsO4-induced thymine glycol lesions, and KMnO4-induced urea and N-substituted urea lesions from double-stranded polynucleotides. As noted previously for the Nth-Spo enzyme (13), ring-fragmented pyrimidines were more efficiently released than thymine glycol by the hNTH1 protein. The major thymine product after KMnO4 treatment of polymers is unsubstituted urea, and 60% of the ethanol-soluble, 14C-labeled material released by hNTH1 was converted to a volatile form by urease treatment, confirming that the base derivative was released in free form by a DNA glycosylase activity. Two-dimensional thin layer chromatography was used to confirm that the product released from the OsO4-treated substrate was thymine glycol (data not shown). hNTH1 was unable to release the oxidized purine derivative 8-hydroxyguanine from a double-stranded oligonucleotide, whereas the E. coli Fpg protein efficiently catalyzed this reaction (data not shown).
Figure 5.
Enzymatic release of urea and thymine glycol by the DNA glycosylase activity of the hNTH1 protein. A poly(dA, [2-14C]dT) polymer was treated with KMnO4 (▪) or OsO4 (•) or was left untreated (◊), and then annealed with a complementary poly(dT) strand before use as enzyme substrate. The DNA was incubated at 37°C with increasing amounts of hNTH1 protein for 30 min under standard conditions. Radioactive material in an aliquot (300 μl) of the ethanol-soluble fraction was measured by scintillation counting.
hNTH1 Protein Has AP Lyase Activity.
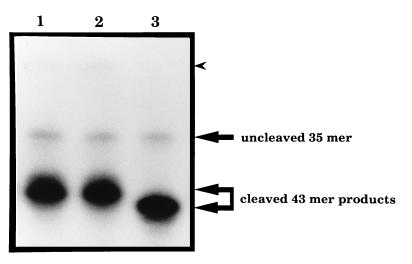
A double-stranded oligonucleotide substrate containing a single AP site in a defined position was prepared by excision of a uracil residue from one strand of the oligonucleotide using uracil-DNA glycosylase. The purified hNTH1 protein exhibited site-specific incision activity on this substrate that occurred only when an AP site was present (data not shown). To confirm that this incision occurred on the 3′ side of AP sites, in the manner of endonuclease III, and not on the 5′ side as per the major hydrolytic AP endonuclease from human cells, HAP1, the products generated by AP site incision by hNTH1, E. coli endonuclease III and HAP1 were compared on acrylamide gels. Fig. 6 shows that the products of oligonucleotide cleavage by hNTH1 and endonuclease III were identical, and different from that generated by HAP1, consistent with hNTH1 incising 3′ to an AP site. Also consistent with hNTH1 acting as an AP lyase, and with the hNTH1 preparation being free of contamination with a hydrolytic AP endonuclease, hNTH1 was shown to lack incision activity on a DNA substrate containing reduced AP sites generated by exposure to NaBH4 (data not shown).
Figure 6.
AP lyase activity of hNTH1. Analysis of the products of the AP site incision activity of hNTH1 (lane 1), E. coli endonuclease III (lane 2), and HAP1 (lane 3) proteins. The positions of the uncleaved 35-mer oligonucleotide, and the products of cleavage of the 43-mer containing a single AP site are indicated on the right. The unannotated arrow head indicates the position where the uncleaved 43-mer would migrate.
The hNTH1 protein was also tested for incision activity on UV- and gamma-irradiated plasmid DNA. The results were indistinguishable from those obtained with the Nth-Spo enzyme on the same substrates (13), and demonstrated that hNTH1 possesses an activity that could incise the backbone of irradiated DNA, but not control unirradiated DNA (data not shown). These incisions most likely occurred at radiation-induced cytosine hydrates in the DNA substrate (23).
DISCUSSION
The gene encoding hNTH1, a human homolog of E. coli endonuclease III, has been identified as OCTS3, a previously unassigned gene lying adjacent to the TSC2 gene on chromosome 16 (16). The OCTS3 gene encodes a 1.0-kb mRNA that is apparently expressed in a wide range of different tissues, as would be expected of a gene encoding a DNA repair enzyme required to counteract the toxic effects of pyrimidine oxidation brought about by endogenous and exogenous DNA damaging species. It may be significant that expression of OCTS3 mRNA is elevated in heart tissue, since the high rate of ATP generation in this tissue is associated with persistent oxidative stress. Because reactive oxygen species are generated at a high rate by normal cellular processes, such as respiration, as well as by ionizing radiation, it seems likely that hNTH1 is an important component of the cellular defense machinery for counteracting cytotoxic and mutagenic damage to the genome.
The predicted sequence of hNTH1 contains a number of motifs that have been conserved in evolution from bacteria through fission yeast to human. Most notable is the Cys-Xaa6-Cys-Xaa2-Cys-Xaa5-Cys motif that binds the iron/sulfur cluster. This domain in the endonuclease III protein has been shown to play a role in recognition of DNA, but not directly in catalysis (6). Interestingly, the S. cerevisiae homolog, NTG1, does not contain this motif, although it recognizes similar DNA damage (15). Based upon a crystallographic analysis of endonuclease III, the highly conserved helix–hairpin–helix motif has been shown to be the binding site for free thymine glycol (6), but recent evidence derived from analysis of a wide range of DNA binding proteins in which this motif is conserved indicates that it may not directly determine substrate specificity (24). Instead, it may be that the solvent-filled pocket lying between the two DNA binding regions defined by the [4Fe-4S] cluster and the helix–hairpin–helix motif interacts with the damaged base. Many of the residues that define this active site pocket are conserved between prokaryotic and eukaryotic endonuclease III-like proteins, including a critical lysine residue (Lys-120 in endonuclease III and Lys-220 in hNTH1; ref. 6 and Fig. 1). The ɛ-amino group of this lysine residue is probably required for the formation of the N-acylimine (Schiff’s base) enzyme-substrate intermediate (25). This reaction intermediate is a characteristic feature of those repair proteins that act as dual DNA glycosylase/AP lyase enzymes. Another highly conserved residue is Asp-138 in endonuclease III (Fig. 1), which has been shown to be essential for the catalytic activity (6), and may activate the ɛ-amino group of Lys-120 (25).
A mammalian DNA glycosylase/AP lyase with substrate specificity similar to that of hNTH1 has been purified previously from calf thymus (26, 27), but the gene encoding the enzyme has not been identified. Breimer (26) purified the enzyme 400-fold by following its urea-DNA glycosylase activity, and showed that the most purified fraction also contained thymine glycol-DNA glycosylase activity and an incision activity specific for AP sites. Hilbert et al. (27) purified a 31-kDa thymine glycol-DNA glycosylase activity with associated AP lyase activity 5000-fold and obtained partial peptide sequences that almost certainly are representative of the bovine counterpart of hNTH1. It is not clear at present if mammalian cells contain more than one counterpart of the E. coli endonuclease III protein, in the same way that E. coli has both endonuclease III and the related endonuclease VIII (14).
The genomic location of OCTS3, adjacent to the TSC2 gene, is potentially interesting because the hamartomatous growths characteristic of tuberous sclerosis have been shown to develop only after a secondary somatic mutation at the TSC locus. These are often large chromosomal deletions leading to loss of heterozygosity of an area that would include an OCTS3 allele (28). However, preliminary characterization of eight constitutional deletions that remove OCTS3, as well as disrupting TSC2 (refs. 16 and 22; unpublished work), shows no clear phenotypic affect of germ-line heterozygous loss of OCTS3. Nevertheless, some hamartoma from such patients may have homozygous loss of OCTS3. It will be interesting, therefore, to study the DNA repair capabilities of cells derived from these growths. The possibility that homozygous loss of OCTS3 (either two germ-line mutations or germ line plus somatic) will be a predisposing factor for malignancies, as with some mismatch repair loci (29–32) should be considered, although no such tumor susceptibility loci (apart from TSC2) have been mapped to this region of chromosome 16p.
In summary, we have identified cDNA clones encoding a human homolog of E. coli endonuclease III and have shown that the human protein possesses a range of enzymatic activities similar to those of its microbial counterparts. Active oxygen is a major cause of endogenous DNA damage (1, 33, 34), and the hNTH1 DNA glycosylase is likely to have a key role in the cellular defense against such lesions. We are now in a position to address whether somatic loss of the gene encoding hNTH1 in patients with tuberous sclerosis plays a role in tumor development.
Acknowledgments
We thank Dr. D. L. Simmons for the cDNA library, Dr. C. Norbury for helpful comments on the manuscript, and the Imperial Cancer Research Fund, the Medical Research Council, and the Tuberous Sclerosis Association for financial support.
Footnotes
References
- 1.Ames B N, Gold L S. Mutat Res. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 2.Martin G M, Austad S N, Johnson T E. Nat Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- 3.Demple B, Harrison L. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 4.Seeberg E, Eide L, Bjoras M. Trends Biochem Sci. 1995;20:391–396. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham R P, Weiss B. Proc Natl Acad Sci USA. 1985;82:474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thayer M M, Ahern H, Xing D, Cunningham R P, Tainer J A. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo C-F, McRee D E, Fisher C L, O’Handley S F, Cunningham R P, Tainer J A. Science. 1992;258:434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- 8.Radman M. J Biol Chem. 1976;251:1438–1445. [PubMed] [Google Scholar]
- 9.Demple B, Linn S. Nature (London) 1980;287:203–208. doi: 10.1038/287203a0. [DOI] [PubMed] [Google Scholar]
- 10.Breimer L H, Lindahl T. J Biol Chem. 1984;259:5543–5548. [PubMed] [Google Scholar]
- 11.Hatahet Z, Kow Y W, Purmal A A, Cunningham R P, Wallace S S. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 12.Bailly V, Verly W G. Biochem J. 1987;242:565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roldan-Arjona T, Anselmino C, Lindahl T. Nucleic Acids Res. 1996;24:3307–3312. doi: 10.1093/nar/24.17.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melamede R J, Hatahet Z, Kow Y W, Ide H, Wallace S S. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 15.Eide L, Bjoras M, Pirovano M, Alseth I, Burdal K, Seeberg E. Proc Natl Acad Sci USA. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Chromosome 16 Tuberous Sclerosis Consortium. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 17.European Polycystic Kidney Disease Consortium. Cell. 1994;77:881–894. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 18.Asahara H, Wistort P M, Bank J F, Bakerian R H, Cunningham R P. Biochemistry. 1989;28:4444–4449. doi: 10.1021/bi00436a048. [DOI] [PubMed] [Google Scholar]
- 19.Walker L J, Robson C N, Black E, Gillespie D, Hickson I D. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boiteux S, O’Connor T R, Lederer F, Gouyette A, Laval J. J Biol Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 21.Barzilay G, Mol C D, Robson C N, Walker L J, Cunningham R P, Tainer J A, Hickson I D. Nature Struct Biol. 1995;2:561–568. doi: 10.1038/nsb0795-561. [DOI] [PubMed] [Google Scholar]
- 22.Brook-Carter P T, Peral B, Ward C J, Thompson P, Hughes J, Maheshwar M M, Nellist M, Gamble V, Harris P C, Sampson J R. Nat Genet. 1994;8:328–332. doi: 10.1038/ng1294-328. [DOI] [PubMed] [Google Scholar]
- 23.Boorstein R J, Hilbert T P, Cadet J, Cunningham R P, Teebor G W. Biochemistry. 1989;28:6164–6170. doi: 10.1021/bi00441a007. [DOI] [PubMed] [Google Scholar]
- 24.Doherty A J, Serpell L C, Ponting C P. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labahn J, Scharer O D, Long A, Ezaz-Nikpay K, Verdine G L, Ellenberger T E. Cell. 1996;86:321–329. doi: 10.1016/s0092-8674(00)80103-8. [DOI] [PubMed] [Google Scholar]
- 26.Breimer L H. Biochemistry. 1983;22:4192–4197. doi: 10.1021/bi00287a005. [DOI] [PubMed] [Google Scholar]
- 27.Hilbert T P, Boorstein R J, Kung H C, Bolton P H, Xing D, Cunningham R P, Teebor G W. Biochemistry. 1996;35:2505–2511. doi: 10.1021/bi952516e. [DOI] [PubMed] [Google Scholar]
- 28.Green A J, Smith M, Yates J R W. Nat Genet. 1994;6:193–196. doi: 10.1038/ng0294-193. [DOI] [PubMed] [Google Scholar]
- 29.Fishel R, Lescoe M K, Rao M R S, Copeland N G, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 30.Parsons R, Li G-M, Longley M J, Fang W-H, Papadopoulos N, Jen J, de la Chapelle A, Kinzler K W, Vogelstein B, Modrich P. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 31.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 32.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, et al. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 33.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 34.Cross C E, Halliwell B, Borish E T, Pryor W A, Ames B N, Saul R L, McCord J M, Harman D. Ann Intern Med. 1987;107:526–545. doi: 10.7326/0003-4819-107-4-526. [DOI] [PubMed] [Google Scholar]