Abstract
RNA biochemical or structural studies often require an RNA sample that is chemically pure, and most protocols for its in vitro production use denaturing polyacrylamide gel electrophoresis to achieve this. Unfortunately, many RNAs do not quantitatively refold into an active conformation after denaturation, creating significant problems for downstream characterization or use. In addition, this traditional purification method is not amenable to studies demanding high-throughput RNA production. Recently, we presented the first general method for producing almost any RNA sequence that employs an affinity tag that is removed during the purification process. Because technical difficulties prevented application of this method to many RNAs, we have developed an improved version that utilizes a different activatable ribozyme and affinity tag that are considerably more robust, rapid, and broadly applicable.
Keywords: RNA purification, affinity tag, activatable ribozyme, native purification
INTRODUCTION
Recent discoveries indicate that the typical cell contains a diverse population of small RNAs, riboregulatory elements, and ribonucleoproteins that play a role in almost every level of the flow of genetic information and even inheritance (for review, see Costa 2007; Prasanth and Spector 2007). Many of these RNAs are poorly characterized, but sequence conservation patterns suggest that part of their function is conferred by their ability to fold into complex shapes. Thus, a crucial part of understanding these RNAs is a biochemical and structural characterization of their secondary and tertiary architecture. However, most investigations still employ methodologies that denature these RNAs during purification following in vitro transcription by T7 RNA polymerase (Milligan et al. 1987; Doudna 1997). The pitfalls of these methods are well known, often leading to misfolded or aggregated material that is unsuitable for further analysis (Uhlenbeck 1995).
Several different methods and protocols recently have been developed to overcome this issue. The first uses chromatography to purify RNA after transcription. Currently, size exclusion chromatography is the most powerful method in that it can resolve fully transcribed RNA from small abortive products, ribozymes used for 5′- or 3′-end processing, plasmid template, and also multimeric species (Lukavsky and Puglisi 2004; Kim et al. 2007). Ion-exchange chromatography also is used, but typically is limited to small (> 50 nucleotides [nt]) oligonucleotides (Anderson et al. 1996; Shields et al. 1999). A second recently developed method uses affinity capture by an immobilized DNA oligonucleotide and a DNAzyme to liberate the product RNA (Cheong et al. 2004). Concurrently, our laboratories developed a “native RNA purification tag” that uses an RNA–protein interaction for immobilization and an imidazole-activated hepatitis delta virus (HδV) ribozyme for elution of the desired RNA (Kieft and Batey 2004). This technique has been successfully employed to solve the structure of the guanine riboswitch (Batey et al. 2004). Unfortunately, none of these techniques has replaced the use of denaturing gel electrophoresis as a dominant purification technique owing to individual idiosyncrasies that prevent their adoption for a broad range of purposes.
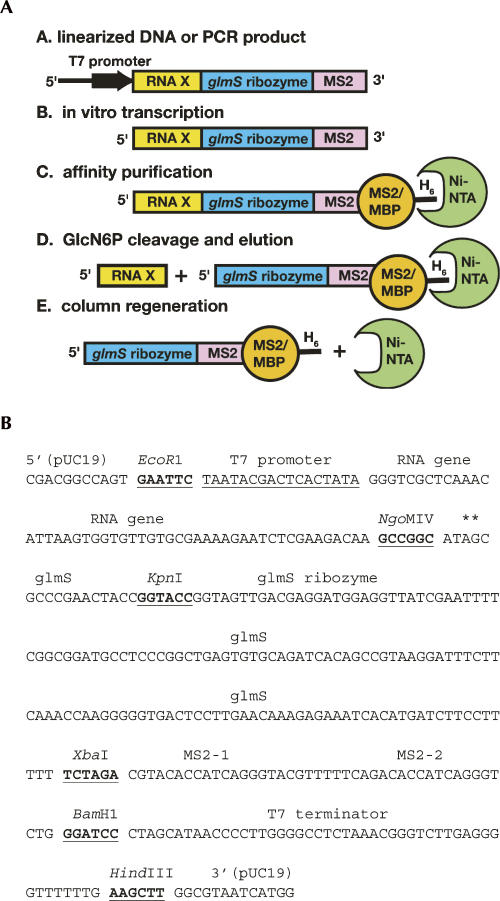
Our goal is to implement a technique for native RNA purification easily accessible to a wide range of researchers with minimal difficulty. To address several technical issues that have arisen with the original native RNA purification tag, we have redeveloped it to use an MS2 coat protein as a means of immobilization (Zhou et al. 2002) and the glmS ribozyme (Winkler et al. 2004) as a means of removing the 3′-affinity tag (Fig. 1A). Furthermore, by employing commercially available nickel-affinity resin, this method can rapidly purify RNA on the nanogram scale using spin columns or up to milligrams on gravity columns using techniques that are familiar to most laboratories. The resultant improved method maintains the high-throughput, parallel purification features of the original method, with increased applicability and reliability.
FIGURE 1.
(A) Scheme for the native purification of any desired sequence (“RNA X”) using the new affinity tag. H6 denotes a 6x histidine tag. (B) Sequence of the RNA expression vector pRAV23 with the relevant restriction sites and elements denoted. The asterisks above the AG step near the NgoMIV site denotes the cleavage site for the glmS ribozyme such that the adenine will be left at the 3′-end of the RNA product. The sequence between the EcoR1 and HindIII sites is inserted into pUC19 to yield pRAV23.
RESULTS AND DISCUSSION
Motivation for redeveloping the original RNA native affinity tag came through feedback from users who experienced technical issues during implementation. First, the TmaM-domain protein, which captures full-length transcript precipitates in low salt concentrations (<200 mM NaCl), is a problem during cation exchange chromatography and the AffiGel-10 coupling steps. While this problem can be overcome by minimizing the time the protein spends at low ionic strength conditions, many users found this issue difficult to resolve. Second, while the TmaM-domain columns can be regenerated and reused, we found that they became ineffective after repeated use. Third, the original activatable ribozyme was the hepatitis delta virus (HδV) ribozyme containing a point mutation (C75U) that causes it to become active only in the presence of 0.2 M imidazole (Perrotta et al. 1999). Complete cleavage requires incubating the RNA on the column with imidazole at 37°–42°C for 2 h. During this time the RNA can significantly degrade, an undesirable feature of a purification process. Finally, certain RNA sequences do not cleave even after prolonged incubation, presumably due to alternate and unpredictable secondary structures at the cleavage site.
Design of the affinity tag
The design of the new affinity tag vectors is based on the previous version of the system, as embodied by the pRAV12 vector (Kieft and Batey 2004). As with pRAV12, these new vectors (called pRAV23 and pRAV24) are derived from pUC19 with all insertions made between the unique EcoRI and HindIII sites. For a new activatable ribozyme, we now employ the recently discovered ribozyme that is activated by glucosamine-6-phosphate (GlcN6P), which controls expression of the glmS gene (Winkler et al. 2004). We chose the ribozyme from Bacillus subtilis as it has been characterized biochemically (McCarthy et al. 2005; Hampel and Tinsley 2006; Jansen et al. 2006; Roth et al. 2006), and the structures of closely related RNAs are known (Klein and Ferre-D'Amare 2006; Cochrane et al. 2007). As the distal loop of the P1 helix is variable and supports different sequences, we placed a unique KpnI site there to allow one to readily insert the desired RNA sequence upstream of the ribozyme (Fig. 1B).
For immobilization, we now employ a hexahistidine-tagged form of the MBP-MS2 coat fusion protein (HMM). This protein has high affinity for a small stem–loop structure (Lim et al. 1994; LeCuyer et al. 1995) and has been used successfully for pull-down and immobilization methods (Zhou et al. 2002; Yu et al. 2005) as well as molecular techniques for visualization of RNA (Bertrand et al. 1998; Rook et al. 2000; Zhang and Simon 2003). Many laboratories have experience with this (or a similar) protein, and it is easily expressed, purified, and manipulated by the user. Furthermore, as our version of this fusion protein has a 6xHis-tag, we can adopt Ni2+-affinity chromatography (Crowe et al. 1994) for immobilization, a very commonly used technique and thus readily accessible by most laboratories.
To adopt the RNA affinity tag for use with the MBP-MS2 protein, we inserted two tandem MS2 coat protein-binding stem–loops between the XbaI and BamHI sites following the glmS ribozyme to create the pRAV23 vector (Fig. 1B). A second vector, pRAV24, is also available in which the original TmaM-domain binding sites have been preserved.
Development of a purification scheme
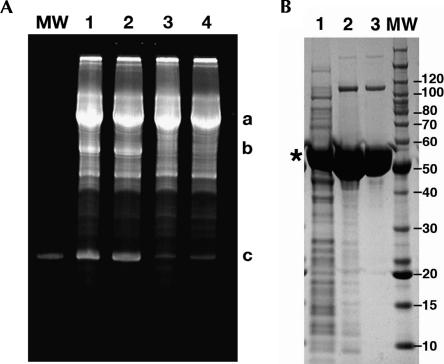
Several issues relating to the new system needed to be addressed prior to its full implementation. First, several groups have noted that the glmS ribozyme is activated at a low level by a number of compounds including Tris buffer (McCarthy et al. 2005; Roth et al. 2006), which is used in many standard reactions, including the PCR reaction used to generate the template and the transcription reaction for RNA synthesis. We have likewise observed that even low quantities of Tris (∼ 10 mM) induce a substantial amount of background cleavage during the timescale of the purification procedure. Thus, we have reformulated the PCR and transcription buffers to use K+-HEPES rather than Tris-HCl, which does not adversely affect the yield of either DNA (data not shown) or RNA product (Fig. 2A). As our enzymes (T7 RNA polymerase and inorganic pyrophosphatase) are stored in a buffer containing only 20 mM sodium phosphate (pH 7.5), these enzymes do not contribute significant amounts of Tris to the final transcription reaction.
FIGURE 2.
(A) Ethidium bromide stained 12% denaturing polyacrylamide gel of transcription of a 94-nt RNA with the 3′ affinity tag attached in different transcription buffers. (Lane 1) Standard Tris transcription buffer with 20 mM DTT; (lane 2) standard Tris transcription buffer with 10 mM DTT; (lane 3) transription with Na-HEPES (pH 8.0) and 20 mM DTT; (lane 4) transcription with Na-HEPES (pH 8.0) and 10 mM DTT. Product “a” is the full-length product, “b” is the cleaved 3′-tag, and “c” is the 94-nt desired product RNA (which is also the size marker in the “MW” lane). (B) 4%–12% SDS-PAGE analysis of expression and purification of the HMM protein. (Lane 1) Soluble fraction of total cellular lysate; (lane 2) elution from the nickel affinity column; (lane 3) peak from the SP-column. (*) The product protein; molecular size standards are to the right of the gel.
The second concern was the use of dithiothreitol (DTT) in transcription reactions. In the typical transcription reaction, 10–20 mM DTT maintains the proper reducing conditions needed for optimal T7 RNA polymerase activity (Milligan et al. 1987; Doudna 1997). However, as we are employing Ni2+-affinity chromatography to immobilize the fully transcribed RNA (through the HMM protein), we needed to minimize [DTT] to prevent reducing of Ni2+ (turning the column brown). We have found that the quantities of DTT in the standard transcription buffer (10 mM) are not sufficient to impact the Ni2+-agarose resin we used (QIAGEN Ni-NTA agarose), although stripping and subsequent recharging of the resin result in some reduction of the Ni2+. Decreasing the DTT concentration of the transcription reaction severely impacts RNA yield, and thus, the only way to address this completely would be to dilute the transcription reaction prior to application to the column. Typically, we do not choose to do this and generally do not observe any adverse impact on the column or the preparation. Obviously, a way around this issue is to use amylose resin for immobilization via the MBP tag. While we do achieve quantitative binding to the resin, we have found that Glc6NP causes a substantial amount of the protein to be released from the column during the cleavage step.
Implementation of the purification protocol
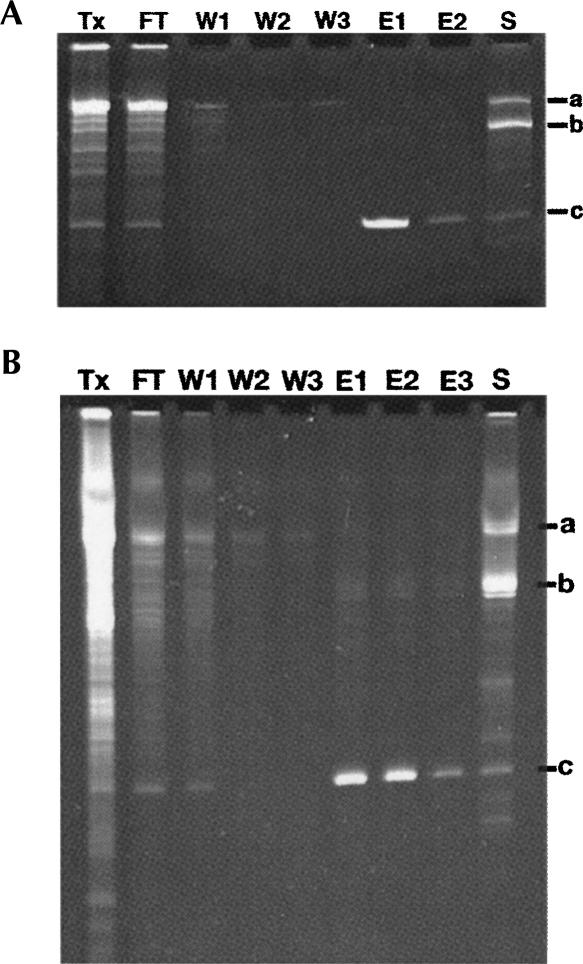
This new affinity tag uses Ni2+-affinity chromatography, making it more accessible to a broader spectrum of users, particularly those who wish to purify only small quantities of RNA (0.1–1 nmol) for molecular biological or biochemical experiments. To this end, we used QIAGEN Ni2+-Spin Columns to create an “RNA miniprep.” This involves performing a small 100 μL T7 polymerase transcription reaction for 1–2 h at 37°C to which is added 150 μg of HMM protein (from 3 to 4 mg/mL stock, prepared as shown in Fig. 2B) and allowed to incubate for 10 min on ice. The protein/RNA mixture is subsequently passed through the spin column that has been equilibrated in Wash Buffer on the lowest spin setting possible (1000 rpm or 93g on an Eppendorf 5415D microfuge) followed by three 600 μL washes with Wash Buffer to remove unbound material. The product RNA is eluted by adding Wash Buffer + 1 mM Glc6NP to the column, giving it a quick pulse through which to pass a little buffer, and incubating it for 10 min at room temperature. As the ribozyme is very rapidly activated by the presence of Glc6NP, most of the RNA is cleaved during this first elution, and a second elution yields only a moderate increase in the yield of RNA. For an RNA of 93 nt (30.5 kDa molecular weight), this yielded 200 μL of 4 μM (0.8 nmol) material. Samples from various steps of this process are shown resolved on a 12% denaturing polyacrylamide gel (stained with ethidium bromide), indicating the quality of RNA eluted from the column, in Figure 3A. Note that this protocol is not optimized for full retention of the RNA (Fig. 3A, lanes 1,2) but, rather, rapid purification, and the amount of RNA obtained from this technique is more than enough for most molecular biological applications.
FIGURE 3.
Affinity purification of a 94-nt RNA on small and large scales analyzed on ethidium bromide stained 12% denaturing polyacrylamide gels. (A) Purification of a 100-μL transcription using a QIAGEN Ni-NTA spin column. (Lane Tx) The raw transcription; (lane FT) the column flowthrough; (lanes W1–W3) the three wash steps; (lanes E1,E2) the two 200-μL elutions; (lane S) the imidazole elution. Bands “a”, “b,” and “c” denote the full-length transcript, 3′-tag, and 94-nt product, respectively. (B) Purification of a 3.25-mL transcription using a 3.0-mL Ni-NTA agarose gravity column. Lanes and bands are marked the same as in A, except that there is a third elution step (lane E3).
For larger-scale production of RNA, the above protocol was modified to use Ni2+-affinity chromatography with gravity flow columns. On the 1–4 mL transcription scale, we use ∼ 3 mL of QIAGEN Ni2+-agarose resin in a 10 mL disposable column (Pierce). We incubate the transcription with 1.6 mg of HMM protein/mL of transcription and the resin for 30 min at 4°C on a rocker; larger volumes of transcription are purified by linearly scaling up the reaction. Following washing and elution of the product with 1 mM GlcN6P, analysis of fractions on a denaturing gel revealed that the product is of high quality (Fig. 3B). To further assess the quality of the resulting RNA, we purified and concentrated the SAM-I riboswitch RNA using this method and crystallized the product (data not shown). The resulting crystals grew under the same conditions and diffracted as well as those obtained from RNA purified using traditional denaturing polyacrylamide gel electrophoresis (Montange and Batey 2006). Thus, like the previous version of the native affinity tag, this new technique yields RNA of sufficient quality to meet rigorous structural and biochemical needs.
As described above, the improved version of our affinity purification system employs both a new activatable ribozyme and a new affinity tag through the use of pRAV23. However, pRAV24 allows the use of the glmS activated ribozyme with our previous TmaM-domain based affinity tag. For this method, the RNA is transcribed as described above (with K+-HEPES buffer), loaded onto the column, and washed as described (Kieft and Batey 2004). To liberate the product RNA, GlcN6P is added as described above, and the column then is regenerated.
Use of the method with a variety of RNAs
The original RNA native affinity tag version did not allow purification of all RNAs, as some sequences failed to cleave from the column. One of these failures was the SAM-I riboswitch RNA, which had been purified by conventional means during a recent crystal structure determination (Montange and Batey 2006). To test if this purification method could be used with a variety of RNAs, we cloned several sequences into the vector and tested them for proper cleavage and elution. In addition to the 94-nt RNA shown in Figure 3, we also tested a 59-nt product RNA containing a pseudoknot sequence and a 66-nt guanine riboswitch sequence. These oligonucleotides were also easily purified using this method, suggesting that the redesigned system can be used with a large variety of RNAs.
Conclusion
In order to create a more robust and simple native RNA purification method, three crucial changes have been made to our original system: (1) the adoption of a ribozyme that nature evolved to be activated by a small molecule ligand, (2) Ni2+ affinity resin as an immobilization matrix, and (3) the use of a protein (MBP-MS2 coat fusion). Most significantly, this protocol has been implemented successfully on the small scale using commercially available spin columns to create what is effectively an “RNA miniprep.” As the vast majority of RNA science is performed on this scale (0.1–1 nmol of RNA), this makes this method accessible and useful to a broad spectrum of the RNA research community.
MATERIALS AND METHODS
Cloning of pRAV vectors
The parental vector for both pRAV23 and pRAV24 is pRAV12 (Kieft and Batey 2004), which is a derivative of pUC19. To construct pRAV24, a DNA sequence was constructed using overlapping oligonucleotides and standard PCR techniques (Sambrook and Russel 2001) that allowed its insertion into pRAV12 between the NcoI and XbaI sites (Fig. 1B). The sequence of the glmS ribozyme is derived from Bacillus subtilis (Winkler et al. 2004) with a change made in the terminal loop of the P1 helix such that a KpnI site was added for cloning purposes. This DNA fragment was placed into pRAV12 using standard molecular cloning techniques, and the resulting vector (pRAV24) was sequence verified. To add the MS2 coat protein affinity sequence, another DNA fragment was generated containing two MS2-coat-protein-binding stem–loops (Zhou et al. 2002) flanked by XbaI and BamH1 sites for cloning. This insert was placed into pRAV24 cut with the same enzymes, and the resulting vector (pRAV23) was sequence verified.
Purification of HMM protein
The HMM (His-tagged MBP-MS2 coat fusion) protein is an ∼ 59-kDa protein containing an N-terminal hexahistidine tag, a central maltose-binding protein (MBP) domain, and a C-terminal MS2 coat protein containing the V29/dIFG mutations (Lim and Peabody 1994) to prevent coat protein multimerization and increase its affinity for RNA. The gene encoding this protein was constructed using standard molecular techniques and inserted between the NdeI and HindIII sites of pET31b (Novagen). The resulting kanamycin-resistant plasmid, pHMM, was sequence verified.
HMM was expressed by transforming pHMM into Rosetta(BL21)/pLysS. Cells were grown in 1 L of LB medium and 10 μg/mL kanamycin at 37°C to an OD600 nm of 0.7, and expression was induced with 0.5 mM IPTG. The cells were allowed to continue to grow for 3 h, harvested by centrifugation, and resuspended in 50 mL of Lysis Buffer (50 mM sodium phosphate at pH 8.0, 300 mM NaCl, 0.5% Tween 20, 10 mM imidazole, and 10% glycerol) and 500 μL of bacterial protease inhibitor cocktail (Sigma). The cells were lysed by sonication, and the cell debris was removed by centrifugation in a JA-20 rotor (Beckman) for 30 min at 35,000g. The supernatant was passed over a Ni2+-affinity column, washed with Lysis Buffer, and eluted using Elution buffer (Lysis Buffer + 250 mM imidazole). Fractions containing protein were pooled and dialyzed against 25 mM Na+-MES (pH 6.0) and 25 mM NaCl overnight at 4°C. The protein was applied to a Hi-Prep 16/10 SP-Sepharose column (GE Healthcare) and eluted with a gradient of 0–1 M NaCl over a 200 mL volume (the protein eluted around 0.2–0.3 M salt). Fractions were pooled and dialyzed into a buffer containing 25 mM Na+-HEPES (pH 7.5), 200 mM NaCl, and 10% glycerol prior to storage at −20°C.
To quantitate the protein, the absorbance at 280 nm was taken and the concentration calculated using a molar extinction coefficient of 83,310 M−1 cm−1. Typical yields (over five individual preparations of the protein on the 2–4 L cell culture size) averaged around 100–120 mg/L culture.
Transcription and purification of RNA
RNA was transcribed from PCR-generated DNA fragments using established protocols for DNA amplification. To amplify the template DNA, two oligonucleotides were used that are directed against the T7 polymerase promoter (5′-GEN, 5′-gcgcgcgaattctaatacgactcactatag-3′) and the 3′-end of the HMM-binding stem–loop sequence (3′-GLM, 5′-cagaccctgatggtgtctgaa-3′). A PCR reaction using 1× HEPES-ThermoPol Buffer (10× consists of 200 mM Na-HEPES at pH 8.6 and 25°C, 100 mM KCl, 10 mM [NH4]2SO4, 20 mM MgSO4, 0.1% Triton X-100), 2 mM total dNTPs, 1 μM each oligonucleotide, 0.1 pg of plasmid DNA, and 10 U of Taq polymerase. This reaction was cycled 32 times using 95°C (30 sec), 55°C (30 sec), and 72°C (45 sec) schemes. (Note that fewer cycles can be used to minimize the potential for spontaneous mutations. A large number of cycles was used here to maximize template amount.) The resulting reaction was used directly for in vitro transcription without any purification.
Transcription was performed in a reaction containing 1× HEPES Transcription Buffer (10× consists of 300 mM Na+-HEPES at pH 8.0, 100 mM DTT, 20 mM spermidine, and 0.1% Triton X-100), 32 mM MgCl2, 4 mM each rNTP, 50 μg/mL T7 RNA polymerase, 1 unit/mL inorganic pyrophosphatase (Sigma), and 8% (v/v) PCR reaction (above) containing the appropriate template. The reaction was incubated for 2 h at 37°C.
To prepare the transcription reaction for affinity purification, 1.6 mg of HMM protein is added directly to the reaction and allowed to incubate for 10 min on ice to allow the protein to bind to the 3′ stem–loops. This reaction then is passed over a column containing ∼ 1 mL of Ni-NTA resin (QIAGEN) per 2 mL of transcription reaction in a gravity column at room temperature; to ensure complete binding of the RNA/protein complex to the column, the flowthrough can be passed through a second time. The column then is washed three times with four column volumes of RNA Column Buffer (50 mM K+-HEPES at pH 7.5, 150 mM NaCl, 10 mM MgCl2, and 10 mM imidazole) to remove impurities such as excess protein, nucleotides, and RNA abortive transcription products. To cleave the product RNA from the tag and elute from the column, two column volumes of RNA Column Buffer + 1 mM glucosamine-6-phosphate (GlcN6P) were added to the column and allowed to pass through. Two subsequent elutions spaced 10 min apart are performed to remove further RNA (usually most of the RNA comes out in the first two elutions). The HMM-3′ tag complex then is removed from the column using Elution Buffer (see above, “Purification of HMM protein”) to regenerate the Ni-NTA resin.
ACKNOWLEDGMENTS
The authors thank David Costantino, Sunny Gilbert, and Crystal Love for technical assistance; Wade Winkler for helpful suggestions; and David Costantino, Jennifer Pfingsten, and Quentin Vicens for providing critical comments on the manuscript. This work was funded through a grant from the National Institutes of Health to R.T.B. (GM 073850) and to J.S.K. (GM 072560) as well as a Development Grant through the University of Colorado Technology Transfer Office (to R.T.B. and J.S.K.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.528007.
REFERENCES
- Anderson, A.C., Scaringe, S.A., Earp, B.E., Frederick, C.A. HPLC purification of RNA for crystallography and NMR. RNA. 1996;2:110–117. [PMC free article] [PubMed] [Google Scholar]
- Batey, R.T., Gilbert, S.D., Montange, R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S.M., Singer, R.H., Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Cheong, H.K., Hwang, E., Lee, C., Choi, B.S., Cheong, C. Rapid preparation of RNA samples for NMR spectroscopy and X-ray crystallography. Nucleic Acids Res. 2004 doi: 10.1093/nar/gnh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane, J.C., Lipchock, S.V., Strobel, S.A. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem. Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, F.F. Non-coding RNAs: Lost in translation? Gene. 2007;386:1–10. doi: 10.1016/j.gene.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Crowe, J., Dobeli, H., Gentz, R., Hochuli, E., Stuber, D., Henco, K. 6xHis-Ni-NTA chromatography as a superior technique in recombinant protein expression/purification. Methods Mol. Biol. 1994;31:371–387. doi: 10.1385/0-89603-258-2:371. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A. Preparation of homogeneous ribozyme RNA for crystallization. Methods Mol. Biol. 1997;74:365–370. doi: 10.1385/0-89603-389-9:365. [DOI] [PubMed] [Google Scholar]
- Hampel, K.J., Tinsley, M.M. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry. 2006;45:7861–7871. doi: 10.1021/bi060337z. [DOI] [PubMed] [Google Scholar]
- Jansen, J.A., McCarthy, T.J., Soukup, G.A., Soukup, J.K. Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme. Nat. Struct. Mol. Biol. 2006;13:517–523. doi: 10.1038/nsmb1094. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Batey, R.T. A general method for rapid and nondenaturing purification of RNAs. RNA. 2004;10:988–995. doi: 10.1261/rna.7040604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, I., McKenna, S.A., Viani Puglisi, E., Puglisi, J.D. Rapid purification of RNAs using fast performance liquid chromatography (FPLC) RNA. 2007;13:289–294. doi: 10.1261/rna.342607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, D.J., Ferre-D'Amare, A.R. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- LeCuyer, K.A., Behlen, L.S., Uhlenbeck, O.C. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry. 1995;34:10600–10606. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- Lim, F., Peabody, D.S. Mutations that increase the affinity of a translational repressor for RNA. Nucleic Acids Res. 1994;22:3748–3752. doi: 10.1093/nar/22.18.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, F., Spingola, M., Peabody, D.S. Altering the RNA binding specificity of a translational repressor. J. Biol. Chem. 1994;269:9006–9010. [PubMed] [Google Scholar]
- Lukavsky, P.J., Puglisi, J.D. Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA. 2004;10:889–893. doi: 10.1261/rna.5264804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, T.J., Plog, M.A., Floy, S.A., Jansen, J.A., Soukup, J.K., Soukup, G.A. Ligand requirements for glmS ribozyme self-cleavage. Chem. Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange, R.K., Batey, R.T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- Perrotta, A.T., Shih, I., Been, M.D. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- Prasanth, K.V., Spector, D.L. Eukaryotic regulatory RNAs: An answer to the “genome complexity” conundrum. Genes & Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Rook, M.S., Lu, M., Kosik, K.S. CaMKIIα 3′ untranslated region-directed mRNA translocation in living neurons: Visualization by GFP linkage. J. Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, A., Nahvi, A., Lee, M., Jona, I., Breaker, R.R. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Russell, D.W. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Shields, T.P., Mollova, E., Ste Marie, L., Hansen, M.R., Pardi, A. High-performance liquid chromatography purification of homogenous-length RNA produced by trans cleavage with a hammerhead ribozyme. RNA. 1999;5:1259–1267. doi: 10.1017/s1355838299990945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck, O.C. Keeping RNA happy. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A., Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Yu, Y., Ji, H., Doudna, J.A., Leary, J.A. Mass spectrometric analysis of the human 40S ribosomal subunit: Native and HCV IRES-bound complexes. Protein Sci. 2005;14:1438–1446. doi: 10.1110/ps.041293005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., Simon, A.E. A novel procedure for the localization of viral RNAs in protoplasts and whole plants. Plant J. 2003;35:665–673. doi: 10.1046/j.1365-313x.2003.01837.x. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Sim, J., Griffith, J., Reed, R. Purification and electron microscopic visualization of functional human spliceosomes. Proc. Natl. Acad. Sci. 2002;99:12203–12207. doi: 10.1073/pnas.182427099. [DOI] [PMC free article] [PubMed] [Google Scholar]