Abstract
T4 RNA ligase 1 (Rnl1) is a tRNA repair enzyme that thwarts a tRNA-damaging host response to virus infection. The 374-aa Rnl1 protein consists of an N-terminal nucleotidyltransferase domain fused to a unique C-terminal domain composed of 10 α helices. We exploited an in vitro tRNA splicing system to demonstrate that Rnl1 has an inherent specificity for sealing tRNA with a break in the anticodon loop. The tRNA specificity is imparted by the C domain, any deletion of which caused the broken tRNA to be sealed as poorly as the linear intron in vitro and also abolished Rnl1 tRNA splicing activity in vivo. Deletion analysis demarcated Rnl1-(1–254) as a minimal catalytic domain of Rnl1, capable of all chemical steps of the nonspecific RNA ligation reaction. Alanine scanning of the N domain identified Ser103, Leu104, Lys117, and Ser118 as important for pRNA ligation in vitro and tRNA repair in vivo.
Keywords: RNA repair, polynucleotide ligase, tRNA breakage, tRNA splicing
INTRODUCTION
Bacteriophage T4 RNA ligase 1 (Rnl1) is a tRNA repair enzyme that the virus uses to evade a tRNA-damaging host response to infection (Amitsur et al. 1987). Rnl1 joins 3′-OH and 5′-PO4 RNA termini via three nucleotidyl transfer steps: (1) Rnl1 reacts with ATP to form a covalent ligase-(lysyl-N)–AMP intermediate plus pyrophosphate; (2) AMP is transferred from ligase–adenylate to the 5′-PO4 RNA end to form an RNA–adenylate intermediate (AppRNA); and (3) Rnl1 catalyzes attack by an RNA 3′-OH on the RNA–adenylate to form a 3′–5′ phosphodiester bond and release AMP (Cranston et al. 1974; Sugino et al. 1977; Uhlenbeck and Gumport 1982). Homologs of Rnl1 are found in certain viruses, bacteria, and eukarya. The best studied of these are the tRNA-splicing ligases of fungi and plants, which are structural and functional homologs of T4 Rnl1 that repair purposeful tRNA breaks in vivo (Phizicky et al. 1992; Sawaya et al. 2003; Schwer et al. 2004; Englert and Beier 2005; Wang et al. 2006a,b).
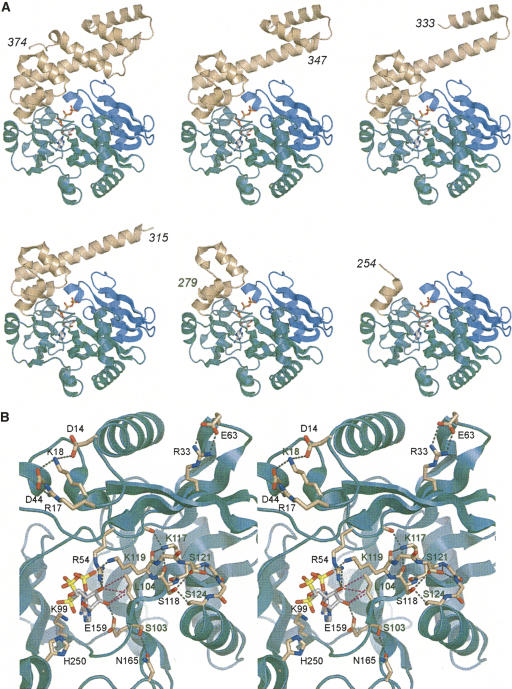
The 374-aa Rnl1 protein consists of two structural domains (El Omari et al. 2006). The N-terminal nucleotidyltransferase domain (Fig. 1A, aa 1–242, colored green and blue) is composed of a central ensemble of β strands and loops that form the nucleotide-binding pocket. The pocket is lined by six peptide motifs (I, Ia, III, IIIa, IV, and V) that define the covalent nucleotidyltransferase enzyme superfamily (Shuman and Lima 2004). The superfamily includes DNA ligases, RNA ligases, and mRNA capping enzymes, all of which catalyze the nucleotidylation of polynucleotide 5′ ends via a covalent enzyme–(lysyl-Nζ)–NMP intermediate. The distinctive feature of Rnl1 is an all-helical C-terminal domain (Fig. 1A, aa 243–374, colored beige) that is unrelated to the C-terminal OB-fold domains of DNA ligases and RNA capping enzymes or to the C domain of the Rnl2 clade of RNA ligases (Nandakumar et al. 2006).
FIGURE 1.
Tertiary structure and active site of T4 Rnl1. (A) The Rnl1 fold (from PDB 2C5U) is shown at top left with AMPCPP in the active site. The adenylyltransferase domain is colored green; the Rnl1-specific N-terminal module is colored blue; the unique all-helical C domain is colored beige. The other images depict the putative structures of the serially deleted versions of Rnl1 subjected to biochemical analysis in the present study. The C terminal residues are indicated for each structural model. (B) Stereoview of the Rnl1 active site and the residues of the N domain that were subjected here to alanine scanning. Hydrogen bonding and ionic interactions are depicted as black dashed lines. The van der Waals interactions of Leu10 with the ribose sugar are depicted as magenta dashed lines.
The requirements for catalysis by the Rnl1 ligase family are emerging from a series of mutational studies (Heaphy et al. 1987; Wang et al. 2003; Wang and Shuman 2005), the most recent of which (Wang et al. 2006b) was guided by the crystal structure of T4 Rnl1 bound to divalent cations and the substrate analog AMPCPP (Fig. 2A; El Omari et al. 2006). Eleven Rnl1 side chains have been identified as essential for activity: Arg54, Lys75, Phe77, Lys99, Asp101, Lys119, Glu159, Glu227, Lys240, Lys242, and Tyr246. Seven of the essential residues are located within Rnl1 counterparts of conserved nucleotidyltransferase motifs I (99 KEDG102), Ia (118SK 119), III (154FTANFEFV161), IV (227 EGYVA231), and V (238HFKIK 242). Lys99 in motif I is the site of covalent AMP attachment to Rnl1 (Thogersen et al. 1985), and it is poised for attack on the α phosphorus of AMPCPP in the Rnl1 crystal structure (Fig. 1B). Lys240 and Lys242 coordinate the α and β phosphates of AMPCPP, while Lys119 coordinates the γ phosphate. Asp101, Glu159, and Glu227 are components of the metal coordination complex that includes the ATP α phosphate. Glu159 also contacts the adenosine ribose 2′-OH. Three other essential residues, Arg54, Lys75, and Phe77, are located upstream of the AMP attachment site within a module of the N domain that defines the Rnl1/tRNA ligase clade (Fig. 1A, colored blue). Lys75 contacts the β phosphate of AMPCPP; Arg54 coordinates the γ phosphate and the ribose 3′-OH (El Omari et al. 2006). Tyr246 is located just distal to motif V, within the first α helix of the C domain, and is the only essential constituent of the C domain that has been identified to date (Wang et al. 2006b). Tyr246 is also a component of the metal coordination complex.
FIGURE 2.
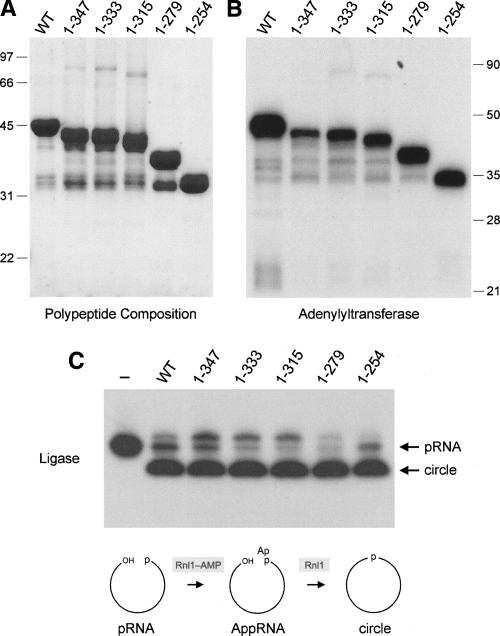
Truncations of the Rnl1 C domain. (A) Aliquots (5 μg) of recombinant wild-type (WT) Rnl1 and the indicated C-terminal truncation mutants were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Adenylyltransferase reaction mixtures (20 μL) containing 50 mM Tris-HCl (pH 8.0), 2 mM DTT, 5 mM MgCl2, 20 μM [α-32P]ATP, and 1 μg WT or mutant Rnl1 were incubated for 12 min at 37°C. The reaction products were analyzed by SDS-PAGE, and the ligase–[32P]AMP adducts were visualized by autoradiography. The positions and sizes (kDa) of marker polypeptides are indicated on the right. (C) RNA ligation reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 2 mM DTT, 20 μM ATP, 1 pmol of pRNA substrate, and 300 ng of WT or mutant Rnl1 were incubated at 37°C for 30 min. The products were resolved by PAGE and visualized by autoradiography. A control reaction lacking Rnl1 is shown in lane “–.” Step 2 (RNA adenylation) and step 3 (phosphodiester bond formation) reactions of the ligation pathway are illustrated schematically at the bottom.
The function of the remainder of the Rnl1 C domain in catalysis and RNA repair is uncharted. El Omari et al. (2006) have speculated that the C domain might comprise a tRNA recognition module, but there are no published experiments to speak to this issue. Here we initiated an analysis of the role of the C domain in Rnl1 function by serially deleting each of the α helices. We find that the C domain confers specificity for tRNA repair, but is not required for sealing of a generic single-stranded 5′-phosphate RNA substrate. Alanine scanning of the C domain identifies a conserved Arg318–Lys319 dipeptide as a candidate tRNA specificity determinant. A new round of alanine scanning of the N domain identifies several essential residues in motifs I and Ia.
RESULTS AND DISCUSSION
Structure-guided truncation of the C domain of Rnl1
To gauge whether and how the C domain contributes to Rnl1 function, we constructed a series of five deletion mutants that serially removed the component α helices (Fig. 1A). Full-length wild-type Rnl1 and mutants Rnl1-(1–347), Rnl1-(1–333), Rnl1-(1–315), Rnl1-(1–279), and Rnl1-(1–254) were produced in bacteria as His10 fusions and then purified from soluble lysates by Ni-agarose chromatography. SDS-PAGE analysis revealed the presence of serially truncated Rnl1 polypeptides of the expected size (Fig. 2A). Each of the Rnl1 preparations, except Rnl1-(1–254), also contained minor polypeptides of ≥ 32 kDa that apparently correspond to proteolytic fragments of Rnl1 that retained the N-terminal His10 tag. The Rnl1-(1–254) polypeptide comigrated with the 32-kDa fragment. We surmise that T4 Rnl1 contains a protease-accessible site close to aa 254 that might correspond to a domain boundary. All of the truncated proteins retained adenylyltransferase activity, as evinced by label transfer from [α-32P]ATP to the Rnl1 polypeptide to form a covalent enzyme–adenylate adduct (Fig. 2B). The size of the major labeled polypeptide decreased incrementally according to the extent of the C-terminal truncation. Minor labeled polypeptides were also observed that corresponded to the putative proteolytic fragments discussed above (Fig. 2B). We conclude that the N-terminal polypeptide from aa 1–254 suffices for the ligase–adenylylation reaction of T4 Rnl1.
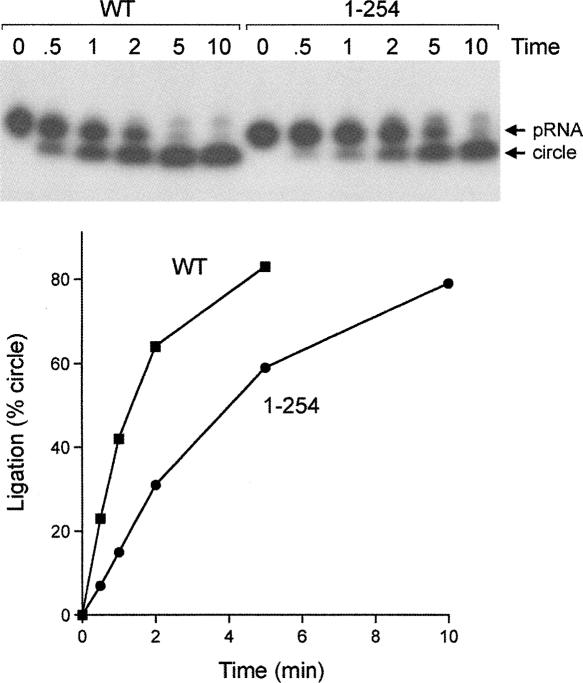
Reaction of wild-type Rnl1 with a 5′ 32P-labeled 18-mer RNA oligonucleotide (pRNA) in the presence of 20 μM ATP resulted in nearly complete conversion of the substrate to a new radiolabeled product, migrating faster than the input 18-mer pRNA strand, that corresponds to a covalently closed 18-mer circle formed by intramolecular ligation of the 5′-PO4 and 3′-OH termini of the pRNA (Fig. 2C). A minor product, migrating ∼ 1 nucleotide (nt) step slower than the input 18-mer, corresponds to the RNA–adenylate (AppRNA) generated by AMP transfer from Rnl1–AMP to the 5′ end of the input 18-mer RNA. The truncation mutants also catalyzed circularization of the substrate, implying that the C-domain is not essential for ligation (Fig. 2C). A finer analysis of the pRNA ligation reaction was performed by following the rate of product formation under conditions of enzyme excess (Fig. 3). RNA circularization by wild-type Rnl1 was virtually complete within 5 min, and there was little RNA–adenylate intermediate detected at earlier times when 20%–60% of the pRNA substrate had been consumed. Rnl1-(1–254) reacted with delayed kinetics but also did not accumulate RNA–adenylate intermediate. The initial rate of sealing by Rnl1-(1–254) was ∼ 40% of the wild-type value (Fig. 3). Thus, the C domain distal to aa 254 makes only a modest contribution to Rnl1 activity on a generic single-stranded RNA substrate. We did not test the effects of more extensive truncations upstream of aa 254 in light of our earlier finding that an alanine mutation of Tyr246 abolished ligase activity (Wang et al. 2006b).
FIGURE 3.
Effect of C domain deletion on the kinetics of RNA sealing. Ligation reaction mixtures (80 μL) containing 8 pmol of labeled pRNA, 20 μM ATP, and 2.4 μg wild-type (WT) Rnl1 or Rnl1-(1–254) were incubated at 37°C. Aliquots (10 μL) were withdrawn and quenched at the times specified above the lanes. The products were resolved by PAGE and visualized by autoradiography. The yield of circular RNA product (as percent of total radiolabeled material) is plotted as a function of time at the bottom.
The C domain is important for the tRNA repair function of T4 Rnl1
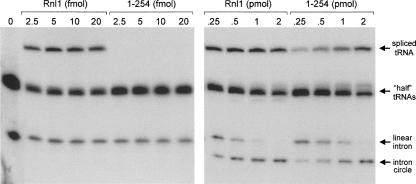
The physiological substrate for T4 Rnl1 is a tRNA containing a single break in the anticodon loop. T4 Rnl1 and T4 polynucleotide kinase–phosphatase (Pnkp) comprise a two-component repair system that heals and seals the tRNA break made by the host anticodon nuclease PrrC in response to bacteriophage infection (Amitsur et al. 1987). Pnkp is a bifunctional enzyme that remodels the ends of the broken tRNA by converting the 2′,3′ cyclic phosphate to a 3′-OH, 2′-OH and by phosphorylating the 5′-OH end to form a 5′-PO4. Rnl1 then joins the 3′-OH and 5′-PO4 RNA ends to form a standard 3′–5′ phosphodiester at the repair junction. To probe the role of the C domain of Rnl1 in a bona fide tRNA repair reaction, we exploited an in vitro tRNA splicing system designed by Englert and Beier (2005). The tRNA substrate containing a single break in the anticodon loop was generated by treating a 32P-labeled intron-containing pre-tRNA with a tRNA splicing endonuclease, which led to quantitative release of a linear 21-nt intron and the formation of two “half-tRNA” molecules: a 37-nt 5′ fragment and a 39-nt 3′ fragment (Fig. 4). Incubation of the broken RNA (140 fmol) with a combination of T4 Pnkp (1 pmol) and limiting amounts of wild-type T4 Rnl1 (2.5–20 fmol) resulted in formation of a mature spliced tRNA but no circularization of the intron (Fig. 4, left panel). Rather, intron circularization required ≥ 100-fold higher amounts of Rnl1, in the range of 0.25 to 2 pmol of ligase (Fig. 4, right panel). This experiment demonstrates that T4 Rnl1 has a strong intrinsic preference for sealing broken tRNA substrates in vitro.
FIGURE 4.
The C domain of Rnl1 confers specificity for tRNA repair in vitro. Reaction mixtures containing 50 mM Tris-acetate (pH 8.0), 10 mM MgCl2, 2 mM DTT, 20 μM ATP, 140 fmol 32P-labeled cleaved tRNA substrate, 1 pmol T4 Pnkp, and increasing amounts of Rnl1 or Rnl1-(1–254) (0, 2.5, 5, 10, or 20 fmol in the left panel and 0.25, 0.5, 1, or 2 pmol in the right panel) were incubated for 30 min at 37°C. The radiolabeled products were resolved by denaturing PAGE and visualized by autoradiography. The identities of the labeled RNAs are indicated by arrows at right.
The instructive findings were that Rnl1-(1–254) failed to catalyze sealing of the tRNA halves at limiting enzyme concentrations that were saturating for wild-type Rnl1 (Fig. 4, left panel), but it did generate spliced tRNA at much higher ligase concentrations (Fig. 4, right panel). Moreover, the tRNA splicing and intron circularization reactions displayed virtually equivalent dependence on the amount of Rnl1-(1–254) added in the range of 0.25 to 2 pmol (Fig. 4, right panel). Based on the level of intron circle as a function of input ligase, we estimate that Rnl1-(1–254) was half as active as wild-type Rnl1, which agrees well with the effect of C-domain deletion on circularization of the 18-mer pRNA substrate. Thus, the effect of deleting the C domain was to abolish the preference for a broken tRNA substrate, without significantly impacting its action on the linear intron. Analyses of tRNA repair in vitro by the Rnl1-(1–347), Rnl1-(1–333), Rnl1-(1–315), and Rnl1-(1–279) proteins revealed them to have the same selective defect in tRNA sealing as Rnl1-(1–254) (not shown).
The enzymatic steps in bacteriophage tRNA repair are broadly similar to those of yeast tRNA splicing. The end-healing and strand-sealing steps of yeast tRNA splicing are performed by a single polypeptide, Trl1, composed of discrete healing and sealing domains (Sawaya et al. 2003). We showed previously that the phage and yeast RNA repair systems are portable in vivo, insofar as a lethal trl1Δ mutation of Saccharomyces cerevisiae can be rescued to normal growth by coexpression of bacteriophage T4 Rnl1 and Pnkp (Schwer et al. 2004). This allows us to test structure–function relationships for Rnl1 by using a plasmid-based functional complementation assay in yeast (Wang et al. 2006). Here we found that, whereas coexpression of wild-type T4 Rnl1 and Pnkp rescued the trl1Δ mutation at 18, 30, and 37°C, the five Rnl1 truncation mutants were unable to support cell growth at any temperature tested (not shown). We surmise that the C domain of Rnl1 is critical for tRNA repair in vitro and in vivo. The simplest interpretation is that lethality in yeast is caused by the loss of tRNA repair activity seen in vitro, albeit subject to the caveat that the Rnl1 C domain truncations might also have reduced the levels of Rnl1 produced in yeast.
Alanine scanning of the C domain
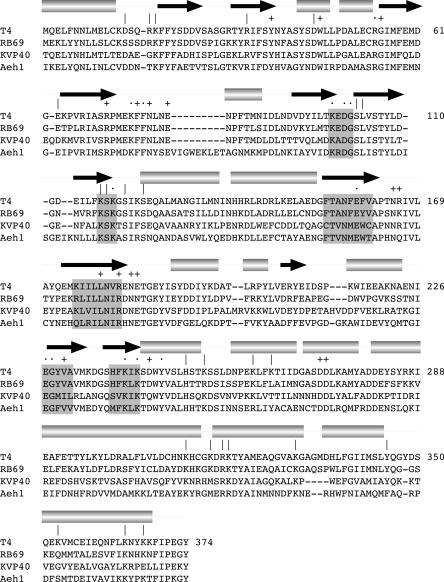
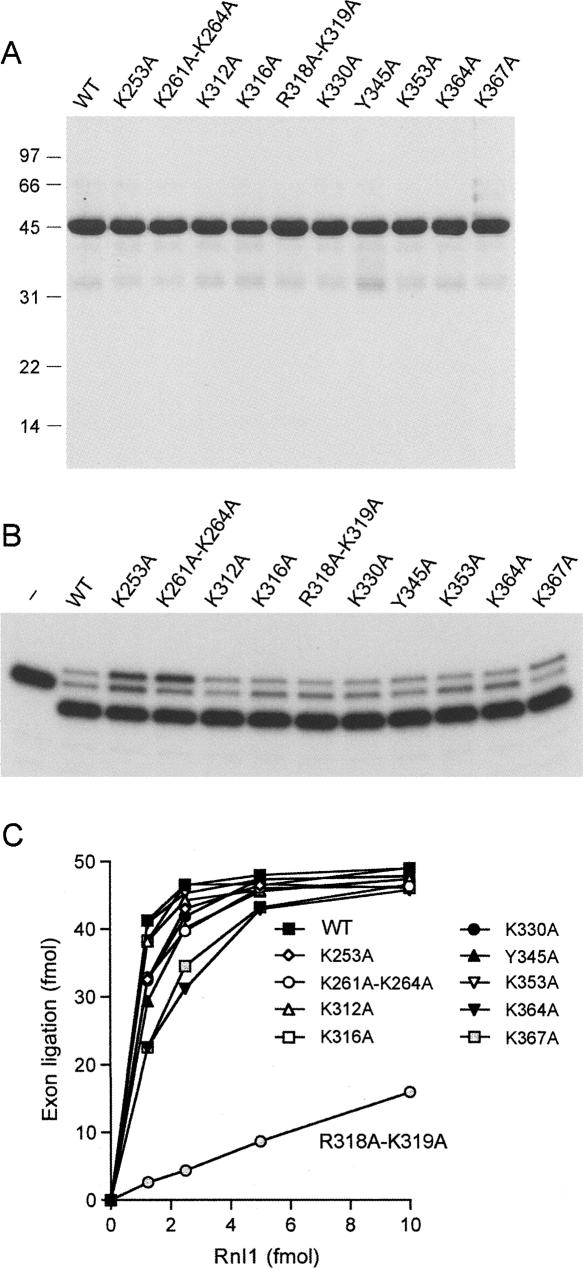
Although the deletion analysis presented above suggests a role for the C domain in tRNA recognition, as initially speculated by El Omari et al. (2006), we cannot discern from the data whether loss of tRNA specificity upon deletion of even one terminal α helix reflects participation of the deleted element in the ligase–tRNA interface or a role for that element in attaining proper folding of the C domain. As an initial step in identifying candidate constituents of a putative tRNA-binding surface, we tested the effect of 10 single-alanine or double-alanine mutations within the C domain of the full-length Rnl1 polypeptide. In choosing which residues to mutate, we focused on lysine and arginine side chains located on the surface of the C domain in the Rnl1 crystal structure as potential ligands for the phosphodiester backbone of the folded tRNA. The basic residues targeted for mutation (Lys253, Lys261–Lys264, Lys312, Lys316, Arg318–Lys319, Lys330, Lys353, Lys364, and Lys367) are indicated by “|” over the aligned viral Rnl1 sequences in Figure 5. The T4 Rnl1 Lys253, Lys261, Lys316, Arg318, Lys319, and Lys364 positions are conserved as basic residues in the Rnl1 homologs from phages RB69, KVP40, and Aeh1 (Fig. 5). In addition, we mutated Tyr345, a conserved aromatic residue. The 10 C-domain Ala-mutant proteins were produced in bacteria as His10 fusions and purified from soluble lysates by Ni-agarose chromatography (Fig. 6A). All of the mutants retained adenylyltransferase activity (data not shown) and pRNA circularization activity (Fig. 6B), as expected given that these functions of Rnl1 survive deletion of the C domain.
FIGURE 5.
Targeted alanine mutagenesis of Rnl1. The amino acid sequence of T4 Rnl1 is aligned to the sequences of the homologous proteins of coliphage RB69, vibriophage KVP40, and Aeromonas hydrophila phage Aeh1. The secondary structure elements of T4 Rnl1 are displayed above the sequence, with β strands indicated by arrows and helices as cylinders. Nucleotidyl transferase motifs I, Ia, III, IIIa, IV, and V are highlighted in shaded boxes. T4 Rnl1 residues identified previously as essential are indicated by “·.” Nonessential residues are indicated by “+.” The amino acids mutated in the present study are denoted by “|.”
FIGURE 6.
Alanine scanning of the Rnl1 C domain. (A) Aliquots (5 μg) of recombinant wild-type (WT) and mutated Rnl1 proteins were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. Positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) RNA ligation reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 2 mM DTT, 20 μM ATP, 1 pmol of pRNA substrate, and 300 ng of WT or mutant Rnl1 were incubated at 37°C for 30 min. The products were resolved by PAGE and visualized by autoradiography. A control reaction lacking Rnl1 is shown in lane “–.” (C) Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 2 mM DTT, 20 μM ATP, 140 fmol 32P-labeled cleaved tRNA substrate, 1 pmol T4 Pnkp, and increasing amounts of WT or mutant Rnl1 (1.25, 2.5, 5, or 10 fmol) were incubated for 30 min at 37°C. The products were resolved by denaturing PAGE and quantified by scanning the gel with a phosphorimager. The extent of tRNA exon ligation is plotted as a function of input Rnl1.
When the Ala mutants were tested for tRNA repair as a function of input enzyme, only one of the proteins, R318A-K319A, displayed a significant loss of activity, i.e., < 10% of wild-type Rnl1 (Fig. 6C). It is conceivable, based on these findings, that Arg318 and/or Lys319 are components of the tRNA interface, especially given that neither side chain makes atomic contacts to another moiety of the Rnl1 protein in the crystal structure. We do not exclude the possibility that one or more of the other targeted basic residues might also contact tRNA. Perhaps the existence of multiple protein–tRNA interactions lessens the effect of losing an individual constituent of the interface. We found that all of the C domain Ala mutants were able to complement the yeast trl1Δ strain when co-expressed with Pnkp (not shown). Apparently, the residual tRNA splicing activity of the R318A-K319A mutant sufficed to support viability in yeast. We noted previously that partially active nucleotidyltransferase mutants of T4 Rnl1 were also able to support yeast growth (Wang et al. 2006b).
New structure-guided alanine scan of the N domain
Previous mutational analyses delineated a suite of catalytically essential functional groups that comprise the Rnl1 active site (Wang et al. 2003, 2006b). The crystal structure of the Rnl1–AMPPCP complex provides a blueprint for additional structure–function studies conducted here. We tested the effects of alanine mutations at 13 individual positions located within the N domain (Fig. 5, indicated by “|”). Residues remote from the ATP-binding site that make bridging contacts between secondary structure elements were mutated to probe whether such contacts are important for Rnl1 folding and function. This category includes Asp14, Arg17, Lys18, and Asp44, which form a salt-bridge network on the enzyme surface (Fig. 1B, top left in the stereoview) and Arg33 and Glu63, which comprise another ion pair (Fig. 1B, top right). Residues Ser103 and Leu104, flanking the Lys99 adenylylation site in motif I (99KEDGSL104), were also targeted in the alanine scan. Ser103 is located on the back side of the active site and makes a tethering contact to Asn165 (Fig. 1B). Leu104 projects into the active site and makes van der Waals contacts with the adenylate ribose (Fig. 1B). Mutations were introduced at positions Lys117, Ser118, Ser121, and Ser124 within and flanking motif Ia (117KSKGSIKS124), which contributes the essential Lys119 moiety to the active site (Wang et al. 2003). Although Lys119 contacts the γ phosphate of ATP in the crystal structure (Fig. 1B), functional studies established that Lys119 (along with Arg54) is dispensable for Rnl1 adenylylation (step 1) but required for the pRNA adenylylation reaction (step 2) of the ligation pathway (Wang et al. 2003). Lys119 is located at the tip of a hairpin loop; the loop appears to be stabilized by a network of side- and main-chain hydrogen bonds involving the presently targeted residues Lys117, Ser118, Ser121, and Ser124 (Fig. 1B). Finally, we targeted His250, a conserved residue (Fig. 5) located near the end of the minimal catalytic domain, which is pointing toward the active site in the crystal structure (Fig. 1B) and thereby deemed a candidate for a role in RNA binding.
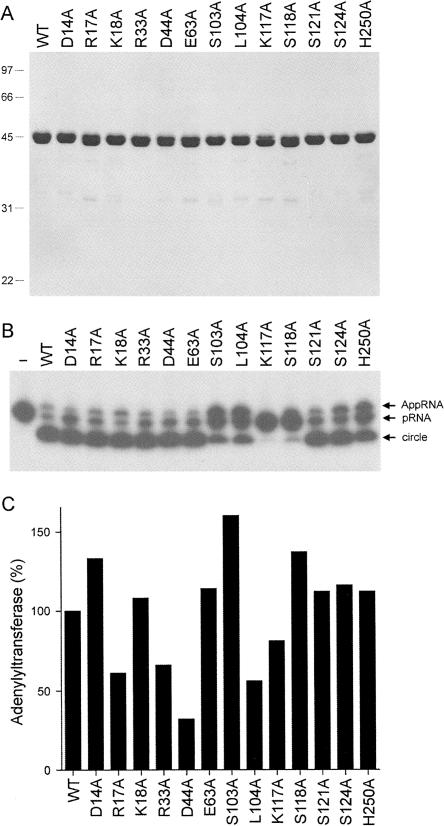
The His10–Rnl1–Ala mutants were produced in bacteria and then purified from soluble lysates by Ni-agarose chromatography (Fig. 7A). Assays for pRNA ligation revealed gross defects in sealing for K117A and S118A (Fig. 7B). S103A, L104A, and H250A displayed partial defects, as judged by the lesser extent of product consumption and/or the accumulation of AppRNA intermediate (Fig. 7B). The other mutants (D14A, R17A, K18A, R33A, D44A, E63A, S121A, and S124A) displayed wild-type–like activity in pRNA circularization in the single-point assay (Fig. 7B). Thus, we surmise that the two salt-bridge networks formed by Asp14, Arg17, Lys18, Asp44, Arg33, and Glu63 are not essential for Rnl1 function.
FIGURE 7.
Alanine scanning of the Rnl1 N domain. (A) Aliquots (5 μg) of recombinant wild-type (WT) and mutated Rnl1 proteins were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) RNA ligation reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 2 mM DTT, 20 μM ATP, 1 pmol of pRNA substrate, and 300 ng of WT or mutant Rnl1 were incubated at 37°C for 30 min. The products were resolved by PAGE and visualized by autoradiography. A control reaction lacking Rnl1 is shown in lane “–.” (C) Adenylyltransferase reaction mixtures (20 μL) containing 50 mM Tris-HCl (pH 8.0), 2 mM DTT, 5 mM MgCl2, 20 μM [α-32P]ATP, and 0.5, 1.0, or 1.5 μg of WT or mutant Rnl1 were incubated for 12 min at 37°C. The reaction products were analyzed by SDS-PAGE. The ligase–[32P]AMP adduct was quantified by scanning the gel. The adenylyltransferase-specific activities (pmol ligase–[32P]AMP formed per μg of protein) were calculated from the slopes of the titration curves and the activities of the mutants were normalized to that of WT Rnl1 (defined as 100%).
Each of the mutants retained adenylyltransferase activity in vitro (Fig. 7C). The instructive point of the adenylyltransferase assay was that the defects of the S103, K117A, and Ser118A proteins in pRNA sealing cannot be attributed to mutational effects on Rnl1 adenylylation, insofar as these mutants formed 70%–140% as much Rnl1–AMP in vitro as the wild-type ligase (Fig. 7C). Note that the extent of label transfer from ATP to Rnl1 in vitro is influenced by the fraction of the protein preparation that is preadenylylated, which accounts for some of the variability in adenylyltransferase (Fig. 7C). We generally impose a criterion of ≤ 10% of wild-type adenylylation, combined with a loss of function in overall ligation, before deeming mutational effects on step 1 to be significant (Wang et al. 2003).
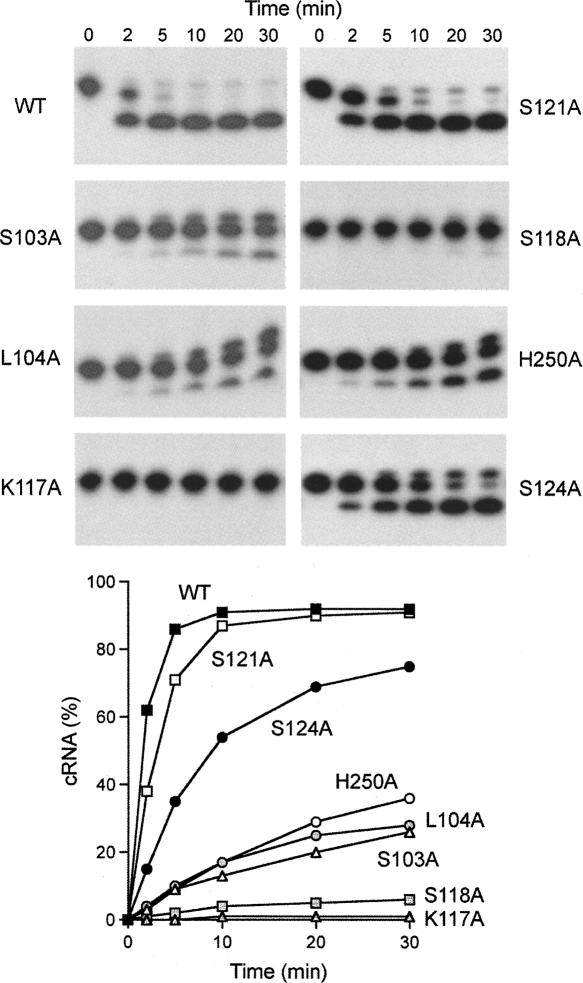
A finer analysis of the pRNA ligation reaction by the motif I, motif Ia, and H250A mutants was performed by following the rate of product formation during a 30-min reaction under conditions of enzyme excess (Fig. 8). Whereas RNA circularization by wild-type Rnl1 was virtually complete within 5 min, the K117A mutant formed no ligated product after 30 min (Fig. 8). S118A formed trace amounts of circle and AppRNA after 30 min. We surmise that Lys117 and Ser118 in motif Ia are essential for catalysis of the RNA adenylylation reaction (step 2). The other motif Ia mutations had more modest effects on the rate of sealing. S124A slowed the rate by a factor of 4 compared to wild-type Rnl1, while S121A reacted at half the wild-type rate (Fig. 8). In the Rnl1 crystal structure, Lys117 makes a trifurcated hydrogen bond to the main-chain carbonyls of Ser118, Lys119, and Gly55 (Fig. 1B). We suspect that Lys117 is essential because these contacts ensure the proper conformation of the motif Ia loop (and perhaps also aid in positioning essential residue Arg54), rather than because the Lys117 side chain (which is not on the enzyme surface) might make direct contact to the RNA substrate. Ser118 makes a bifurcated hydrogen bond to the Ser121 main-chain carbonyl and to Ser124 Oγ (Fig. 1B). Because the S124A mutation had a much milder effect than the S118A change, we suspect that the Ser118 hydrogen bond to the Ser121 carbonyl is the major contribution of Ser118 in attaining an active conformation of the motif Ia loop. The benign effects of the S121A mutation indicate that the bifurcated hydrogen bonds from Ser121 Oγ to the main-chain amides of Ile122 and Lys123 seen in the crystal (Fig. 1B) are not important for Rnl1 activity in vitro.
FIGURE 8.
Kinetics of pRNA ligation by N domain mutants. Ligation reaction mixtures containing labeled pRNA and Rnl1 as specified were incubated at 37°C. Aliquots were withdrawn and quenched at the times specified above the lanes. The products were resolved by PAGE and visualized by autoradiography. The yield of circular RNA product (as percent of total radiolabeled material) is plotted as a function of time at the bottom.
The rates of pRNA circularization by the S103A, L104A, and H250A mutants were 5%, 6%, and 6% of the wild-type rate, respectively (Fig. 8). S103A and L104A appeared to generate higher levels of the AppRNA intermediate (relative to ligated circle) than did the other proteins analyzed in parallel. Because His250 makes no contacts to ATP or other Rnl1 constituents in the crystal structure but is located above the adenylate phosphate (Fig. 1B), it emerges from the mutational study as a candidate for interaction with one of the reactive RNA termini. Ser103 could be playing a tethering role via its hydrogen bond to Asn165, but it might also make contact to RNA from its position on the protein surface (Fig. 1B). We presume that the effects of L104A reflect a role in positioning the adenylate in the Rnl1–AMP active site. In the case of other polynucleotide ligases, the AMP is implicated in recognizing the 5′-PO4 end of the nucleic acid substrate (Odell et al. 2000; Nandakumar and Shuman 2004; Nair et al. 2007).
When tested for tRNA repair activity in vivo in yeast, the K117A allele failed to support growth under FOA selection at 18, 30, or 37°C. Thus the K117A change was lethal in vivo. The S103A, L104A, S118A, S121A, S124A, and H250A alleles all yielded FOA-resistant colonies at 30°C. The resulting strains were then tested for growth on rich medium (YPD agar) at 18, 30, and 37°C. The S118A, S103A, and L104A strains were temperature sensitive (no growth at 37°C). The S121A, S124A, and H250 strains grew at all temperatures (not shown). Thus, the hierarchy of mutation effects on pRNA sealing in vitro assay correlated with the severity of growth phenotypes in vivo.
Conclusions
We have exploited the crystal structure of Rnl1 (El Omari et al. 2006) to functionally dissect the active site and probe the role of the unique C domain in tRNA repair. An in vitro tRNA repair system (Englert and Beier 2005) allowed us to demonstrate (for the first time, to our knowledge) that T4 Rnl1 has an inherent specificity for sealing tRNA with a break in the anticodon loop. The broken tRNA is preferred ∼ 100-fold compared to the linear intron liberated by the tRNA splicing endonuclease. The Pnkp-healed linear intron is analogous to the synthetic linear pRNA used as a generic substrate for Rnl1 in our studies and to the linear poly(A)20–40 substrate exploited by Hurwitz and colleagues in their original identification and characterization of T4 Rnl1 as an RNA-circularizing enzyme (Silber et al. 1972; Cranston et al. 1974). We find here that the tRNA specificity of Rnl1 is conferred by the C domain, any deletion of which levels the playing field so that the broken tRNA is now sealed as well (or rather as poorly) as the linear intron. Our deletion analysis demarcates Rnl1-(1–254) as a minimal catalytic domain of Rnl1, capable of all chemical steps of the pRNA ligation reaction. A new round of alanine scanning of 13 residues within the minimal N domain identified Ser103, Leu104, Lys117, and Ser118 as important for pRNA ligation in vitro and tRNA repair in vivo.
Our initial efforts to identify candidate tRNA specificity determinants by alanine scanning of the C domain suggest that the conserved Arg318-Lys319 dipeptide might play such a role. The present study opens up a number of avenues for further study of Rnl1 specificity, including footprint analysis of Rnl1–tRNA contacts, alterations of the tRNA structure to delineate features important for preferential sealing by Rnl1, and (guided by the results of such studies) attempts to cocrystallize Rnl1 with a broken tRNA or tRNA-like substrate.
MATERIALS AND METHODS
Recombinant T4 Rnl1
Stop codons and amino acid substitution mutations were introduced into the rnl1 gene by PCR amplification with mutagenic primers (Wang et al. 2003). The mutated genes were digested with NdeI and BamHI and then inserted into pET16b. The plasmid inserts were sequenced completely to exclude the acquisition of unwanted changes during amplification and cloning. Wild-type and mutant pET-RNL1 plasmids were transformed into Escherichia coli BL21(DE3). Induction of Rnl1 production with IPTG and purification of His10–Rnl1 from soluble bacterial extracts by Ni-agarose chromatography were performed as described previously (Wang et al. 2003). Protein concentrations were determined with the Bio-Rad dye reagent using bovine serum albumin as the standard.
RNA ligase assay
An 18-mer oligoribonucleotide (5′-AUUCCGAUAGUGACUACA) was 5′ 32P-labeled using T4 polynucleotide kinase and [γ-32P]ATP. The labeled 18-mer was purified by electrophoresis through a 20% polyacrylamide gel. RNA ligation reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 8.0), 2 mM DTT, 10 mM MgCl2, 1 pmol of 5′ 32P-labeled 18-mer RNA (pRNA), 20 μM ATP, and 300 ng Rnl1 were incubated for 30 min at 37°C. The reactions were quenched by adding 5 μL of 95% formamide and 20 mM EDTA. For kinetic analysis of RNA sealing, reaction mixtures (80 μL) containing 50 mM Tris-HCl (pH 8.0), 2 mM DTT, 10 mM MgCl2, 8 pmol of labeled pRNA, 20 μM ATP, and 2.4 μg Rnl1 were incubated at 37°C. The reactions were initiated by adding enzyme. Aliquots (10 μL) were withdrawn at the time specified and quenched immediately with formamide and EDTA. The samples were analyzed by electrophoresis through a 18% polyacrylamide gel containing 7 M urea in 45 mM Tris-borate and 1 mM EDTA. The ligation reaction products were visualized by autoradiography of the gel and quantified with a Fujifilm BAS-2500 imager.
Adenylyltransferase assay
Reaction mixtures (20 μL) containing 50 mM Tris-HCl (pH 8.0), 2 mM DTT, 5 mM MgCl2, 20 μM [α-32P]ATP, and wild-type or mutant Rnl1 as specified were incubated for 12 min at 37°C. The reactions were quenched with SDS, and the products were analyzed by SDS-PAGE. The ligase–[32P]AMP adduct was visualized by autoradiography of the dried gel and, where indicated, quantified by scanning the gel with a Fujifilm BAS-2500 imager.
Assay of tRNA repair in vitro
The intron-containing pre-tRNA is a chimera consisting of the mature tRNA sequence of plant pre-tRNATyr plus the intron and anticodon of Methanocaldococcus pre-tRNATrp (Englert and Beier 2005). This pre-tRNA was generated by in vitro transcription of BstN1-cut plasmid pNtY9-T7-M1 by T7 RNA polymerase in the presence of [α-32P]ATP as described (Keppetipola et al. 2007). The labeled pre-tRNA was cleaved within the anticodon loop by treatment with Methonocaldococcus jannaschii tRNA splicing endonuclease as described (Englert and Beier 2005; Keppetipola et al. 2007). The tRNA cleavage products were phenol-extracted, precipitated with ethanol, resuspended in 100 μL of 10 mM Tris-HCl (8.0), 1 mM EDTA, and stored at −20°C. The tRNA repair reaction mixtures (10 μL) containing 50 mM Tris-acetate (pH 8.0), 10 mM MgCl2, 2 mM DTT, 20 μM ATP, 140 fmol radiolabeled cleaved tRNA substrate, 1 pmol of T4 Pnkp, and increasing amounts of Rnl1 as specified were incubated for 30 min at 37°C. The reactions were quenched by adding 10 μL of 95% formamide/50 mM EDTA. The samples were heated at 95°C for 2 min and then analyzed by electrophoresis through a 15-cm 12.5% polyacrylamide gel containing 7 M urea in 45 mM Tris-borate/1 mM EDTA. The products were visualized by autoradiography.
Test of Rnl1 tRNA repair function in vivo by plasmid shuffle
The trl1Δ haploid strain YRS1 (Sawaya et al. 2003) was co-transformed with a CEN TRP1 RNL1 plasmid bearing a wild-type or mutated version of T4 Rnl1 under the control of the yeast TPI1 promoter and a CEN HIS3 PNKP plasmid expressing bacteriophage T4 Pnkp under the control of the yeast SLU7 promoter. Transformants were selected on medium lacking tryptophan and histidine. Two individual colonies were transferred to fresh selective medium. The isolates were then streaked on agar medium containing 0.75 mg/mL 5-FOA. The plates were incubated at 18, 30, and 37°C. Lethal mutations were those that did not allow formation of FOA-resistant colonies after 7–10 d at any of the temperatures tested. Other mutated alleles supported FOA-resistant colony formation at one or more of the growth temperatures. Individual colonies were picked from the FOA plate, transferred to yeast extract/peptone/dextrose (YPD) medium, and then tested for growth on YPD agar at 18, 30, and 37°C.
ACKNOWLEDGMENTS
We thank Markus Englert and Hildburg Beier for the generous gift of plasmids containing the hybrid pre-tRNA and archaeal splicing endonuclease genes. This work was supported by NIH grant GM42498. S.S. is an American Cancer Society Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.591807.
REFERENCES
- Amitsur, M., Levitz, R., Kaufman, G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranston, J.W., Silber, R., Malathi, V.G., Hurwitz, J. Studies on ribonucleic acid ligase: Characterization of an adenosine triphosphate–inorganic pyrophosphate exchange reaction and demonstration of an enzyme–adenylate complex with T4 bacteriophage-induced enzyme. J. Biol. Chem. 1974;249:7447–7456. [PubMed] [Google Scholar]
- El Omari, K., Ren, J., Bird, L.E., Bona, M.K., Klarmann, G., LeGrice, S.F.J., Stammers, D.K. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- Englert, M., Beier, H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy, S., Singh, M., Gait, M.J. Effects of single amino acid changes in the region of the adenylylation site of T4 RNA ligase. Biochemistry. 1987;26:1688–1696. doi: 10.1021/bi00380a030. [DOI] [PubMed] [Google Scholar]
- Keppetipola, N., Nandakumar, J., Shuman, S. Reprogramming the tRNA splicing activity of a bacterial RNA repair enzyme. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, P.A., Nandakumar, J., Smith, P., Odell, M., Lima, C.D., Shuman, S. Structural basis for nick recognition by a minimal pluripotent DNA ligase. Nature Struct. Mol. Biol. 2007 doi: 10.1038/nsmb1266. (in press) [DOI] [PubMed] [Google Scholar]
- Nandakumar, J., Shuman, S. How an RNA ligase discriminates RNA damage versus DNA damage. Mol. Cell. 2004;16:211–221. doi: 10.1016/j.molcel.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Nandakumar, J., Shuman, S., Lima, C.D. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive the reaction forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- Odell, M., Sriskanda, V., Shuman, S., Nikolov, D.B. Crystal structure of eukaryotic DNA ligase–adenylate illuminates the mechanism of nick sensing and strand joining. Mol. Cell. 2000;6:1183–1193. doi: 10.1016/s1097-2765(00)00115-5. [DOI] [PubMed] [Google Scholar]
- Phizicky, E.M., Consaul, S.A., Nehrke, K.W., Abelson, J. Yeast tRNA ligase mutants are nonviable and accumulate tRNA splicing intermediates. J. Biol. Chem. 1992;267:4577–4582. [PubMed] [Google Scholar]
- Sawaya, R., Schwer, B., Shuman, S. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J. Biol. Chem. 2003;278:43298–43398. doi: 10.1074/jbc.M307839200. [DOI] [PubMed] [Google Scholar]
- Schwer, B., Sawaya, R., Ho, C.K., Shuman, S. Portability and fidelity of RNA-repair systems. Proc. Natl. Acad. Sci. 2004;101:2788–2793. doi: 10.1073/pnas.0305859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman, S., Lima, C.D. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Silber, R., Malathi, V.G., Hurwitz, J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl. Acad. Sci. 1972;69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino, A., Snopek, T.J., Cozzarelli, N.R. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J. Biol. Chem. 1977;252:1732–1738. [PubMed] [Google Scholar]
- Thogersen, H.C., Morris, H.R., Rand, K.N., Gait, M.J. Location of the adenylylation site in T4 RNA ligase. Eur. J. Biochem. 1985;147:325–329. doi: 10.1111/j.1432-1033.1985.tb08753.x. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck, O.C., Gumport, R.I. T4 RNA ligase. Enzymes. 1982;15:31–58. [Google Scholar]
- Wang, L.K., Ho, C.K., Pei, Y., Shuman, S. Mutational analysis of bacteriophage T4 RNA ligase 1. Different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation step of the ligation reaction. J. Biol. Chem. 2003;278:29454–29462. doi: 10.1074/jbc.M304320200. [DOI] [PubMed] [Google Scholar]
- Wang, L.K., Shuman, S. Structure–function analysis of yeast tRNA ligase. RNA. 2005;11:966–975. doi: 10.1261/rna.2170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.K., Schwer, B., Englert, M., Beier, H., Shuman, S. Structure–function analysis of the kinase–CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res. 2006a;34:517–527. doi: 10.1093/nar/gkj441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.K., Schwer, B., Shuman, S. Structure-guided mutational analysis of T4 RNA ligase 1. RNA. 2006b;12:2126–2134. doi: 10.1261/rna.271706. [DOI] [PMC free article] [PubMed] [Google Scholar]