Abstract
A series of nucleobase-modified siRNA duplexes containing “rare” nucleosides, 2-thiouridine (s2U), pseudouridine (Ψ), and dihydrouridine (D), were evaluated for their thermodynamic stability and gene silencing activity. The duplexes with modified units at terminal positions exhibited similar stability as the nonmodified reference. Introduction of the s2U or Ψ units into the central part of the antisense strand resulted in duplexes with higher melting temperatures (Tm). In contrary, D unit similarly like wobble base pair led to the less stable duplexes (ΔTm 3.9 and 6.6°C, respectively). Gene-silencing activity of siRNA duplexes directed toward enhanced green fluorescent protein or beta-site APP cleaving enzyme was tested in a dual fluorescence assay. The duplexes with s2U and Ψ units at their 3′-ends and with a D unit at their 5′-ends (with respect to the guide strands) were the most potent gene expression inhibitors. Duplexes with s2U and Ψ units at their 5′-ends were by 50% less active than the nonmodified counterpart. Those containing a D unit or wobble base pair in the central domain had the lowest Tm, disturbed the A-type helical structure, and had more than three times lower activity than their nonmodified congener. Activity of siRNA containing the wobble base pair could be rescued by placing the thio-nucleoside at the position 3′-adjacent to the mutation site. Thermally stable siRNA molecules containing several s2U units in the antisense strand were biologically as potent as their native counterparts. The present results provide a new chemical tool for modulation of siRNA gene-silencing activity.
Keywords: siRNA, thermodynamic stability, duplex asymmetry, 2-thiouridine, chemical modification
INTRODUCTION
Exogenous control of gene expression for functional studies and potentially for therapeutic applications became realistic by the use of RNA interference (RNAi) technology. Shortly after its discovery by Fire et al. (1998) in the Caenorhabditis elegans system, the RNAi machinery was proven to be present and effective in mammalian cells (Elbashir et al. 2001a). Sequence-specific gene silencing may be induced by synthetic, short, double-stranded RNAs (short interfering RNAs, siRNAs) identified in Drosophila melanogaster embryo extracts (Zamore et al. 2000; Elbashir et al. 2001b; Ketting et al. 2001). Most commonly used siRNA duplexes consist of two 21-nucleotide (nt) complementary strands terminated on their 3′-ends with 2-nt overhangs (Elbashir et al. 2001b; Nykanen et al. 2001; Tang et al. 2003). The siRNA duplex is incorporated into nucleoprotein complex RISC (RNA-induced silencing complex) (Martinez et al. 2002). One of the two strands is selected as a guide strand and the other one, designated as the sense or passenger strand, is endonucleolytically cleaved by the RISC (Matranga et al. 2005; Rand et al. 2005). The remaining guide (antisense) strand directs the RISC complex to the complementary mRNA target. Assembly of the active RISC complex is an asymmetrical process in which the selection of a guide siRNA strand is determined by the relative thermodynamic stability of the 3′- and 5′-ends of the siRNA duplex (Khvorova et al. 2003; Schwarz et al. 2003).
Even though siRNAs have been successfully used for gene silencing in vivo, there is still a need to improve their pharmacokinetic features, which predetermine therapeutic utility. One approach to reach this goal is the introduction of chemical modifications into siRNA duplexes (Manoharan 2004; Fougerolles et al. 2005; Uprichard 2005; Nawrot and Sipa 2006). Research efforts in this field are focused on improving three of the most important siRNAs features: (1) cell membrane permeability, (2) in vivo stability, and (3) specificity of their silencing action. Insufficient cross-membrane cellular uptake limits the application of nonmodified oligonucleotides, including siRNA, in medicine. Interestingly, Soutschek and colleagues reported improvement in in vivo cell membrane trafficking of siRNA with a cholesteryl group attached at the 3′-end (Soutschek et al. 2004).
As far as stability is concerned, chemical modifications introduced into internucleotide phosphate bonds, or at the C2′ position of the ribose ring, have kindled some hopes for enhanced resistance of siRNA to hydrolysis by cellular nucleases. Encouraging results have been reported concerning replacement of internucleotide phosphates with phosphorothioates (Li et al. 2005) as well as with boranophosphates (Hall et al. 2004). Phosphorothioate-derived oligonucleotides have been reported to stimulate the physical cellular uptake of siRNA in human cells (Overhoff and Sczakiel 2005). The studies performed so far with siRNAs modified at the C2′ position included introduction of fluorine (Layzer et al. 2004; Dowler et al. 2006), LNA units (LNA = locked nucleic acid) (Braasch et al. 2003; Elmen et al. 2005), as well as 2′-O-alkyl modifications (2′-O-Me, 2′-O-MOE) (Prakash et al. 2005). Recently 4′-thioribose appeared to be another interesting example of a stabilizing modification (Hoshika et al. 2005; Dande et al. 2006).
The last of the above-mentioned problems with the therapeutic application of siRNA is related to the induction of so-called “off target effects,” first documented by Jackson and colleagues with cDNA-microarray technology (Jackson et al. 2003). Recent reports (Fedorov et al. 2006; Jackson et al. 2006) provide some evidence that chemical modification of the guide strand of the siRNA duplex can suppress such unintended gene silencing.
Base-modified siRNAs have been investigated so far to a very limited extent (Parrish et al. 2000; Chiu and Rana 2003). The paper by Parrish et al. (2000) describes induction of RNAi in C. elegans by dsRNAs containing 4-thiouridine, 5-bromo-, 5-iodo-, 5-(3-aminoallyl)-uridine, or inosine. While these experiments did not demonstrate any remarkable differences in silencing activity regardless of the introduced modifications, the paper by Chiu and Rana (2003) describes reduced silencing activity of duplexes containing 5-bromouridine, 5-iodouridine, 2,6-diaminopurine, and N-3-methyl-uridine. The patent literature also claims several base-modified nucleosides as components of small interfering nucleic acids, albeit no details are provided with regard to properties of such modified siRNAs (Leake et al. 2004).
In these studies we evaluated the thermodynamic stability and gene-silencing activity of base-modified siRNAs containing three naturally occurring modified nucleosides: 2-thiouridine (s2U), pseudouridine (Ψ), and dihydrouridine (D). As has been already demonstrated, the first two nucleosides, due to the preferred C3′-endo conformation of the ribose ring (Agris et al. 1992; Davis et al. 1998), improve the thermodynamic stability of the s2U- (Kumar and Davis 1997; Luyten and Herdewijn 1998; Testa et al. 1999; Shohda et al. 2000) and the Ψ-containing double-stranded RNA helices (Davis et al. 1998). Moreover, the s2U unit, when present in the anticodon sequence of transfer RNA, enhances its specificity due to preferred base pairing with A and restricted wobble base pairing with G (Agris et al. 1992). Introduction of the s2U unit into the siRNA structure also seems to be profitable due to increased hydrophobicity of sulfur-modified molecules which might exhibit elevated cellular uptake (Cummins et al. 1995). The dihydrouridine unit destabilizes the RNA structure as it prefers the C2′-endo ribose ring conformation and induces the B-DNA-type sugar ring conformation (Stuart et al. 1996). In addition, the D nucleoside contains a nonaromatic base ring that excludes stabilizing base stacking with neighboring nucleotides within the RNA strand (Westhof et al. 1988; Davis 1998). In these studies modified nucleosides were introduced into functionally important regions of the siRNA duplex, e.g., at position 10 of the antisense strand, which is opposite the cleaved internucleotide bond of a target mRNA sequence (Elbashir et al. 2001c), or at the 3′-ends of the sense and antisense strands (Khvorova et al. 2003; Schwarz et al. 2003). The results presented here throw some light on the area of siRNA structure–activity relationships and provide a new chemical tool for modulation of siRNA-silencing potency.
RESULTS
Dual fluorescence reporter system
For silencing activity studies we validated the quantitative reporter system established by Chiu and Rana, based on measurement of the relative fluorescence intensity of the enhanced green fluorescent protein (EGFP) and red fluorescent protein (RFP), expressed in HeLa cells from the pEGFP-C1 and pDsRed1-N1 plasmids, respectively (Chiu and Rana 2002, 2003). Our modifications of the model system included optimization of transfection conditions with commercially available pDsRed2-N1 plasmid or pBACE1–GFP fusion plasmid, provided by Dr Weihong Song, The University of British Columbia, Vancouver, Canada, and a 96-well plate format, compatible with the Synergy HT plate reader (details in Materials and Methods). Correlation between fluorescence intensity and the level of mRNA of fluorescent proteins was confirmed by RT-PCR (data included in Supplemental Material).
It was suggested by Layzer et al. (2004) that transient transfection performed in the Chiu and Rana system may lead to some false positive results of siRNA activity. To eliminate the possible influence of the decay of EGFP gene expression in the time course of the experiment, we performed a time-dependent experiment (HeLa cells cotransfected with pEGFP-C1 and pDsRed2-N1 plasmids) and observed only negligible decrease of the fluorescence intensity over 228 h after transfection (data included in Supplemental Material). Therefore, we conclude that the decrease of the fluorescence level, observed in our 36-h assay, results mostly from the siRNA activity.
Selection of model siRNA for chemical modification
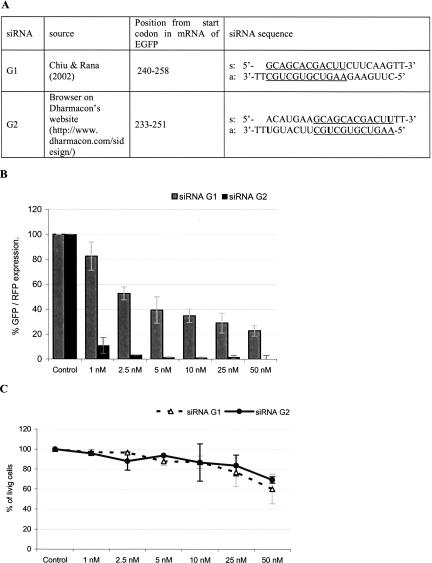
The sequence of siRNA duplex G1, selected by Chiu and Rana (2002), was not suitable for our studies due to the lack of uridine units in positions selected for modifications (at position 19 of the sense strand and at positions 10 and 19 of the antisense strand). Therefore, we selected the G2 siRNA sequence shifted 7 nt upstream from that of G1 (Fig. 1A) and determined the silencing activity in a validated dual fluorescence assay (Materials and Methods). The G2 duplex induced remarkable GFP gene silencing, even at concentrations as low as 1 nM, while the activity of G1 under the same conditions was only moderate (Fig. 1B). The dramatic difference in the silencing potency between G1 and G2 is not surprising in the light of previous reports that a shift of the target sequence by a few nucleotides can strongly affect the siRNA duplex activity (Holen et al. 2002; Harborth et al. 2003). Both siRNA duplexes exhibited noticeable cytotoxicity (MTT assay) at the highest concentrations tested (50 nM), but only a negligible effect was observed at low concentrations (1–5 nM). The cytotoxic “off-target” effects were concentration-dependent and could be diminished by using siRNA in as low as 1 nM concentrations (Persengiev et al. 2004).
FIGURE 1.
Silencing activity of siRNA duplexes directed toward EGFP mRNA. (A) Sequences of two siRNA molecules, indicated as G1 and G2 (common region is underlined). (B) Silencing activity of G1 and G2 used in 1–50 nM concentrations. (C) HeLa cells viability 36 h after transfection with siRNAs in concentrations given above. The means ± standard deviation is given.
Chemically modified siRNAs
All the syntheses of RNA oligonucleotides containing s2U, Ψ, and D modified units (Fig. 2) were performed on the synthesizer by the phosphoramidite approach, according to the procedures described previously (Guenther et al. 1994; Agris et al. 1995). Since during the iodine-assisted P(III)→P(IV) oxidation step oligonucleotides containing s2U units undergo a side-reaction leading to the loss of sulfur, we performed this step using tert-butyl hydroperoxide instead of the I2/pyridine/water mixture (Kumar and Davis 1995; Sochacka 2001). The chemical stability of oligomers containing dihydrouridine is another important issue during deprotection/purification steps. According to our experience the conditions used (MeNH2/EtOH/DMSO and aqueous sodium bicarbonate quenching) were sufficiently mild to avoid dihydrouracil ring opening, the reported side-reaction during the routine synthesis (Chaix et al. 1989). The structure of all oligonucleotides tested was confirmed by MALDI-TOF mass spectrometry and their purity was assessed by PAGE analysis. MALDI-TOF mass spectra for the representative oligomers (sD19, as2U19, aΨ10, and as2Uall 17 base pairs [bp]) are given in the Supplemental Material.
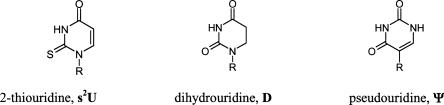
FIGURE 2.
Structures of naturally occurring rare nucleosides used for modification of siRNA duplexes. R = ribose residue.
Both strands of the G2 siRNA duplex were modified with one of the three selected nucleosides, at positions considered as important for the silencing activity (Table 1). We assumed that the introduction of the Ψ and s2U units, due to their well-documented positive influence on the A-type RNA helix structure (Kumar and Davis 1997; Davis et al. 1998; Testa et al. 1999; Diop-Frimpong et al. 2005), would enhance siRNA duplex stability and silencing activity. In contrast, dihydrouridine disturbs the structure of a single-stranded RNA (Dalluge et al. 1996; Stuart et al. 1996). According to our best knowledge there have been no data demonstrating the influence of dihydrouridine on the structure of double-stranded RNA, nor any detailed reports on the influence of the rare nucleobases, used in the present investigation, on the silencing activity of siRNA duplexes.
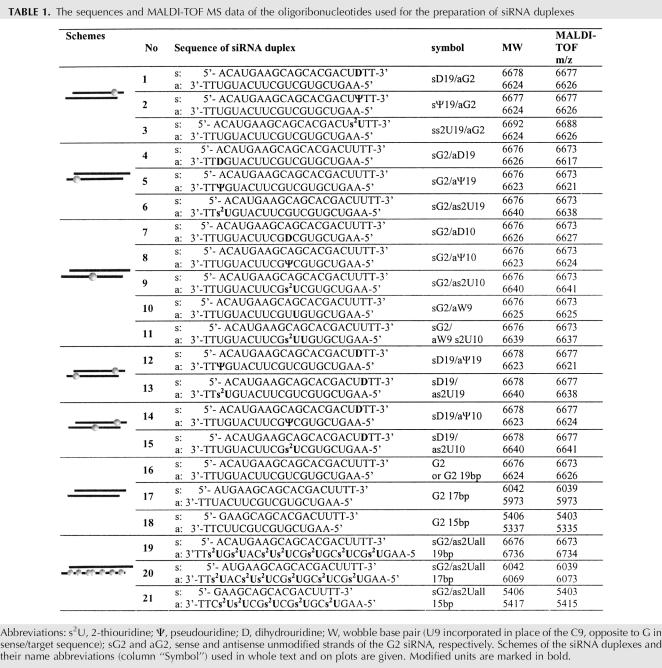
TABLE 1.
The sequences and MALDI-TOF MS data of the oligoribonucleotides used for the preparation of siRNA duplexes
At first, s2U, Ψ, and D units were introduced into the 3′-terminal parts of the sense (duplexes 1–3) or the antisense (duplexes 4–6) strands at positions 19 from the 5′-ends (Table 1), as the relative thermodynamic stability of the duplex ends decides which end of the siRNA duplex unwinds more easily and, subsequently, which strand is incorporated into the RISC complex (Khvorova et al. 2003; Schwarz et al. 2003; Tomari et al. 2004). The modified units were also incorporated into the central position of the antisense strand (nucleotide 10 from its 5′-end) (Table 1, duplexes 7–11). This position is situated opposite of the cleavage site of the mRNA and is known to be crucial for duplex activity (Elbashir et al. 2001c). Recent data further support the importance of the central domain of the siRNA duplex since the first step of releasing the passenger strand occurs via its nucleolytic cleavage at this part (Matranga et al. 2005; Rand et al. 2005). Doubly modified duplexes 12–15 were obtained by annealing the modified strands mentioned above.
We also prepared a series of G2 siRNA molecules possessing 17 or 15 double-stranded tracts—each strand with two thymidine-nucleotide overhangs (duplexes 17 and 18, respectively). In modified versions of the duplexes 16, 17, and 18, all uridine units in the antisense strands were substituted with the 2-thio-analog yielding siRNA molecules containing 7, 6, and 5 s2U units (Table 1, duplexes 19–21, respectively). We assumed that such s2U-modified siRNA duplexes would exhibit higher specificity and enhanced affinity toward the target sequence.
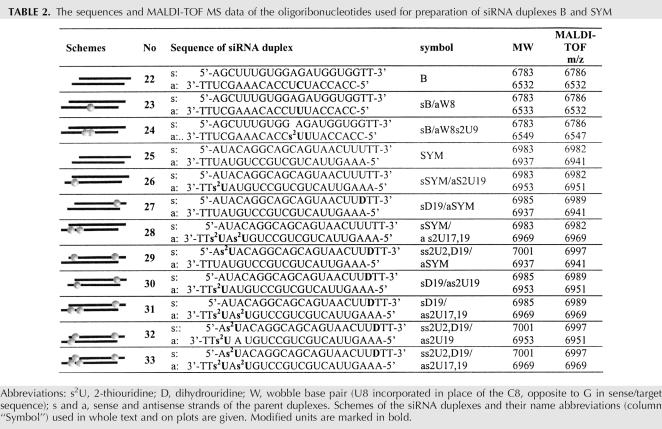
With the aim of verifying the results obtained for variants of the G2 duplex we screened two other sets of modified duplexes originating from two distinct siRNA molecules—siRNA B (Table 2, duplexes 22–24) and siRNA SYM (Table 2, duplexes 25–33).
TABLE 2.
The sequences and MALDI-TOF MS data of the oligoribonucleotides used for preparation of siRNA duplexes B and SYM
Hybridization studies
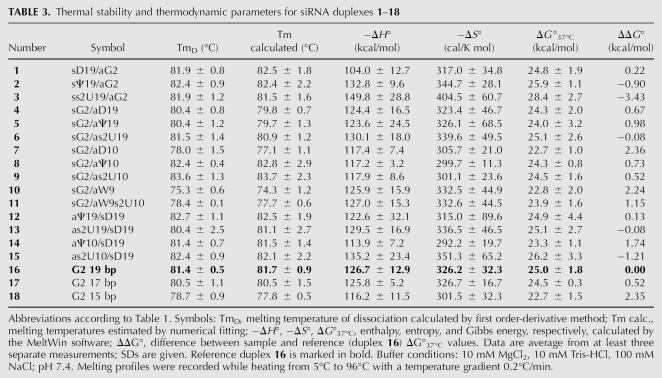
The melting temperatures (Tm) for siRNA duplexes 1–21, relative to duplex G2 (Table 1), were determined from the UV-monitored thermal dissociation profiles (Table 3). The numerical fitting of the curves, based on the two-state van't Hoff model, was used to determine the melting temperatures (Tm calculated) and the standard thermodynamic parameters ΔH°, ΔS°, and ΔG° for duplexes 1–18 (Table 3; Breslauer 1994). However, for highly modified duplexes 19–21 no plateau was observed in the corresponding melting profiles; therefore, melt curves could not be determined. In most cases of the series 1–15 we did not observe remarkable fluctuations in Tm or ΔG in comparison to the reference G2 duplex (16). These differences were much smaller than those reported earlier for shorter, 4–6-bp RNA duplexes (Luyten and Herdewijn 1998; Testa et al. 1999), probably due to the negligible contribution of one or two modified bases to the 19-bp duplex stability. Nonetheless, in some cases observed changes were meaningful.
TABLE 3.
Thermal stability and thermodynamic parameters for siRNA duplexes 1–18
The most noticeable decrease of duplex stability, with ΔTm in the range of 3°C–5°C, was observed for the duplexes modified with a D unit or with the wobble base pair in the central part of the antisense strand (duplexes 7 and 10, respectively). The decreased stability of the sG2/aD10 duplex (7) was expected to be due to the presence of a nonaromatic nucleobase disrupting base stacking. Moreover, dihydrouridine preferentially adopts the B-DNA type of sugar ring conformation (C2′-endo) and thus causes pronounced changes in the A-type RNA structure (Westhof et al. 1988; Stuart et al. 1996; Davis 1998). Even less stability was observed for the sG2/aW9 duplex (10), originating from the stronger structural perturbation of base stacking with the Watson–Crick base pairs situated at both sides of the wobble mismatch (Mizuno and Sundaralingam 1978; Varani and McClain 2000).
Some thermodynamic stabilization was observed for the duplexes modified with the s2U and Ψ units in the central position (duplexes 9 and 8, respectively), but only in the increase of the Tm (>2°C for s2U and ∼1°C for Ψ), while ΔG values were virtually the same as for the G2 counterpart. Such a negligible effect may be explained by the presence of two G:C base pairs at each side of the modification site, limiting the influence of a single-base modification. The stabilizing effect of an s2U unit on the siRNA duplex is well illustrated in the case of duplex 11, in which the thio-modification is situated next to the wobble base pair. In this case Tm and ΔG values exhibit considerable differences in comparison to corresponding values for parent duplex 10 (ΔTm = 3°C and ΔΔG° = 1.1 kcal/mol).
Modifications introduced at the ends of the duplexes (at positions 19 of the sense or antisense strands) have very little effect on duplex stability (ΔTm ± 1°C, ΔΔG° values for duplexes 1, 2, 4–6, 12, and 13), with only one exception of ΔΔG° = −3.43 kcal/mol for the duplex ss2U19/aG2 (3). We cannot rationally explain this result in the light of reported data that modifications introduced at the ends of a duplex should have relatively little influence on its overall stability (Nawrot et al. 2004).
Duplexes sG2/as2Uall 19 bp, sG2/as2Uall 17 bp, and sG2/as2Uall 15 bp (19–21) with antisense strands fully modified with s2U were thermally very stable (no transitions completed below 96°C). We assume that such exceptional thermodynamic stability results from increased affinity of s2U-modified oligomers for its complementary strands, originating from the preferential C3′-endo sugar ring puckering (Scheit and Faerber 1975; Agris et al. 1992; Kumar and Davis 1997; Testa et al. 1999). Similarly, the same structural feature makes LNA-modified siRNA duplexes thermodynamically very stable (Elmen et al. 2005). Melting temperatures for duplexes 19–24 could be determined in buffer with no magnesium cations (0.1 mM EDTA, 10 mM Tris-HCl, 100 mM NaCl) and with greater temperature gradient per minute (see Supplemental Table S1 for thermodynamic data of duplexes 1–21 in these conditions).
Circular dichroism spectra of modified siRNA duplexes
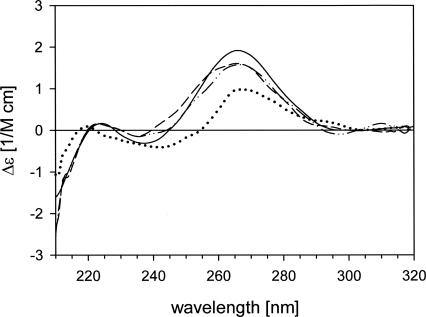
Circular dichroism (CD) spectra (Fig. 3) were measured for six selected siRNA duplexes that exhibited the biggest differences in thermodynamic stability or appreciable silencing activity in comparison to the reference G2 duplex. The CD spectrum of the G2 duplex (16) indicates a typical A-shaped structure of the double-stranded RNA with the maximum of the positive Cotton effect at 268 nm and crossover points at 245 and 290 nm. The most significant differences from this spectrum are seen in the CD spectrum of the sG2/aD10 duplex (7) (a shift of the crossover point from 245 to 255 nm and less negative band at 210 nm). The differences observed suggest that the structure of siRNA duplex 7 is distorted from the A-type to the B-type helix typical for double-stranded DNA. Such a change was expected since, as mentioned above, a D nucleoside destabilizes the RNA structure (Dalluge et al. 1996; Stuart et al. 1996). Therefore, its insertion in the central part of the siRNA duplex has significant structural consequences.
FIGURE 3.
Circular dichroism (CD) spectra of duplex G2 19 bp (16 ——) and siRNA duplexes modified at the central domain changing the duplex structure sG2/aD10 (7 ·········), sG2/aW9 (10 – – –), and sG2/aW9s2U10 (11 −··−··−). Reagents and conditions: 10 mM Tris-HCl, 100 mM NaCl, and 10 mM MgCl2 buffer (pH 7.4), Temp. 25°C, duplexes concentration = 1 μM. Sequences of the corresponding duplexes are given in Table 1.
The CD spectrum of the sG2/aW9 duplex (10) also differs considerably from the reference one, showing a crossover point shifted from 245 to 238 nm. This change may reflect the difference between the structure of the U:G wobble base pair and classical G:C or A:U Watson–Crick base pairs (Saenger 1984). The duplex undergoes structural rearrangement when the s2U unit is placed at position 10 of the antisense strand (3′-adjacent), as can be concluded from the spectrum of the sG2/aW9s2U10 duplex (11). This effect may be explained by the influence of the nearest-neighbor base pairs on a wobble base-pair structure (Gray et al. 1992) and supports the thermodynamic studies mentioned above. Duplexes containing exclusively Ψ or s2U units in position 10 of the antisense strand (duplexes 8 and 9, respectively) have CD spectra similar to that of the reference duplex 16. This result confirms that these modified units do not disturb the helical structure of the double-stranded RNA. No remarkable changes were observed in the shape of the CD spectra of the 3′-terminally modified duplexes (data not shown). This observation is consistent with data that show the insignificant influence of the terminal units on the duplex structure (Nawrot et al. 2004). Representative of this conclusion is the spectrum of the biologically most active modified siRNA duplex 13 (sD19/as2U19).
Cytotoxicity of the base-modified siRNA duplexes
Because chemically modified siRNA duplexes may induce severe cytotoxic effects (Amarzguioui et al. 2003; Czauderna et al. 2003), we checked the influence of duplexes 1–21 on cell viability using the MTT test (Supplemental Fig. S6). We assumed that the cytotoxicity of the siRNA duplexes containing the naturally occurring s2U, Ψ, and D units would be similar to that of the parent duplexes. The toxicity of the duplexes 1–21 used at the concentration of 1 nM in HeLa cells was negligible. Fully modified 19-bp duplex sG2/as2Uall (19) showed the biggest, but still minor, effect on cell viability (6% reduction), as compared to the control (cells transfected with plasmids only). The modified series of siRNAs B (duplexes 22–24) and siRNA SYM (duplexes 25–33) did not show any notable cytotoxicity in concentrations used in the study (data not shown).
Effect of base modification on gene silencing activity of siRNA
Modification of duplex termini
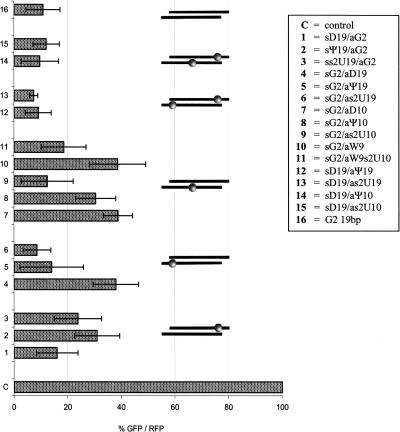
We expected that Ψ or s2U units introduced at position 19 of the sense strand would “close” the 5′-end of the corresponding duplexes (Davis et al. 1998; Testa et al. 1999), and thus would reduce their silencing activity. Indeed, data presented in Figure 4 show that the siRNAs modified within the s2U and Ψ units in position 19 of the sense strand, as in duplexes sΨ19/aG2 (2) and ss2U19/aG2 (3), have lower silencing potency (31% and 24% of EGFP expression in control cells, respectively) compared to the activity of the reference duplex G2 (16), which lowered EGFP expression to 11% of the control value.
FIGURE 4.
Gene-silencing activity of siRNAs modified with s2U, Ψ, or D units at central and terminal domains of G2 duplex. SiRNA activity was tested in a dual fluorescence assay, as described in Materials and Methods. HeLa cells were transfected with siRNA duplexes in 1 nM concentration. A positive control: cells transfected with pDsRed2-N1 and pEGFP-C1 plasmids only. Percentage of EGFP protein expression was normalized to RFP protein level. Error bars correspond to standard deviations of results of at least three independent transfection experiments (±SD). siRNA symbols and abbreviations according to Table 1.
On the other hand, when these modifications were introduced at position 19 of the antisense strand (on the 3′-end of the duplex), they would be expected to maintain or slightly increase duplex potency. The results are in accordance with our assumptions, as the activity of duplexes sG2/aΨ19 (5) and sG2/as2U19 (6) is noticeably enhanced (14% and 9% of control) as compared to duplexes 2 and 3, respectively, and similar to the native G2 duplex.
The dihydrouridine unit, incorporated at the 3′-end of the siRNA sense strand, should result in the “opening” of this terminus, and thus have a positive influence on the silencing activity. In contrast, its introduction on the 3′-end of the guide strand might cause the opposite effect. Unexpectedly, the sD19/aG2 duplex (1), containing a D unit at its 5′-end (in respect to the guide strand), was slightly less active (16% of control EGFP) as compared to the reference G2 duplex. In agreement with our expectations a threefold suppression of the silencing potency was observed for the sG2/aD19 duplex (4). Similar effects were reported for the siRNA molecules containing single chemical modifications or mismatches, including wobble base pairs, at the 3′- and/or 5′-terminal positions of the duplexes (Chiu and Rana 2003; Schwarz et al. 2003; Holen et al. 2005; Dande et al. 2006; Dowler et al. 2006).
Modification of position 10 of the antisense strand
We intended to check the impact of base modifications in the central part of the duplex on its activity, due to the direct involvement of this domain in RISC-associated cleavage of the target mRNA (Elbashir et al. 2001c). We assumed that s2U and Ψ nucleosides would strengthen antisense siRNA/mRNA interactions and, possibly result in higher potency of such modified constructs. However, only the sG2/as2U10 duplex (9) exhibited tolerance for s2U modification, showing similar activity (12% of control EGFP) to the parent G2 duplex (Fig. 4). This observation is interesting in light of earlier reported data about modifications of the central part of the antisense strand of siRNA duplex not being well tolerated (Hamada et al. 2002; Saxena et al. 2003; Du et al. 2005; Holen et al. 2005). The remaining two duplexes sG2/aD10 (7) and sG2/aΨ10 (8) exhibited three- to fourfold suppressed silencing activity. Interestingly, noticeable enhancement of activity (> 50%) was achieved by introduction of the s2U unit at the 3′-adjacent position of the wobble base pair in the sG2/aW9 (10) duplex (see sG2/aW9s2U10 duplex, 11). It is worth noticing that observed loss and subsequent recovery of silencing activity of siRNAs modified in central positions (duplexes 10 and 11, respectively) correlate with changes in their thermodynamic stability (Table 3) and helical structure (Fig. 3, CD spectra).
We decided to verify this last intriguing result by screening the activity of siRNA B and its modified variants (Table 2, entries 22–24). Duplex B was designed toward mRNA of human and mouse BACE1 protein and thus expression of fusion plasmid pBACE–GFP was monitored in a dual fluorescence assay. Nonmodified duplex B in 5 nM concentration decreased the level of BACE–GFP expression down to ∼ 30% of the control. Its variant mutated in position 8 (C→U) of the antisense strand (duplex BaW8, 23) was half as active. Introduction of the s2U unit at the 3′-adjacent position to the G–U base pair gave duplex 24 (B aW8s2U9), exhibiting silencing activity comparable with that of the nonmodified duplex 22 (Fig. 5). Therefore, we confirmed that introduction of the s2U unit next to the wobble base pair leads to the recovery of most of the siRNA duplex silencing activity. The observed effect probably originates from the improved A-type helical structure of the s2U-containing RNA duplex.
FIGURE 5.
Gene-silencing activity of siRNA B modified with wobble and/or s2U unit at central domain. SiRNA activity was tested in a dual fluorescence assay, as described in Materials and Methods. HeLa cells were transfected with siRNA duplexes in 5 nM concentration. A positive control: cells transfected with pDsRed2-N1 and pBACE–GFP plasmids only. Percentage of BACE–GFP protein expression was normalized to RFP protein level. Error bars correspond to standard deviations of results of at least three independent transfection experiments (±SD). siRNA symbols and abbreviations according to Table 2.
Doubly modified siRNAs
Although we did not observe any pronounced increase in the activity of duplexes 1–9 containing single s2U, Ψ, or D mutations in the G2 sequence, we wondered whether simultaneous introduction of a dihydrouridine unit at the 3′-end of the passenger strand and s2U or Ψ at the 3′-end of the guide strand would result in improved duplex asymmetry and therefore in enhancement of siRNA silencing efficiency. As shown in Figure 4 duplexes sD19/as2U19 (13) and sD19/aΨ19 (12) exhibited slightly increased activity (9% and 7% expression of the EGFP in control cells) in comparison with the reference G2 duplex (11% of EGFP). Moreover, at lower concentrations (down to 0.1 nM) duplexes 12 and 13 exhibited a tendency for improved silencing potency over the reference duplex (data not shown).
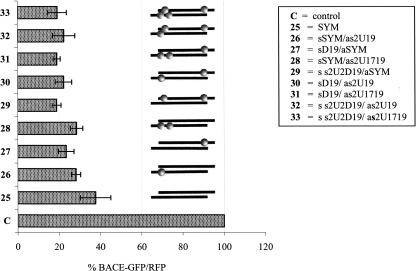
These promising data were verified by structure–activity relationship studies of the siRNA duplex SYM (25), containing at each terminus the same signature of Watson–Crick hydrogen bonds (three A–U bp, followed by one G–C bp and one A–U bp) (Table 2). We assumed that such a duplex, with similar thermodynamic stability of both ends, should be an accurate model to study asymmetry inducing modifications. All modified duplexes, screened in this test, containing a D nucleoside at their 5′-end and s2U unit(s) at their 3′-end with respect to the guide strand (Table 2, sequences 26–33), showed 25%–50% higher BACE1 silencing activity in comparison with the unmodified symmetrical duplex 25 (Fig. 6). Introduction of a single dihydrouridine unit at the 3′-end of the sense strand (as in 27) induced stronger enhancement of the silencing activity (40% more than nonmodified 25) than a single or double 2-thiouridine at the 3′-end of the antisense strand. Duplexes 26 and 28 were 25% more active than reference 25. Simultaneous introduction of s2U and D units in the same sense strand caused the strongest of the observed improvements (duplex 29 was ∼ 50% more active than the parent one, 25). Introduction of further s2U units at the 3′-end of the duplex, as in duplexes 31–33, did not cause any further increase of the silencing activity (Fig. 6).
FIGURE 6.
Gene-silencing activity of siRNA SYM modified with D and/or s2U units at terminal domains. SiRNA activity was tested in a dual fluorescence assay, as described in Materials and Methods. HeLa cells were transfected with siRNA duplexes in 1 nM concentration. A positive control: cells transfected with pDsRed2-N1 and pBACE–GFP plasmids only. Percentage of BACE–GFP protein expression was normalized to RFP protein level. Error bars correspond to standard deviations of results of at least three independent transfection experiments (±SD). siRNA symbols and abbreviations according to Table 2.
In our opinion this set of results strongly supports the observation of the positive influence of nucleobase modifications, introduced at the duplex termini, on siRNA activity caused by its increased thermodynamic asymmetry.
Duplexes of various lengths with antisense strands fully modified with s2U
Taking into account the improved affinity of s2U-modified strands for complementary templates and, consequently, their enhanced target specificity, we also prepared a series of siRNA duplexes with 19, 17, or 15 bp (16–18) and their modified counterparts (19–21), containing antisense strands fully modified with seven, six, or five s2U units, respectively. As shown by hybridization studies, duplexes aG2/as2Uall 19 bp (19), aG2/as2Uall 17 bp (20), and aG2/as2Uall 15 bp (21) exhibit extremely high thermodynamic stability (the relevant melting profiles did not reach a plateau up to 96°C). We asked the question whether such stable duplexes can be effectively unwound by the dsRNA helicase engaged in the RISC complex (Chiu et al. 2005). It has already been reported that thermodynamically stable duplexes containing LNA-modified units are active in RNAi-induced gene silencing (Braasch et al. 2003; Elmen et al. 2005), although their ethylene bridged nucleic acid analogs completely lost their silencing potency (Hamada et al. 2002). Martinez and Tuschl (2004) proved that a 15-nt sequence of the antisense strand is required for effective cleavage catalyzed by the RISC complex. On the basis of these data we assumed that shortened s2U-modified siRNA duplexes might still exhibit remarkable silencing activity.
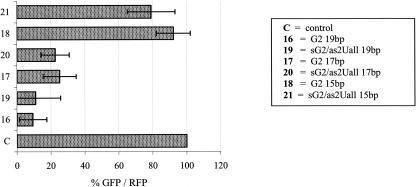
Indeed, as we demonstrate here, the duplexes 19, 20, and 21 are as active in EGFP silencing as the parent G2 duplexes of the same length (Fig. 7). Trimming of the siRNA duplexes 16 and 19 on their 3′-ends (with respect to the guide strands) by two nucleotides caused a twofold decrease in the silencing activity. Further shortening of the length of the duplexes decreased their efficiency dramatically, for both the unmodified (G2 15 bp, 18) and s2U-modified (sG2/as2Uall 15 bp, 21) cases. Interestingly, the shortest modified duplex 21 was slightly more active (79% of control EGFP) than its counterpart (92% of control EGFP), which might originate from its enhanced binding affinity to the target mRNA.
FIGURE 7.
Silencing activity of siRNA duplexes G2 varying in length: 19 bp (16), 17 bp (17), 15 bp (18), and their modified analogs containing, in the antisense strand, seven (sG2/as2Uall 19 bp, 19), six (sG2/as2Uall 17 bp, 20), or five (sG2/as2Uall 15 bp, 21) s2U units, respectively. Silencing activity of siRNA duplexes 19–21 in HeLa cells was determined in a dual fluorescence assay, with siRNAs used at 20 nM concentration. Error bars correspond to the standard deviations of results of at least three independent transfection experiments (±SD).
Theoretical prediction of siRNA asymmetry
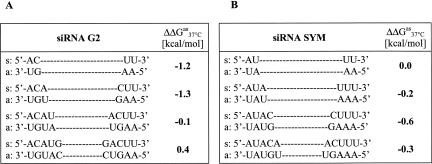
The nearest-neighbor model and the free energy values (Freier et al. 1986) were used to calculate the ΔGas 37°C for the 3′- and 5′-ends (in respect to the guide strand) in G2 and SYM siRNA duplexes. To assess the relative thermodynamic stability of the duplex ends, the differences between a free Gibbs' energy of the 3′- and 5′-ends of the duplex (ΔΔGas 37°C) were calculated for two, three, four, and five consecutive base pairs (Fig. 8). For two and three consecutive base pairs in G2 duplex the ΔΔGas 37°C values were −1.2 and −1.3 kcal/mol, respectively, indicating higher thermodynamic stability of the 3′-terminus. Interestingly, the stabilities determined for 4-bp ends were almost identical (ΔΔGas 37°C = −0.1 kcal/mol), while those calculated for 5-bp strands indicated the opposite duplex regiostability, as the 3′-end was thermodynamically less stable than the 5′-end counterpart. The respective ΔΔGas 37°C values for SYM duplex were smaller, suggesting similar thermodynamic stability of both SYM termini, independently of the length of the evaluated strand.
FIGURE 8.
The differences between a free Gibbs' energy (ΔΔGas 37°C) calculated for the 3′- and 5′-ends of different length in the siRNA duplexes G2 (A) and SYM (B). The duplex polarity is assigned according to the antisense strand polarity.
DISCUSSION
In this paper we demonstrate that siRNA duplexes containing naturally occurring rare nucleosides such as 2-thiouridine, pseudouridine, and dihydrouridine may serve as efficient gene expression inhibitors. As expected, their activity strongly depends on the site of modification. Incorporated modified units affect the thermodynamic stability of the siRNA duplexes and, therefore, modulate their specificity and silencing activity. These assumptions were made on the basis of the reported importance of the relative thermodynamic stability of duplex termini during the RISC assembly process (Khvorova et al. 2003; Schwarz et al. 2003). The biochemical explanation for these observations has emerged from structural studies in a Drosophila melanogaster system (Tomari et al. 2004). Thus, selection of the siRNA guide strand is initiated by binding of a duplex to the heterodimeric protein complex Dcr-2/R2D2 (dicer and dsRNA binding protein, respectively). Dicer preferentially binds to a more easily accessible 5′-end of the strands in a duplex. At the same time the R2D2 protein binds near the end with the greatest double-stranded character, choosing it as the 5′-end of the passenger strand. Therefore, the structural properties of the dicer and R2D2 proteins determine the fate of the siRNA strands. Gregory et al. (2005) provided the evidence for an analogous mechanism operating in human cells. These studies demonstrate the ability of the hdicer–TRBP–hAgo2 complex to specifically load the guide strand of siRNA to an active RISC. While our studies concerned mainly the thermodynamic stability of the duplex ends, the above-mentioned studies indicate the importance of nucleic acid–protein interactions in the siRNA silencing potency. Such interactions involve mostly hydrogen bonding between amino acid residues and the sugar-phosphate RNA backbone and, therefore, are not altered by nucleobase modifications.
In our studies, we observed that, within a set of s2U, Ψ, and D-modified duplexes G2 (duplexes 1–15), the most active were duplexes with a s2U unit incorporated at the 3′-end of the guide strand, especially in combination with the sense strand modified with a dihydrouridine unit in position 19. This observation was confirmed with the SYM duplex (25), possessing five subsequent Watson–Crick base pairs at both duplex ends of the same pattern (the same order of U–A and C–G base pairs). Thermodynamic symmetry of this model duplex was confirmed by the rather low value of the ΔΔGas 37°C parameters (Fig. 8).
In the modified SYM duplexes 25–33, the presence of the s2U unit(s) at the 3′-end and/or that of the dihydrouridine unit at the 5′-end resulted in a remarkable increase of siRNA duplex activity (25%–50%). Therefore, we assume that such an “opening” of the 5′-end of the siRNA by the D modification and its “closing” at the 3′-end by the s2U unit(s) are advantageous for duplexes containing symmetrical sequences at their termini (with respect to base-pairing interactions) and exhibiting moderate or low silencing activity. One can imagine the usefulness of such modifications for duplexes of moderate or low activity, designed for fixed sequences, e.g., SNP-detecting siRNAs. Furthermore, our calculations indicate that siRNA duplex asymmetry is defined mostly by the relative thermodynamic stability of its three terminal base pairs. This conclusion is based on the observation that, for the highly active G2 duplex, the calculated ΔΔGas 37°C values are meaningful when not more than three base pairs are taken into account. These data may help in the rational design (including chemical modification) of siRNA duplexes.
Modifications at the duplex termini have an impact on siRNA activity probably not only because of induction of duplex asymmetry but as a result of other effects, which should be taken into account, including interactions between guide strand and complementary mRNA as well as between RNA and proteins of RNAi machinery. For example, the expected lower potency of the sG2/aD19 duplex 4, containing a dihydrouridine unit in position 19 of the antisense strand, may result from weakened binding of this modified strand to the target mRNA. The dihydrouridine unit changes the ribose ring conformation, and therefore may also influence binding of the modified 3′-end of the antisense strand to the PAZ domain of the Argonaute protein (Lingel et al. 2003; Yan et al. 2003; Ma et al. 2004).
We did not modify the 5′-end of the guide strand, because previous studies reported that modification of this end of the duplex impairs siRNA-silencing activity (Chiu and Rana 2003). The recent structural studies by Ma et al. (2005) provide an explanation for the functional importance of the 5′-end of the antisense strand, as it participates in nucleation with messenger RNA. In this process the PIWI domain of the Argonaute protein binds to the siRNA antisense strand via interactions between the phosphate groups of the RNA chain and the amino acid residues of the protein. In light of these new data we suppose that modifications within the nucleobases at the 5′-end of the antisense strand should not alter interactions with RISC. It was recently shown by Jackson et al. (2006) and Fedorov et al. (2006) that subtle chemical modifications can limit siRNA off-target effects. These studies demonstrate that a single 2′-O-Me modification introduced at position 2 of the antisense strand does not interfere with duplex potency and, importantly, limits unintended “off-target” silencing. According to the authors a possible explanation for the observed effect is the alteration of the binding of the guide strand to the PIWI domain of the RISC by the 2′-O-methyl group. In our opinion it is worth noticing that the 2′-O-Me modification also decreases the free energy of hybridization (due to the C3′-endo conformation) and thus should enhance the binding affinity to mRNA within the seed region. Our s2U modification exhibits similar hybridization features, however, should not cause any steric hindrance when bound to the RISC. Therefore, using the s2U unit might be valuable in further studies on the mechanism of limiting nonintended silencing by modification at position 2 of the guide strand.
In addition, sulfur in position 2 of the nucleobase ring enhances favorable base stacking, causing further enhancement of affinity for the target sequence (Cummins et al. 1995). Another feature of the s2U modification is its restricted wobble base pairing, resulting in improved specificity for the complementary template (Agris et al. 1992). This feature might be beneficial for intended target recognition by limiting the affinity for “off-target” transcripts. Therefore, in our opinion, further studies on duplexes containing an s2U modification at the 5′-ends of their antisense strands are worthwhile.
We intended to check also the impact on activity of base modifications positioned in the central part of the siRNA duplex, due to the direct involvement of this domain in RISC-associated cleavage of the target mRNA (Elbashir et al. 2001c). The structure of this region of siRNA is also crucial, as demonstrated recently, due to the cleavage of the sense strand of siRNA before its dissociation from the duplex (Matranga et al. 2005; Rand et al. 2005). Our results show that 2-thiouridine modifications within the central part of the antisense strand, which improve duplex stability, do not interfere with either the helical structure or the silencing potency. However, it is not clear why pseudouridine at position 10 of the guide strand led to reduced activity of the sG2/aΨ10 construct, when thermodynamic and CD data suggested that the structure of this duplex is not distorted (see Table 3 and Supplemental Fig. S5). In contrast, dihydrouridine placed at position 10 or the C→U mutation at position 9 as in G2 relative duplex 10 or at position 8 as in duplex 23 of the antisense strand (leading to wobble base pairing with both the sense strand of siRNA and a target mRNA sequence) caused much lower silencing activity. The presence of the D or wobble base pair in the central domain of the guide strand probably significantly disturbs the A-type helical structure of the guide/mRNA complex. Furthermore, wobble base pairing at the central position of the siRNA duplex may also disturb the efficiency of the cleavage of the sense strand and, in this way, disturbs RISC assembly (Matranga et al. 2005; Rand et al. 2005). Similar observations of reduction of the silencing activity of siRNA modified with wobble base pairs in the center of the duplex were reported at the time our project was underway (Holen et al. 2005). Interestingly, the noticeable recovery of activity of both wobble-mutated duplexes 10 and 23 could be achieved by introduction of the s2U unit at the 3′-adjacent position of the wobble base pair. Improvement of silencing activity of the duplex 11 correlates with its enhanced thermodynamic stability (Table 3) and with the minimal disruption of its structural integrity, as it could be concluded from the corresponding CD spectrum (Fig. 3). In our opinion this observation strongly supports the idea that the conformational changes induced by the s2U nucleoside leading to A-type helical structure are advantageous for the siRNA duplex silencing activity.
It is interesting to notice that the unimpaired silencing activity of duplexes 19–21, which contain antisense strands fully modified with s2U units and which are extremely stable to thermodynamic denaturation, suggests that the proteins of the RISC complex possess an unusual ability to facilitate unwinding and dissociation of such thermodynamically very stable siRNA duplexes.
The usefulness of similar 2-thiosubstituted nucleobase modifications as candidates for application in RNAi was recently mentioned by the Egli group (Diop-Frimpong et al. 2005). Their structural studies show that a DNA duplex modified with a 2′-O-[2(methoxy)ethyl]-2-thiothymidine unit exhibits favorable features for base pairing and induces only small structural perturbations within the A-form double-stranded helix. Therefore, combinations of chemical modification involving sulfur in position 2 of the nucleobase and 2′-modification at the ribose moiety are potential objects for future studies on chemically modified siRNA duplexes.
In conclusion, we have demonstrated that the naturally occurring modified nucleosides, 2-thiouridine, pseudouridine, and dihydrouridine, when introduced into an siRNA duplex, modulate its silencing potency. The extent of this effect depends on the modification position and is most advantageous when the s2U, Ψ, and D nucleosides participate in an enhancement of the siRNA duplex asymmetry. Therefore, the modified nucleosides described here may be considered as useful units for improving siRNA activity and specificity.
MATERIALS AND METHODS
Preparation of siRNAs
Oligoribonucleotides were prepared according to the routine phosphoramidite approach (Caruthers 1985) using standard LCA CPG glass support and commercially available nucleoside phosphoramidites (Glen Research). Phosphoramidites of suitably protected modified units (s2U, Ψ, D) were prepared as described previously (Guenther et al. 1994; Agris et al. 1995). Oligonucleotide synthesis was performed on a 1 μM scale on an Applied Biosystems 394 instrument under conditions recommended by the synthesizer supplier, except for the synthesis of s2U-containing oligomers. In these cases the oxidation step was performed with 1 M tBuOOH (Fluka) in acetonitrile/methylene chloride (95/5) for 6 min (Kumar and Davis 1995; Sochacka 2001). Oligonucleotides were cleaved from the solid support as 5′-DMT-derivatives, and then deprotected and purified according to the procedure described elsewhere (McSwinggen and Beigelman 2003). Briefly, support-bound oligonucleotides were treated with 33% ethanolic methylamine and DMSO, 1:1 mixture (v:v) for 15 min at 65°C and then with TEA × 3HF for 15 min at 65°C. The reaction mixture was frozen for 30 min at − 20°C, quenched with cold 1.5 M ammonium bicarbonate and poured into a conditioned SepPak (Waters) cartridge. Shorter oligomers were eluted with 14% CH3CN in 50 mM NaOAc and 50 mM NaCl. The remaining oligomer was treated with 2% aqueous TFA for 15 min at room temperature and washed with water, 1 M NaCl, and water. The product was eluted from the cartridge with 30% aqueous solution (aq.) CH3CN, and after partial solvent removal the RNA aqueous solution was frozen and kept at − 20°C. The concentration of oligomers was determined spectrophotometrically by UV absorbance at λmax in water using the extinction coefficients calculated according to the method published earlier (Brown and Brown 1991). The structure of the oligonucleotides was confirmed by MALDI-TOF mass spectrometry (data given in Table 1) and their purity was assessed by PAGE (20% acrylamide/7 M urea, 32P-labeled).
Assembly of siRNA duplexes was done in water by mixing equimolar amounts of complementary sense and antisense oligonucleotides (final duplex concentration was 20 μM), heating the mixture at 95°C for 2 min, and slow (3 h) cooling down to room temperature. Purity of the resulting duplexes was confirmed by PAGE (20% polyacrylamide, nondenaturing conditions, 32P-labeled), and formation of duplexes was monitored by 4% agarose electrophoresis.
Melting profiles and thermodynamic calculations
All absorption measurements were carried out in a 1-cm path length cell with a UV/Vis 916 spectrophotometer equipped with a Peltier Thermocell (GBC). Complementary oligonucleotides were mixed in 10 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2 buffer (pH 7.4) at final concentration of duplexes of 1 μM, heated at 96°C, and strands association was achieved by cooling down to 5°C, with a temperature gradient of 0.4°C/min. Melting profiles were recorded while heating from 5°C to 96°C with a temperature gradient of 0.2°C/min. The melting temperatures were calculated by the first-order derivative method. Thermodynamic parameters were obtained by fitting of the recorded curves (MeltWin software, version 3.5). Each set of experimental data was fitted twice.
CD spectra recording
CD spectra were recorded on a CD6 dichrograph (Jobin-Yvon) at 25°C in the same buffer as in melting experiments at a duplex concentration of 1 μM using a 5-mm path length cell, 2-nm bandwidth, and 1–2-sec integration time. Each spectrum was smoothed with a 25-point algorithm (included in the manufacturer's software, version 2.2) after averaging of at least three scans.
Dual fluorescence model
Cell cultures and transfections
HeLa (human cervix carcinoma) cells were cultured in RPMI 1640 medium (Gibco; BRL) supplemented with 10% heat-inactivated FBS (Gibco, BRL, Paisley) and with antibiotics (penicillin 100 U/mL, streptomycin 100 μg/mL, Polfa) at 37°C and 5% CO2. Twenty-four hours prior to the experiment, cells were plated in 96-well black wall plates with a transparent bottom (Perkin-Elmer) at a density of 15,000 cells per well. One hour before treatment, the cell medium was replaced with one free of antibiotics (100 μL/well). The cells were cotransfected with plasmid DNA (pDsRed2-N1, 15 ng/well, and pEGFP-C1, 30 ng/well, BD Biosciences or p-BACE–GFP; Qing et al. 2004), 70 ng/well, provided by Dr Weihong Song, The University of British Columbia, Vancouver, Canada, and siRNA (1–50 nM final concentrations) dissolved in OPTI-MEM medium (50 μL/well, Gibco) and complexed with transfection reagent (Lipofectamine 2000, Invitrogen) in the proportion 1 μL of lipofectamine per 1 μg of nucleic acid. The cells were incubated in the transfection mixture for 5 h and then the mixture was replaced with fresh, culturing medium with antibiotics. After 36-h incubation at 37°C in atmosphere of 5% CO2 cells were washed three times with phosphate saline buffer (PBS) and lysed overnight with NP-40 buffer (150 mM NaCl, 1% IGEPAL, 50 mM Tris-HCl [pH 7.0], 1 mM PMSF) at 37°C. Cell lysates were used for fluorescence determination.
Quantification of siRNA activity
Fluorescence values of enhanced green fluorescent protein (EGFP) and red fluorescent protein (RFP) fluorophores were measured in cell-culturing plates using Synergy HT (BIO-TEK) reader, and data quantification was done with KC4 software. Excitation and emission wavelengths for both fluorescent proteins were as follows (the bandwidth is given after the slash): λEx = 485/20 nm and λEm = 528/20 nm for GFP and λEx = 530/25 nm and λEm = 590/30 nm for RFP. The siRNA activity was calculated as a ratio of GFP to RFP fluorescence values. Each transfection experiment was performed in eight wells. A mean value of fluorescence was calculated after discarding the most extreme values. The mean value of background fluorescence (CL, cells treated with lipofectamine only) was subtracted from the mean sample fluorescence value. The level of GFP fluorescence of control transfected cells (treated with pDsRed2-N1 and the corresponding pEGFP-C1 or p-BACE–GFP plasmids only, control) was taken as a reference (100%). Each siRNA activity value given on the plots is an average of mean values from at least three independent experiments and the standard deviations are given as error bars.
Cytotoxicity
The cytotoxicity of siRNA duplexes for HeLa cells was measured using the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; Sigma) assay (activity of the mitochondrial respiratory chain) (Hansen et al. 1989). Cells were plated and transfected as described above (siRNAs at 1 nM concentration). As a background control, the cells were treated with lipofectamine only. After 36 h of incubation at 37°C and 5% CO2, MTT solution (5 mg/mL) in PBS was added to each well and incubated for additional 2 h at 37°C. Finally, 95 μL of lysis buffer (20% SDS, 50% aqueous dimethylformamide, pH 4.5) was added to each well and incubated overnight at 37°C. Absorbance of a given sample was measured at 570 nm, with the reference wavelength 630 nm (plate reader Synergy HT, BIO-TEK). The percentage of living cells (PLC) was calculated from the equation: PLC = (ASpl − ACL)/(AMock − ACL) × 100% where ASpl is the absorbance of a given sample of cells treated with siRNA, ACL is the background absorbance (CL control), AMock is the absorbance of the cells untreated with siRNAs (control transfected cells). Data points represent means of at least four measurements.
SUPPLEMENTAL DATA
The following Supplemental Material is available upon request from the corresponding author (e-mail: bnawrot@bio.cbmm.lodz.pl): correlation of the fluorescence intensity and the level of mRNA of EGFP and RFP; time course of the expression of pEGFP-C1 plasmid in HeLa cells; MALDI-TOF MS of oligonucleotides sD19, as2U19, aΨ10, and as2Uall 17 bp; CD spectra of duplexes 8, 9, and 13; cytotoxicity of siRNA duplexes 1–21; thermal stability of siRNA duplexes 1–21 in buffer with no magnesium ions added.
ACKNOWLEDGMENTS
The authors thank Professor Wojciech J. Stec for scientific inspiration, continuous support, and critical reading of this manuscript. Dr. Weihong Song, The University of British Columbia, Vancouver, Canada, is acknowledged for his kind gift of fusion plasmid p-BACE–GFP. We kindly appreciate financial support from the State Committee for Scientific Research in Poland and ICGEB grant CRP/POL04-01.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.538907.
REFERENCES
- Agris, P.F., Sierzputowska-Gracz, H., Smith, W., Malkiewicz, A., Sochacka, E., Nawrot, B. Thiolation of uridine carbon-2 restricts the motional dynamics of the transfer RNA wobble position nucleoside. J. Am. Chem. Soc. 1992;114:2652–2656. [Google Scholar]
- Agris, P.F., Malkiewicz, A., Kraszewski, A., Everett, K., Nawrot, B., Sochacka, E., Jankowska, J., Guenther, R. Site-selected introduction of modified purine and pyrimidine ribonucleosides into RNA by automated phosphoramidite chemistry. Biochimie. 1995;77:125–134. doi: 10.1016/0300-9084(96)88115-6. [DOI] [PubMed] [Google Scholar]
- Amarzguioui, M., Holen, T., Babaie, E., Prydz, H. Tolerance of mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch, D.A., Jensen, S., Liu, Y., Kaur, K., Arar, K., White, M.A., Corey, D.R. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- Breslauer, K.J. Methods in molecular biology. In: Agrawal S., editor. Protocols for oligonucleotide conjugates. Vol. 26. Humana Press; Totowa, N.J: 1994. pp. 437–472. [Google Scholar]
- Brown, T., Brown, D.J.S. Modern machine-aided methods of oligodeoxy-ribonucleotide synthesis. In: Eckstein F., editor. Oligonucleotides and Analogues: A Practical Approach. IRL Press; Oxford, UK: 1991. pp. 1–24. [Google Scholar]
- Caruthers, M.H. Gene synthesis machines: DNA chemistry and its uses. Science. 1985;230:281–285. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
- Chaix, C., Duplaa, A.M., Molko, D., Teoule, R. Solid phase synthesis of the 5′-half of the initiator t-RNA from B. subtilis . Nucleic Acids Res. 1989;17:7381–7393. doi: 10.1093/nar/17.18.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, Y.L., Rana, T.M. RNAi in human cells: Basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- Chiu, Y.L., Rana, T.M. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, Y.L., Dinesh, C.U., Chu, C.Y., Ali, A., Brown, K.M., Cao, H., Rana, T.M. Dissecting RNA-interference pathway with small molecules. Chem. Biol. 2005;12:643–648. doi: 10.1016/j.chembiol.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Cummins, L.L., Owen, S.R., Risen, L.M., Lesnik, E.A., Freier, S.M., McGee, D., Guinosso, C.J., Cook, P.D. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna, F., Fechtner, M., Dames, S., Aygun, H., Klippel, A., Pronk, G.J., Giese, K., Kaufmann, J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalluge, J.J., Hashizume, T., Sopchik, A.E., McCloskey, J.A., Davis, D.R. Conformational flexibility in RNA: The role of dihydrouridine. Nucleic Acids Res. 1996;24:1073–1079. doi: 10.1093/nar/24.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dande, P., Prakash, T.P., Sioufi, N., Gaus, H., Jarres, R., Berdeja, A., Swayze, E.E., Griffey, R.H., Bhat, B.P. Improving RNA interference in mammalian cells by 4′-thio-modified small interfering RNA (siRNA): Effect on siRNA activity and nuclease stability when used in combination with 2′-O-alkyl modifications. J. Med. Chem. 2006;49:1624–1634. doi: 10.1021/jm050822c. [DOI] [PubMed] [Google Scholar]
- Davis, D. Biophysical and conformational properties of modified nucleosides in RNA (magnetic resonance studies) In: Grosjean H., Benne B., editors. Modification and editing of RNA. ASM Press; Washington, DC: 1998. pp. 85–102. [Google Scholar]
- Davis, D.R., Veltri, C.A., Nielsen, L. An RNA model system for investigation of pseudouridine stabilization of the codon-anticodon interaction in tRNALys, tRNAHis, and tRNATyr. J. Biomol. Struct. Dyn. 1998;15:1121–1132. doi: 10.1080/07391102.1998.10509006. [DOI] [PubMed] [Google Scholar]
- Diop-Frimpong, B., Prakash, T.P., Rajeev, K.G., Manoharan, M., Egli, M. Stabilizing contributions of sulfur-modified nucleotides: Crystal structure of a DNA duplex with 2′-O-[2-(methoxy)ethyl]-2-thiothymidines. Nucleic Acids Res. 2005;33:5297–5307. doi: 10.1093/nar/gki823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler, T., Bergeron, D., Tedeschi, A.L., Paquet, L., Ferrari, N., Damha, M.J. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-β-D-arabinonucleic acid (FANA) Nucleic Acids Res. 2006;34:1669–1675. doi: 10.1093/nar/gkl033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Q., Thonberg, H., Wang, J., Wahlestedt, C., Liang, Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir, S., Lendeckel, W., Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendecel, W., Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001c;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen, J., Thonberg, H., Ljungberg, K., Frieden, M., Westergaard, M., Xu, Y., Wahren, B., Liang, Z., Orum, H., Koch, T., et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov, Y., Anderson, E.M., Birmingham, A., Reynolds, A., Karpilow, J., Robinson, K., Leake, D., Marshall, W.S., Khvorova, A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S.Q., Montgomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabitis elegans . Nature. 1998;391:806–813. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fougerolles, A., Manoharan, M., Meyers, R., Vornlocher, H.-P. RNA interference in vivo: Toward synthetic small inhibitory RNA-based therapeutics. Methods Enzymol. 2005;392:278–296. doi: 10.1016/S0076-6879(04)92016-2. [DOI] [PubMed] [Google Scholar]
- Freier, S.M., Kierzek, R., Jaeger, J.A., Sugimoto, N., Caruthers, M.H., Neilson, T., Turner, D.H. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, D.M., Ratliff, R.L., Vaughan, M.R. Circular dichroism spectroscopy of DNA. Methods Enzymol. 1992;211:389–406. doi: 10.1016/0076-6879(92)11021-a. [DOI] [PubMed] [Google Scholar]
- Gregory, R.I., Chendrimada, T.P., Cooch, N., Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Guenther, R.H., Bakal, R.S., Forrest, B., Chen, Y., Sengupta, R., Nawrot, B., Sochacka, E., Jankowska, J., Kraszewski, A., Malkiewicz, A., et al. Aminoacyl-tRNA synthetase and U54 methyltransferase recognize conformations of the yeast tRNAPhe anticodon and T stem/loop domain. Biochimie. 1994;76:1143–1151. doi: 10.1016/0300-9084(94)90043-4. [DOI] [PubMed] [Google Scholar]
- Hall, A.H.S., Wan, J., Shaughnessy, E.E., Ramsay Shaw, B., Alexander, K.A. RNA interference using boranophospate siRNAs: Structure–activity relationships. Nucleic Acids Res. 2004;32:5991–6000. doi: 10.1093/nar/gkh936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, M., Ohtsuka, T., Kawaida, R., Koizumi, M., Morita, K., Forukawa, H., Imanishi, T., Miyagishi, M., Taira, K. Effects on RNA interference in gene expression (RNAi) in cultured mammalian cells of mismaches and the introduction of chemical modifications at the 3′-ends of siRNAs. Antisense Nucleic Acid Drug Dev. 2002;12:301–309. doi: 10.1089/108729002761381285. [DOI] [PubMed] [Google Scholar]
- Hansen, M.B., Nielsen, S.E., Berg, K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Vandenburgh, K., Manninga, H., Scaringe, S.A., Weber, K., Tuschl, T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- Holen, T., Amarzguioui, M., Wiiger, M.T., Babaie, E., Prydz, H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen, T., Moe, S.E., Sorbo, J.G., Meza, T.J., Ottersen, O.P., Klungland, A. Tolerated wobble mutations in siRNAs decrease specificity, but can enhance activity in vivo. Nucleic Acids Res. 2005;33:4704–4710. doi: 10.1093/nar/gki785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshika, S., Minakawa, N., Kamiya, H., Harashima, H., Matsuda, A. RNA interference induced by siRNA modified with 4′-thioribonucleotides in cultured mammalian cells. FEBS Lett. 2005;579:3115–3118. doi: 10.1016/j.febslet.2005.04.073. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Burchard, J., Leake, D., Reynolds, A., Schelter, J., Guo, J., Johnson, J.M., Lim, L., Karpilow, J., Nichols, K., et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans . Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kumar, R.K., Davis, D.R. Synthesis of oligoribonucleotides containing 2-thiouridine: Incorporation of 2-thiouridine phosphoramidite without base protection. J. Org. Chem. 1995;60:7726–7727. [Google Scholar]
- Kumar, R.K., Davis, D.R. Synthesis and studies on the effect of 2-thiouridine and 4-thiouridine on sugar conformation and RNA duplex stability. Nucleic Acids Res. 1997;25:1272–1280. doi: 10.1093/nar/25.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layzer, J.M., McCaffrey, A.P., Tanner, A.K., Huang, Z., Kay, M.A., Sullenger, B.A. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake, D., Reynolds, A., Khvorova, A., Marshall, W., and Scaringe, S. 2004. Stabilized polynucleotides for use in RNA interference. US Patent No. 20040266707.
- Li, Z.Y., Mao, H., Kallick, D.A., Gorenstein, D.G. The effects of thiophosphate substitutions on native siRNA gene silencing. Biochem. Biophys. Res. Commun. 2005;329:1026–1030. doi: 10.1016/j.bbrc.2005.02.071. [DOI] [PubMed] [Google Scholar]
- Lingel, A., Simo, B., Izaurralde, E., Sattler, M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Luyten, I., Herdewijn, P. Hybridization properties of base-modified oligonucleotides within the double and triple helix motif. Eur. J. Med. Chem. 1998;33:515–576. [Google Scholar]
- Ma, J.B., Ye, K., Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.B., Yuan, Y.R., Meister, G., Pei, Y., Tuschl, T., Patel, D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan, M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Tuschl, T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes & Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleveage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Matranga, C., Tomari, Y., Shin, C., Bartel, D.P., Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- McSwiggen, J. and Beigelman, L. 2003. RNA interference mediated treatment of Alzheimer's disease using short interfering nucleic acids (siNA). European Patent No. EP1423404.
- Mizuno, H., Sundaralingam, M. Stacking of Crick Wobble pair and Watson–Crick pair: Stability rules of G–U pairs at ends of helical stems in tRNAs and the relation to codon–anticodon Wobble interaction. Nucleic Acids Res. 1978;5:4451–4461. doi: 10.1093/nar/5.11.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrot, B., Sipa, K. Chemical and structural diversity of siRNA molecules. Curr. Top. Med. Chem. 2006;6:913–925. doi: 10.2174/156802606777303658. [DOI] [PubMed] [Google Scholar]
- Nawrot, B., Michalak, O., Janicka, M., Maszewska, M., Wojcik, M., Nowak, G., Mikolajczyk, B., Stec, W.J. Novel nucleic acid analogs with a chimeric phosphinate/phosphate backbone, synthesis and biophysical properties. Arkivoc. 2004:151–175. (iii) [Google Scholar]
- Nykanen, A., Haley, B., Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Overhoff, M., Sczakiel, G. Phosphorothioate-stimulated uptake of short interfering RNA by human cells. EMBO Rep. 2005;6:1176–1181. doi: 10.1038/sj.embor.7400535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, S., Fleenor, J., Xu, S., Mello, C., Fire, A. Functional anatomy of a dsRNA trigger: Differential requirement for the two trigger strands in RNA interference. Mol. Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Persengiev, S.P., Zhu, X., Green, M.R. Nonspecific, concentration-dependent stimulation and represion of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, T.P., Allerson, C.R., Dande, P., Vickers, T.A., Sioufi, N., Jarres, R., Baker, B.F., Swayze, E.E., Griffey, R.H., Bhat, B. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- Qing, H., Zhou, W., Christensen, M.A., Sun, X., Tong, Y., Song, W. Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 2004;18:1571–1573. doi: 10.1096/fj.04-1994fje. [DOI] [PubMed] [Google Scholar]
- Rand, T.A., Petersen, S., Du, F., Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Saenger, W. Forces stabilizing association between bases: Hydrogen bonding and base stacking. In: Cantor C.R., editor. Principle of nucleic acids structure. Springer-Verlag; New York: 1984. pp. 116–158. [Google Scholar]
- Saxena, S., Jonsson, Z.O., Dutta, A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- Scheit, K.H., Faerber, P. The effects of thioketo substitution upon uracil-adenine interactions in polyribonucleotides. Synthesis and properties of the alternating polynucleotides poly[r(A-s2U)] and poly[r(A-s2s4U)] Eur. J. Biochem. 1975;50:549–555. doi: 10.1111/j.1432-1033.1975.tb09895.x. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Shohda, K., Okamoto, I., Wada, T., Seio, K., Sekine, M. Synthesis and properties of 2′-O-methyl-2-thiouridine and Oligoribonucleotides containing 2′-O-methyl-2-thiouridine. Bioorg. Med. Chem. Lett. 2000;10:1795–1798. doi: 10.1016/s0960-894x(00)00342-5. [DOI] [PubMed] [Google Scholar]
- Sochacka, E. Efficient assessment of modified nucleoside stability under conditions of automated oligonucleotide synthesis: Characterization of the oxidation and oxidative desulfurization of 2-thiouridine. Nucleosides Nucleotides Nucleic Acids. 2001;20:1871–1879. doi: 10.1081/NCN-100107198. [DOI] [PubMed] [Google Scholar]
- Soutschek, J., Akinc, A., Bramlage, B., Charisse, K., Constein, R., Donoghue, M., Elbashir, S., Geick, A., Hadwiger, P., Harborth, J., et al. Therapeutic silencing of an endogenous gene by systhemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Stuart, J.W., Basti, M.M., Smith, W., Forrest, B., Guenther, R., Sierzputowska-Gracz, H., Nawrot, B., Malkiewicz, A., Agris, P.F. Structure of trinucleotide Dp(acp)3UpA with coordinated Mg2+ demonstrates that modified nucleosides contribute to regional conformations of tRNA. Nucleosides Nucleotides. 1996;15:1009–1028. [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes & Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa, S.M., Disney, M.D., Turner, D.H., Kierzek, R. Thermodynamics of RNA–RNA duplexes with 2- or 4-thiouridines: Implications for antisense design and targeting a group I intron. Biochemistry. 1999;38:16655–16662. doi: 10.1021/bi991187d. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Matranga, C., Haley, B., Martinez, N., Zamore, P.D. Protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Uprichard, S.L. The therapeutic potential of RNA interference. FEBS Lett. 2005;579:5996–6007. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani, G., McClain, W.H. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof, E., Dumas, P., Moras, D. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr. A. 1988;44:112–123. [PubMed] [Google Scholar]
- Yan, K.S., Farooq, A., Hen, A., Zeng, L., Zhou, M.M. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:469–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., Bartel, D.P. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]