Abstract
In plants, worms, and fungi, RNA-dependent RNA polymerases (RDRs) amplify the production of short-interfering RNAs (siRNAs) that mediate RNA silencing. In Arabidopsis, RDR6 is thought to copy endogenous and exogenous RNA templates into double-stranded RNAs (dsRNAs), which are subsequently processed into siRNAs by one or several of the four Dicer-like enzymes (DCL1→4). This reaction produces secondary siRNAs corresponding to sequences outside the primary targeted regions of a transcript, a phenomenon called transitivity. One recognized role of RDR6 is to strengthen the RNA silencing response mounted by plants against viruses. Accordingly, suppressor proteins deployed by viruses inhibit this defense. However, interactions between silencing suppressors and RDR6 have not yet been documented. Additionally, the mechanism underlying transitivity remains poorly understood. Here, we report how several viral silencing suppressors inhibit the RDR6-dependent amplification of virus-induced and transgene-induced gene silencing. Viral suppression of primary siRNA accumulation shows that transitivity can be initiated with minute amounts of DCL4-dependent 21-nucleotide (nt)-long siRNAs, whereas DCL3-dependent 24-nt siRNAs appear dispensable for this process. We further show that unidirectional (3→5′) transitivity requires the hierarchical and redundant functions of DCL4 and DCL2 acting downstream from RDR6 to produce 21- and 22-nt-long siRNAs, respectively. The 3→5′ transitive reaction is likely to be processive over >750 nt, with secondary siRNA production progressively decreasing as the reaction proceeds toward the 5′-proximal region of target transcripts. Finally, we show that target cleavage by a primary small RNA and 3→5′ transitivity can be genetically uncoupled, and we provide in vivo evidence supporting a key role for priming in this specific reaction.
Keywords: Dicer, RNA-dependent RNA polymerase, transitivity, siRNA, viral suppressors
INTRODUCTION
In several organisms including plants, nematodes, and fungi, proteins known as RNA-dependent RNA (RDR) polymerases amplify the accumulation of short-interfering RNAs (siRNAs), the products of RNase III enzymes in the Dicer family that mediate RNA silencing (Cogoni and Macino 1999; Dalmay et al. 2000b; Mourrain et al. 2000; Smardon et al. 2000; Sijen et al. 2001). Although RDRs were among the first factors to be genetically implicated in RNA silencing pathways, their effects remained largely unexplained until the discovery, in plants and Caenorhabditis elegans, of a phenomenon known as “transitivity,” which diagnoses RDR activities in vivo (Voinnet et al. 1998; Sijen et al. 2001; Vaistij et al. 2002). In transitivity, an initial pool of small RNAs, called “primary” small RNAs, directed against one region of a transcript induces the production of “secondary” siRNAs corresponding to regions outside the primary RNA target sites. Like primary siRNAs, the secondary siRNAs produced via RDR activities are thought to direct RNA-induced silencing complexes (RISCs) to promote sequence-specific events reducing transcription, stability, or translation of target RNA molecules (Hammond et al. 2000; Ekwall 2004). RDR-mediated amplification systems most likely explain the remarkable potency and the persistence of RNAi in nematodes, of quelling in fungi, and of post-transcriptional gene silencing (PTGS) in plants, although the molecular mechanisms involved are not fully understood.
Although RDRs were primarily implicated in transgene-induced silencing, they also have important cellular functions. For instance, the C. elegans EGO-1 is essential for germline development, in addition to its role in germline-specific RNAi (Smardon et al. 2000). In Schizosaccharomyces pombe, Rdp1 is required to produce pericentromeric small RNAs that are important for heterochromatin formation required for proper chromosome segregation at meiosis (Zaratiegui et al. 2007). Similarly, Arabidopsis RDR2, one of the six RDR paralogs, is required for biogenesis of very diverse and abundant siRNA species involved in maintenance of genome integrity and transposon taming through heterochromatin formation (Xie et al. 2004). Arabidopsis RDR6 controls aspects of leaf development and juvenile-adult phase transitions through the synthesis of endogenous trans-acting siRNAs (tasiRNAs) that function in a manner similar to exogenous siRNAs, by promoting cleavage of target transcripts (Peragine et al. 2004; Vazquez et al. 2004; Allen et al. 2005). RDR6 also has roles during virus-induced gene silencing (VIGS). In VIGS, double-stranded RNA (dsRNA) produced upon virus replication, or derived from stem–loop structures within single-stranded viral RNA (ssRNA), is processed by Dicer-like (DCL) enzymes into siRNAs (Blevins et al. 2006; Bouche et al. 2006; Deleris et al. 2006). These molecules are then thought to guide an antiviral RISC to promote cleavage of viral transcripts (Pantaleo et al. 2007). Production of antiviral siRNAs can be further enhanced by the action of RDR6, which presumably copies some viral ssRNAs into dsRNA, thereby amplifying the VIGS response (Schwach et al. 2005; Wassenegger and Krczal 2006). Consistent with this model, Cucumber mosaic virus (CMV) accumulates to higher levels in the Arabidopsis rdr6 mutant (Mourrain et al. 2000), and, likewise, reducing the levels of the Nicotiana benthamiana (Nb) RDR6 ortholog induces hypersusceptibility to several plant viruses (Muangsan et al. 2004; Qu et al. 2005; Schwach et al. 2005). A common response of plant viruses to antiviral silencing is the production of suppressor proteins that target various stages of the silencing mechanism (for review, see Voinnet 2005; Li and Ding 2006). However, their potential effect on plant RDR functions has not been investigated yet.
Here, we describe a series of experiments involving transgenic Arabidopsis plants that express various RNA silencing suppressors isolated from unrelated viruses and investigate their effects on VIGS amplification and secondary siRNA synthesis at endogenous and transgene loci. Using an original RNA silencing model system, we also address several key mechanistic issues relating to the mode of operation of RDR6 during unidirectional (3→5′) transitivity in Arabidopsis.
RESULTS AND DISCUSSION
Viral-encoded silencing suppressors inhibit RDR6-dependent processes that restrict PVX accumulation in Arabidopsis
To compare side-by-side the effects of distinct viral silencing suppressors on RDR-mediated amplification of VIGS, we sought to develop a genetically tractable system resembling the situation encountered during authentic virus infections. Because accumulation of Potato virus X (PVX) is naturally restricted by the activity of NbRDR6 (Qu et al. 2005; Schwach et al. 2005), we used the RDR6-dependent AMPLICON (AMP) system in Arabidopsis (Dalmay et al. 2000a). In the AMP line, a transgene expressing GFP-tagged and replicating PVX RNA (PVX-GFP) is both an initiator and a target of RNA silencing (Fig. 1A; Dalmay et al. 2000a). Transgenic and replicating PVX-GFP RNA levels are vastly reduced, and the plants appear uniformly red under UV illumination, owing to chlorophyll fluorescence (Fig. 1B, upper panel; Fig. 1C, lane 2). Viral-derived siRNAs remain undetectable (Fig. 1C, lane 2).
FIGURE 1.
Viral-encoded suppressors reactivate PVX-GFP accumulation in the AMP and AMPXGFP Arabidopsis lines. (A) Schematic of GFP silencing in the Arabidopsis AMP and AMPXGFP lines. Expression of the AMP transgene, driven by the constitutive 35S promoter, leads to the production of viral RNAs (➀). These molecules are used as templates by the viral replicase to produce replicative double-stranded RNA intermediates (RI) that, presumably, are processed by Dicer into siRNAs (➁). In the AMPXGFP system, these molecules can direct the cleavage of viral RNAs, as in AMP, but also of GFP mRNAs deriving from a separate transgene driven by the 35S promoter. RI-derived siRNAs can also promote RDR6 activity using viral RNAs or GFP mRNAs as templates (➂) to amplify RNA silencing through the production of secondary siRNAs (➃ and ➄). (B) GFP expression is restored in AMP (upper panel) and AMPXGFP (lower panel) plants expressing viral-encoded suppressors, and in the rdr6 mutant background. The red color is from chlorophyll autofluorescence under UV light, and indicates an absence of GFP accumulation. (C) RNA blot analyses of high- and low-molecular-weight RNAs using DNA probes against GFP (upper panel) or the PVX-specific coat protein (CP) sequences. RNA extracted from GFP transgenic plants was used as a control in lane 1. (rRNA) Ethidium bromide staining of ribosomal RNAs; (gRNA) PVX genomic RNA; (sgRNA) subgenomic RNA; (CP) PVX coat protein.
Introducing the rdr6 mutation in the AMP line promoted the appearance of multiple green fluorescent foci in leaves (Fig. 1B, upper panel). Presumably, virus replication had resumed in these foci because genomic and subgenomic PVX-GFP RNA species as well as 21-nt viral-derived siRNAs were detected (Fig. 1C, lane 6). We crossed the AMP line with transgenic lines expressing the P19, P38, and P15 proteins from Tomato bushy stunt virus (TBSV), Turnip crinkle virus (TCV), and Peanut clump virus (PCV), respectively. All of the suppressors reactivated GFP expression from replicating PVX, and accumulation of 21-nt-long viral-derived siRNAs (Fig. 1B, upper panel; Fig. 1C, lanes 3–5). To study the impact of silencing suppressors on trans-acting RDR6 activities, a GFP transgene was introduced into the AMP line and in the corresponding crosses involving P19, P38, and P15. In this dual system (AMPXGFP), PVX-GFP replication triggers the RDR6-dependent silencing of the introduced GFP transgene (Fig. 1A). The resulting self-sustaining GFP siRNAs constitute the vast majority of detectable small RNAs, and they, in turn, silence PVX-GFP replication (Fig. 1B, lower panel; Fig. 1C, lane 7, GFP probe). Consequently, virus-specific siRNAs are below the detection limit (Fig. 1C, lane 7, CP probe). Introducing the rdr6 mutation in the AMPXGFP line led to a homogenous green fluorescent phenotype owing to resumed GFP transgene expression (Fig. 1B, lower panel). Additionally, PVX-GFP replication and accumulation of virus-derived siRNAs were restored, as described (Fig. 1C, lane 11; Dalmay et al. 2000b). As shown in Figure 1C, lanes 8–10, the silencing suppressors had similar (Fig. 1C, P19, lane 8) or stronger effects (Fig. 1C, P38 and P15, lanes 9,10), although none promoted the uniform green fluorescent phenotype found in the rdr6 background (Fig. 1B, lower panel).
Collectively, these results indicate that P19, P38, and P15 could inhibit the cis- and trans-acting inhibitory effects of the RDR6-dependent mechanism that normally restricts PVX-GFP accumulation in Arabidopsis. These observations extend previous findings made with the HcPro suppressor protein of potyviruses, which suppressed silencing from a GUS-tagged PVX AMPLICON in tobacco (Mallory et al. 2002). Collectively, the results suggest that viral silencing suppressors are potent inhibitors of VIGS amplification pathways in natural infection contexts. They also indicate that suppression of amplified VIGS paradoxically correlates with enhanced accumulation of viral-derived siRNAs, suggesting that a dynamic equilibrium between silenced and active viral replication states is created. It is at present unclear if these states are spatially separated within plant tissues, but this could explain the mosaic of green fluorescent (active) and red fluorescent (silenced) foci observed upon expression of most silencing suppressors in the AMP and AMPXGFP lines (Fig. 1B).
P19, P38, and HcPro suppress RDR6-directed transitive RNA silencing, whereas P15 and P25 do not interfere with this process
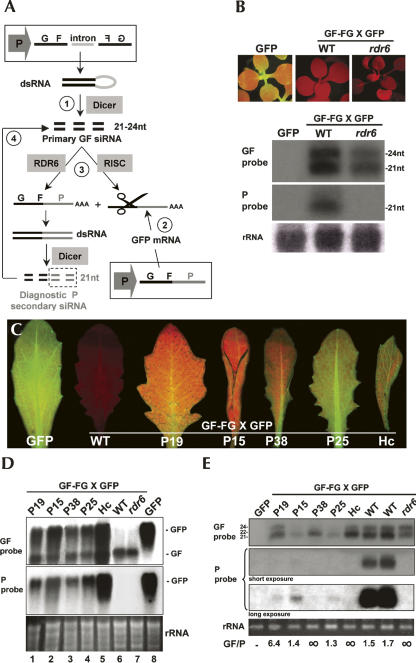
The intrinsic complexity of the AMP and AMPXGFP systems precluded the discrimination between primary (RDR6-independent) and secondary (RDR6-dependent) siRNAs, such that a bona fide effect of the suppressors on RDR6 activity could not be ascertained. Moreover, P19, P38, P15, and HcPro are known to interfere with siRNA accumulation and/or siRNA-directed RISC activity (Chapman et al. 2004; Dunoyer et al. 2004; Lakatos et al. 2004), both of which are likely involved in the overall VIGS amplification process. To overcome these caveats, we used the GF-FGXGFP line (Fig. 2A), in which discrete 21-nt and 24-nt primary siRNAs are processed by DCL4 and DCL3, respectively, from inverted-repeat transcripts corresponding to the 5′ part (“GF”) of a separate GFP transgene (Dunoyer et al. 2006). Transitive RNA silencing was measured by the relative accumulation of RDR6-dependent secondary siRNAs (exclusively 21 nt in length) with sequence of the non-overlapping 3′ part (“P”) of the GFP transgene (Fig. 2B). Due to the robust nature of the silencing initiated by the GF-FG construct, GFP silencing was as extensive in wild-type (WT) plants as it was in rdr6 mutant plants, and both accumulated similar levels of GF-FG inverted-repeat transcripts (Fig. 2D, cf. lanes 6 and 7). As expected, P siRNAs did not accumulate in plants carrying the rdr6 mutation, leading to a GF/P ratio with an infinite value (Fig. 2E). The approximate 1.6 mean value of the GF/P ratio found in plants with intact RDR6 was used as a quantitative reference for the efficacy of transitivity in the GF-FGXGFP system (Fig. 2E).
FIGURE 2.
P19, P38, and HcPro, but not P15 and P25, interfere with RDR6-dependent transitivity. (A) Schematic of GFP silencing in the Arabidopsis GF-FG X GFP line. Expression of the GF-FG transgene leads to accumulation of primary GF siRNAs, which cleave the mRNAs from a separate GFP transgene and guide RDR6 to amplify RNA silencing. RDR6-mediated transitivity is diagnosed by the production of “P” secondary siRNAs that derive from the GFP mRNAs used as templates by RDR6. (B) GFP expression in GF-FG X GFP and GF-FG X GFP/rdr6 plants is uniformly silenced (upper panels). RNA blot analyses of low-molecular-weight RNAs using DNA probes against the GF (upper panel) or P (lower panel) regions of the GFP sequence. (C) Reactivation of GFP expression in GF-FG X GFP plants expressing different suppressors of RNA silencing. (D) RNA blot analyses of high-molecular-weight RNAs using DNA probes against the GF (upper panel) or P (lower panel) regions of the GFP sequence. Note the detection of the hairpin arms (GF) by the GF-specific, but not by the P-specific, probe. (E) RNA blot analyses of the low-molecular-weight fraction assayed in D. GF/P values indicate the ratio between the amount of GF siRNAs and P siRNAs for each sample, as calculated by quantitative PhosphorImaging. (rRNA) Same as in Figure 1. RNA extracted from an Arabidopsis line expressing GFP constitutively (as shown in B and C) was used as a control in the Northern analyses presented in D and E.
In addition to P19, P15, and P38, used above, the analysis in the GF-FGXGFP system included the HcPro protein of Turnip mosaic virus (TuMV) (Dunoyer et al. 2004). We also tested the PVX-encoded P25 silencing suppressor (Voinnet et al. 2000; Dunoyer et al. 2004), which could not be assayed in AMP and AMPXGFP because both systems generate siRNAs targeted against the P25 sequence. First, we confirmed that all of the suppressors were expressed upon their introduction into the GF-FGXGFP line (Supplemental Data 1). This was also evident from the typical morphological defects associated with expression of some of those proteins, also observed in the AMP and AMPXGFP systems (Figs. 1B, 2C). Although none of the suppressors inhibited accumulation of the long GF transcripts producing primary siRNAs, they all restored green fluorescence and GFP mRNA accumulation in leaves (Fig. 2C; Fig. 2D, lanes 1–5). GF- and P-specific probes were used in Northern analyses of small RNAs isolated from those plants to assay for transitivity. Both P25 and P15 caused a loss of 24-nt-long primary siRNAs (Fig. 2E). They also strongly reduced (by at least 10-fold), but did not eliminate, the accumulation of primary, 21-nt siRNAs (Fig. 2E). Despite those reduced primary siRNA levels, longer exposures revealed that the GF/P ratios were similar to that found in GF-FGXGFP plants, suggesting that neither P25 nor P15 inhibits the activity of RDR6 (Fig. 2E). We also conclude from those data that drastically reduced levels of primary 21-nt-long siRNAs are sufficient to trigger efficient transitivity, and that primary 24-nt siRNAs are dispensable for this process.
P19 and HcPro altered the accumulation of 24-nt-long GF primary siRNAs (Fig. 2E) and had moderate effects on the 21-nt-long species, consistent with previous reports (Dunoyer et al. 2004). However, accumulation of secondary P siRNAs was either strongly reduced (P19) or abolished (HcPro) (Fig. 2E). The levels of 24-nt-long GF primary siRNAs were reduced in P38-expressing plants, and there was a shift in the size of 21-nt-long siRNAs, which migrated as a 22-nt-long species. Additionally, P secondary siRNAs were below the detection limit (Fig. 2E). We conclude that, unlike the P15 and P25 proteins, P19, P38, and HcPro strongly inhibit secondary siRNA production. The effects of P19 might be explained by its previously demonstrated ability to bind hairpin-derived, 21-nt-long siRNAs (Dunoyer et al. 2004), presumably precluding their involvement in the transitivity process. The size shift of the 21-nt GF primary siRNAs in P38-expressing plants is diagnostic of the DCL4-antagonizing property of this suppressor, as inactivating DCL4 activity promotes the surrogate action of DCL2, which generates 22-nt siRNAs (Bouche et al. 2006; Deleris et al. 2006; Fusaro et al. 2006). Since the levels of the 22-nt-long GF siRNAs accumulating in the P38-transgenic plants were much higher than those of 21-nt-long GF siRNAs that permitted transitivity in P25- and P15-transgenic plants, we infer that either 22-nt-long siRNAs generally cannot recruit RDR6 for transitivity or that their biological activity is impaired by P38. We cannot formally exclude, at this stage, an additional effect of P38 downstream from RDR6. The effects of HcPro on transitivity remain, at present, ill defined, but they might be linked to the known ability of the protein to suppress siRNA-guided RISC activity (Chapman et al. 2004; Dunoyer et al. 2004), possibly by binding to siRNAs (Lakatos et al. 2006).
The effects of silencing suppressors on accumulation of endogenous trans-acting siRNAs
We further tested the accumulation of endogenous trans-acting siRNAs in the various suppressor-transgenic lines used in the above study. tasiRNA biogenesis involves an initial miRNA-guided cleavage of primary tasiRNA transcripts, which sets a defined point for RDR6-directed complementary strand synthesis, followed by a phased DCL4-dependent processing reaction that generates mature tasiRNAs. These molecules then incorporate into an RISC to guide cleavage of specific mRNAs that contain tasiRNA-complementary sequences (Vazquez et al. 2004; Allen et al. 2005; Yoshikawa et al. 2005). We reasoned that the involvement of RDR6 in the tasiRNA biosynthetic pathway could potentially be exploited to further ascertain the effects of some silencing suppressors on transitivity, as assessed in the GF-FGXGFP experimental system (Fig. 2).
As expected from previous reports (Chapman et al. 2004; Dunoyer et al. 2004), P15 did not affect the levels of the miRNAs required (miR390 for TAS3; miR173 for TAS1 and TAS2) or not (miR171) for biogenesis of the three tasiRNAs tested in this study, whereas HcPro promoted the overaccumulation of most miRNAs tested (Fig. 3A, lanes 2 and 5, respectively). Surprisingly, P19 inhibited miR173 accumulation, whereas it had little effect on miR390 and miR171 levels (Fig. 3A, lane 1), as expected from previous work (Chapman et al. 2004; Dunoyer et al. 2004). P19, P15, and HcPro were previously shown to prevent miRNA-directed functions, which partly accounts for the recurrent developmental defects caused by those factors (Fig. 2C). This shared property likely explained the strong inhibitory effects of the three suppressors on accumulation of TAS1 tasiR255, TAS2 tasiR752, and TAS3 tasiR5′D7 (Fig. 3A, lanes 1,2,5). Accordingly, accumulation of the ARF3 transcripts, a target of tasiR5′D7, was significantly higher in the P19-, P15-, and HcPro-expressing plants than in WT plants, and it was comparable to, or higher than those found in rdr6 mutant plants (Fig. 3B). Since tasiRNAs are crucially implicated in developmental phase transitions, leaf development, and polarization (Peragine et al. 2004), their inhibition by P19, P15, and HcPro may well contribute to the recurrent developmental aberrations caused by those proteins in transgenic (Fig. 2C) and, perhaps, in authentic infection contexts.
FIGURE 3.
Effects of silencing suppressors on accumulation of endogenous, RDR6-dependent trans-acting siRNAs and RDR6-independent miRNAs. (A) Low-molecular-weight RNA analyses in transgenic plants expressing the five silencing suppressors, using DNA oligonucleotide probes specific for endogenous small RNAs. (rRNA) Same as in Figure 1. (B) Quantitative-RT PCR analyses of ARF3 (At2g33860) accumulation in WT and suppressor-expressing plants. The primers flanked the second complementary sequence of the TAS3 tasiRNA5′D7, as found in the ARF3 mRNA.
Unlike P19, P15, and HcPro transgenic plants, the P38- and P25-expressing plants used in this and other studies did not show developmental defects, contained normal miRNA levels, and had intact miRNA-guided functions (data not shown; Fig. 3A, lanes 3,4; Dunoyer et al. 2004). Nonetheless, P38 reduced the tasiRNA levels (Fig. 3A, lane 3) and caused a threefold increase in ARF3 levels (Fig. 3B), an effect likely attributable to the DCL4-antagonizing properties of this suppressor. In contrast, neither the tasiRNA nor the miRNA levels were affected by P25, and, accordingly, the ARF3 levels were comparable to those found in WT plants (Fig. 3A, lane 4; Fig. 3B). We conclude that P25 affected none of the steps required for tasiRNA biogenesis or action. This analysis therefore confirms that the PVX-encoded P25 protein does not interfere with RDR6-directed transitivity (Fig. 2E), a result consistent with the reversal of the AMP- and AMPXGFP-mediated silencing in the rdr6 Arabidopsis mutant (Fig. 1), and with the observation that PVX accumulation is naturally restricted by the action of the N. benthamiana RDR6 ortholog despite production of P25 (Qu et al. 2005; Schwach et al. 2005). P25 therefore appears to exert specific inhibitory effects on the accumulation of hairpin-derived primary siRNAs, which resembles the effects of specific dcl4 mutant alleles that retain normal tasiRNA biogenesis and action, yet abolish accumulation of hairpin-derived, 21-nt siRNAs (Dunoyer et al. 2005).
Processing of the dsRNA products of transitivity involves the hierarchical and redundant activities of DCL4 and DCL2
Having established the effects of silencing suppressors on transitive RNA silencing, we sought to determine some of the parameters that influence this process in vivo. In plants, the only available mechanistic data relate to the production of endogenous tasiRNAs, which involves a phased, DCL4-dependent processing reaction initiated by one or two discrete miRNA-directed cuts (Allen et al. 2005; Yoshikawa et al. 2005; Axtell et al. 2006). However, it is unclear if this mechanism is a peculiarity of the tasiRNA pathway, or if it applies broadly to silencing amplification, notably during virus-induced and transgene-induced silencing, which were first associated with transitivity (Voinnet et al. 1998; Vaistij et al. 2002). Among the outstanding questions relating to transitivity in those systems are the nature of the DCL(s), if any, that can act downstream from RDR6 action, the processivity of the amplification reaction, and whether siRNA priming, as opposed to siRNA-directed cleavage events, initiates the amplification process.
Out of the four Arabidopsis DCLs, the most likely candidates for the enzymes potentially acting downstream from RDR6 during transgene-induced silencing are DCL2 and DCL4 (Dunoyer et al. 2005; Wassenegger and Krczal 2006). However, together with DCL3, the two enzymes are also required for synthesis of primary siRNAs used to initiate transitivity in hairpin-based RNA silencing systems (Dunoyer et al. 2005; Fusaro et al. 2006), preventing the use of the GF-GFxGFP line to address the issue. Therefore, we sought to exploit a system in which initiation of transitivity does not rely on the siRNA products of DCL2, DCL3, or DCL4. We previously described a transgenic GFP sensor construct reporting the inhibitory activity of the endogenous and DCL1-dependent miR171 (Parizotto et al. 2004). The construct (herein referred to as “GFP171.1”) carries, within its 3′-UTR, the authentic and fully complementary target sequence of miR171, as found in the SCARECROW-LIKE 6 (SCL6)-II, -III, and -IV transcripts (Fig. 4A). We reported that GFP expression was uniformly silenced in 100% of WT Arabidopsis GFP171.1 transformants, owing to an RDR6-dependent transitive RNA silencing process initiated by miR171, which causes production and movement of secondary GFP-specific siRNAs (Parizotto et al. 2004). These molecules promote extensive GFP silencing that masks the miR171-directed cleavage pattern (Fig. 4C, middle leaf; Fig. 4E, lane 2). A bona fide cleavage pattern was only revealed in an rdr6 mutant background (Fig. 4C, right leaf; Parizotto et al. 2004), where GFP expression was restricted to the vascular tissue. This was complementary to the miR171 transcription profile, which is mostly epidermal and excluded from the veins (Fig. 4B). The expected 3′-cleavage fragment resulting from miR171-directed cleavage was also evident in the rdr6 mutant background (Fig. 4E, lane 3). Therefore, the vein-centered green fluorescent phenotype in the GFP171.1/rdr6 plants faithfully reports the RNA cleavage activity of miR171 and simultaneously diagnoses the loss of miR171-directed transitivity. Note that unlike the GF-FG system, which monitors 5→3′ transitivity, the GFP171.1 system reports 3→5′ transitivity exclusively.
FIGURE 4.
Dissection of transitive RNA silencing induced by a single, endogenous miRNA. (A) Schematic of RDR6-dependent transitive RNA silencing triggered by miR171 in line GFP171.1. Annealing of the miR171-guided RISC promotes cleavage of the GFP 171.1 mRNA. In addition, RDR6 produces dsRNA using the GFP 171.1 mRNA as a template. The dsRNA is then processed by DCL(s) into secondary siRNAs that can target GFP 171.1 mRNA and spread into neighboring cells. (B) Transverse section of leaves from an Arabidopsis line expressing GFP under the control of the miR171 promoter (two upper left panels) or from the GFP171.1 line (two upper right panels) in the rdr6 mutant background. The diagrams below schematize the pattern of GFP expression in each case. (Black) High GFP expression; (white) a lack of GFP expression; (gray) faint GFP expression. Note that the miR171 transcription pattern mirrors that of miR171 activity. (epid) Upper and lower epidermis; (p.p.) palisade parenchyma; (l.p.) lacuna parenchyma; (v) vein. (C) Uniform GFP expression in plants transformed with a GFP transgene devoid of the miR171 target site (left) and complete lack of GFP expression upon transformation of the GFP 171.1 construct (middle) in WT plants. The vein-specific pattern corresponding to miR171 activity is evident in the vast majority of the GFP171.1 transformants carrying the rdr6 mutation (right). (D) GFP expression in dcl2, dcl3, dcl4, and dcl2-dcl4 plants transformed with GFP 171.1. In each case, 30 transformants were chosen randomly and inspected individually under UV light. The number of plants exhibiting the phenotype shown is indicated on the upper right corner in each case. (E) Northern analyses of high- (upper panel) and low- (lower panel) molecular-weight RNAs from GFP 171.1 in the various mutant backgrounds. A probe specific to miR171 was used to confirm normal accumulation of the miRNA in each background. U6 hybridization was used as a control for RNA loading. (F) Northern analyses of low-molecular-weight RNAs accumulating in GFP 171.1 plants with a WT genetic background. The DNA probes were specific to different sections of the GFP 171.1 transcript, as indicated on the scheme. s/n values indicate, for each probe, the ratio (quantified by PhosphorImaging) between the signal intensities for GFP171.1/WT (left signal) and GFP171.1/rdr6 (right signal). (G) Schematic of the GFP171.14 construct carrying three nucleotide mismatches in the 5′ part of the miR171-complementary sequence. Most of the GFP171.14/WT transformants display the vein-centered GFP pattern of the GFP171.1/rdr6 and GFP171.1/dcl2-dcl4 plants (left panel). (Right panel) Northern analyses of high- (upper panel) and low- (middle panel) molecular-weight RNAs accumulating in the GFP171.1/WT, GFP171.1/rdr6, and GFP 171.14/WT plants. The DNA probe used was specific for the GFP coding sequence. (rRNA) (lower panel) Same as in Figure 1.
We transformed the GFP171.1 construct into Arabidopsis plants carrying single or double mutations in the DCL2, DCL3, and DCL4 genes. None of the single dcl3 or dcl4 mutations affected the miR171-directed transitivity as all the primary transformants were uniformly red under UV light. This was also observed for most of the dcl2 transformants. In all cases, red plants accumulated low GFP mRNA levels (data not shown; Fig. 4D,E, lanes 4,5), and the levels of secondary GFP-specific siRNAs were as high as in the GFP171.1 line (Fig. 4E, cf. lanes 4,5 and 2). A typical size shift in the 21-nt-long GFP siRNAs was observed in the dcl4 mutant background, indicating that DCL2 had substituted DCL4 to generate active 22-nt-long siRNAs (Fig. 4E, lane 5). This observation suggested that inactivating both DCL2 and DCL4 was likely necessary to alleviate transitivity. Agreeing with this hypothesis, only the double dcl2-dcl4 mutant recapitulated the effects of the rdr6 mutation among the dcl combination mutants tested: in 90% of the transformants, there was vascular-restricted GFP expression (Fig. 4D, right panel), loss of secondary GFP siRNAs, and the expected miR171-generated 3′-cleavage fragment was detected, as in the rdr6 background (Fig. 4E, lane 6). We conclude that the hierarchical and redundant activities of DCL4 and DCL2 account for transitivity in the GFP171.1 system and that both enzymes can process the dsRNA products of RDR6.
Transitive RNA silencing in line GFP171.1 is a unidirectional and processive reaction that can span >750 nt
Unlike in hairpin-triggered silencing systems (e.g., GF-FG) that generate heterogeneous primary siRNA species (Llave et al. 2002), transitivity is initiated in GFP171.1 by a single small RNA species (miR171), at a defined position within the GFP171.1 transcript (Fig. 4A). This unique feature granted an evaluation of the extent of secondary siRNA production, both 5′ and 3′ of the miR171 target site. Contiguous DNA fragments with similar G:C contents and spanning the entire sequence of the GFP171.1 transcript were used in Northern analyses to assay for siRNA accumulation (Fig. 4F). The same filter was stripped and re-probed following each hybridization. A blank corresponding to RNA extracted from GFP171.1/rdr6 plants, run in parallel, was used to calculate a signal-to-noise ratio (s/n) that provided an intrinsic measurement of the intensity of each hybridization signal (Fig. 4F). A strong siRNA signal was detected just upstream of the miRNA target site, but it progressively faded toward the 5′-end of the GFP171.1 transcript, and it was barely above background level with the 5′-most proximal probe (Fig. 4F). No signal was detected with a probe spanning the region located 3′ of the miR171 cleavage site, although the same probe could readily detect the GFP171.1 3′-cleavage fragment in Northern analysis of high-molecular-weight RNA from GFP171.1/rdr6 plants (data not shown). We conclude that, unlike in the GF-FGXGFP system (Fig. 2), transitivity occurs mostly 3→5′ in the GFP171.1 line, and that RDR6 and/or DCL4 are likely processive over >750 nt. Together with the progressive loss of siRNA production toward the 5′-end of the GFP171.1 transcript, these results suggest that secondary siRNAs do not engage in further rounds of amplification to generate tertiary siRNAs, at least not to the extent seen with primary siRNAs.
Small RNA-guided cleavage and transitive RNA silencing can be genetically uncoupled
Several models for transitivity in plants entail that cleavage of target transcripts by primary small RNAs is a prerequisite for RDR6-mediated transitivity, as appears to be the case for biogenesis of some tasiRNAs (Allen et al. 2005). Alternatively, primary small RNAs have been proposed to act as primers for RDRs, but this model has been challenged by the observation that spreading of secondary siRNA production often occurs both 5′ and 3′ of primarily targeted regions in plants (Voinnet et al. 1998; Vaistij et al. 2002; Allen et al. 2005). The finding that miR171-directed transitivity occurs mostly, if not exclusively, 3→5′ in line GFP171.1 (Fig. 4F) prompted us to experimentally assess the second possibility. To that aim, we re-engineered the GFP171.1 sensor construct to carry three nucleotide mismatches corresponding to the 3′-end of miR171 (Fig. 4G, diagram). The results of previous transient expression experiments suggested that this modification was unlikely to affect annealing and cleavage by miR171 (Parizotto et al. 2004). However, it was predicted to strongly inhibit a putative priming event initiated by miR171. The modified construct, referred to as GFP171.14, was introduced into WT Arabidopsis plants, and transformants were scored for GFP expression. Most (80%) of them exhibited a vein-restricted green fluorescent pattern that was indistinguishable from that of the GFP171.1/rdr6 plants (Fig. 4G, left panel; Fig. 4C, rightmost panel). Moreover, secondary GFP siRNAs were at or below detection level, and the 3′-fragment diagnostic of miR171-directed cleavage could be detected, as in GFP171.1/rdr6 plants (Fig. 4G). We conclude that the three mutations in the GFP171.14 sensor uncoupled miR171-directed cleavage from miR171-directed transitivity. The results also strongly suggest that priming, as opposed to cleavage, is the factor that determines the onset of transitivity in the GFP171.1 system.
Conclusions
Our study indicates that inhibition of RDR6-dependent processes is a feature of many viral-encoded silencing suppressors, consistent with the observation that RDR6 activity restricts accumulation of several viruses in plants (Mourrain et al. 2000; Muangsan et al. 2004; Qu et al. 2005; Schwach et al. 2005). Also agreeing with the antiviral role of RDR6, the hierarchical and redundant activities of DCL4 and DCL2, previously implicated in biogenesis of primary siRNAs from viral dsRNAs (Bouche et al. 2006; Deleris et al. 2006), are required downstream from RDR6 during transgene-induced and, presumably, virus-induced gene silencing. In principle, alteration of RDR6 functions by viruses could be achieved through direct inhibition of its RNA polymerase activity. However, given the strong interactions found between viral suppressors and siRNAs produced by DCL4 and DCL2 (Lakatos et al. 2006), it is likely that RDR6 suppression occurs mostly upstream or downstream of dsRNA synthesis. This was clearly exemplified here with P15 and P25, which affected silencing amplification without altering the efficacy of transitivity per se (Fig. 2E). Rather, both proteins dramatically reduced the levels of primary siRNA required to trigger this process in the GF-FGXGFP system, which reports 5→3′ transitivity. Note that the effects of silencing suppressors on 3→5′ transitivity (GFP171.1 system) were not tested here because P15, HcPro, and P19 were shown to inhibit miR171-guided functions in Arabidopsis, a feature that would have confounded the interpretation of the results obtained in the GFP171.1 system (Dunoyer et al. 2004).
The precise role of primary siRNAs in transitivity has been debated. Some models, for instance, infer that siRNA-directed cleavage is key to initiate this process. Axtell and colleagues further proposed that multiple cuts by primary small RNAs might enhance the formation of aberrant RNAs (abRNAs, lacking both caps and tails) from targeted transcripts, providing preferential templates to RDR6 (Axtell et al. 2006). This model is supported by the apparent conservation of two miRNA hits in some, but not all, tasiRNA primary transcripts, and by the observation that accumulation of uncapped mRNAs, indeed, promoted the RDR6-dependent silencing of a transgene in Arabidopsis (Gazzani et al. 2004). The “two-hit model” of Axtell et al. (2006) also offers a straightforward explanation to the bidirectional (3→5′ and 5→3′) nature of transitivity during hairpin-triggered RNAi (as is probably the case in the GF-FGXGFP system) and biogenesis of some tasiRNAs in plants. However, an obvious requirement of this model is the presence of at least two separate small RNA hits within a target transcript, a situation not applicable to the GFP171.1 sensor RNA, which contains a single target sequence for a single primary small RNA (miR171). In fact, the results obtained with the modified GFP171.14 sensor show that cleavage by miR171, on the one hand, and transitivity, on the other, can be genetically uncoupled. Therefore, the two molecular events are not always linked. The results also strongly suggest the involvement of priming in the GFP171.1 transitive system, which agrees with the unidirectional spread (3→5′) of secondary siRNA synthesis in this case.
One way to reconcile the data is to propose that, perhaps, transitivity activated by unique small RNA species might be mostly primed, cleavage-independent, and unidirectional, while transitivity associated with multiple primary small RNA cuts might be mostly unprimed, bidirectional, and strongly dependent on cleavage to produce the abRNA templates of RDR6. These two separate modes of RDR6 operation might have arisen out of specific biological needs, but, in any case, we do not consider them as being necessarily mutually exclusive: both processes might, for instance, operate during virus infections. Hence, aborted viral replication products that commonly accumulate during infections could be perceived as abRNAs and could trigger unprimed, bidirectional transitivity, while isolated siRNA species, typically produced from discrete stem–loop structures within single-stranded viral transcripts (Molnar et al. 2005), could prime unidirectional transitivity.
A recent analysis of transitivity in C. elegans involved a system very similar to the GFP171.1 device employed here in Arabidopsis. In this system, prevalently unidirectional (3→5′) transitivity was triggered by a unique siRNA produced from a transgenic, short-hairpin construct (Sijen et al. 2007). The results of this and of another recent study (Pak and Fire 2007) suggest that transgene-induced secondary siRNA synthesis might proceed in a Dicer-independent manner in Caenorhabditis elegans, through a reaction whereby RDRs produce 19–24-nt-long complementary RNAs directly from their templates, without priming requirements (Sijen et al. 2007). Our data, suggest, on the contrary, that single primary siRNA species likely act as primers for unidirectional secondary siRNA production in Arabidopsis, by promoting synthesis of long dsRNA molecules through a processive reaction that can extend up to at least 750 nt. In addition, the activities of DCL4, and of its surrogate DCL2, are necessary to generate secondary siRNAs downstream from RDR6 action. Incidentally, the DCL4–RDR6 consortium has been also implicated in synthesis and amplification of silencing movement between cells, further supporting a previously proposed model whereby extensive silencing spread results from reiterated short-distance movement events involving the 21-nt siRNA products of DCL4 (Dunoyer et al. 2004). These and additional drastic differences in the mechanism of transitivity in plants and worms were not initially anticipated because the RDRs from the two organisms are similar in terms of amino acid sequence and structure (Wassenegger and Krczal 2006). It will be of great interest to identify what specific features of plant and worm RNA silencing pathways underlie the contrasted mode of operation of RDRs, and how these features relate to the cellular functions of those proteins in the two organisms.
MATERIALS AND METHODS
Plant material
Arabidopsis lines AMP, AMPXGFP, AMPXGFP/rdr6, GF-FGXGFP, and GF-FGXGFP/rdr6 were described previously (Dalmay et al. 2000a,b; Dunoyer et al. 2006), as was the Arabidopsis line reporting the transcriptional activity of miR171 (Parizotto et al. 2004). Binary vectors for silencing suppressors (Dunoyer et al. 2004) were used to transform WT Arabidopsis (ecotype Col-0) using the floral dip method (Bechtold and Pelletier 1998). Stable T2 lines were established from representative transformants for each suppressor, based on mRNA expression levels (see Supplemental Data 1), and crossed to the various GFP-silenced lines. The GFP 171.1 and GFP 171.14 constructs were introduced into WT and mutant plants (all in the Col-0 background) using the floral dip method (Bechtold and Pelletier 1998). All single and combination silencing mutants were based on dcl2-1, dcl3-1, dcl4-2, rdr6-15, and dcl2-dcl4 as described previously (Deleris et al. 2006). Plants were germinated under selective media in standard growth chamber conditions for 3 wk and subsequently transferred into soil under short-days conditions before analyses.
Analysis of nucleic acids
Total RNA from mature leaves was extracted using Trizol reagent (Invitrogen), precipitated with 100% isopropanol, and redissolved in 50% formamide. Northern analyses of low- and high-molecular-weight RNAs were performed with 10 μg of total RNA, as described previously (Dunoyer et al. 2004). DNA radiolabeled probes used for detection of the different portion of the GFP RNA or the PVX RNAs were made by random priming reactions (Promega) in the presence of [α-32P]dCTP. Trans-acting siRNA and miRNA probes were complementary DNA oligonucleotides end-labeled with [γ-32P]ATP using the T4 polynucleotide kinase (New England Biolabs). An LNA probe (Exiqon) was used to detect TAS3 tasiRNA5′D7 as described in Allen et al. (2005). Ethidium bromide staining of total RNA before transfer, or detection of U6 snRNA after transfer, was used to confirm equal loading. Sequences of the primers used to produce the PCR fragments analyzed in Figure 4F are as follows, from 5′ to 3′ fragments:
GFP1: 59-AGTAAAGGAGAAGAACTTTTCACT-39;
GFP2: 59-GTATCTTGAAAAGCATTGAACAC-39;
GFP3: 59-GTGTTCAATGCTTTTCAAGATAC-39;
GFP4: 59-TTCCGTCCTCCTTGAAATCGA-39;
GFP5: 59-ACATCCTCGGCCACAAGTTG-39;
GFP6: 59-GGTAAAAGGACAGGGCCATC-39;
GFP7: 59-GATGGCCCTGTCCTTTTACC-39;
miR171:1REV: 59-CGCGGTTATAGTTAAATC-39;
miR171:2FWD: 59-GGCTCATCAGGATCCTTA-39;
35ST:REV: 59-ACTCACACATTATTTATGGA-59:
Quantitative RT-PCR analyses
Reactions were performed as described previously (Dunoyer et al. 2005), and involved 1 μg of total RNA. DNase I and reverse transcription reactions (Superscript; Promega) were performed according to the manufacturer's instructions. Actin-specific amplifications were used to adjust the amount of cDNA within each sample. Primer sequences used to amplify the ARF3 transcript (At2g33860) were as follows:
ARF3-FWD: 5′-GCCACAGACTTTGAGGAATCA-3′;
ARF3-REV: 5′-GGTTATTGAGTAGGCGGGAAC-3′.
GFP171.1 and GFP171.14 plasmid constructs
Construct GFP171.1 was described in Parizotto et al. (2004). To engineer the GFP171.14, the ER-mGFP5 cDNA was PCR-amplified with forward and reverse primers carrying the BamHI restriction site. Reverse primers contained the miR171 target sequence carrying three mismatches in its 5′-end. Primer sequences are available upon request. PCR products were mobilized into BamHI-linearized pBin61 binary vector (Parizotto et al. 2004). The integrity of the resulting plasmids was confirmed by sequencing before transformation into Agrobacterium strain GV3101. Pictures were taken under a Nikon SMZ15000 dissecting microscope coupled to a 100W epi-fluorescence module.
SUPPLEMENTAL DATA
All Supplemental Materials are available upon request to Olivier Voinnet; e-mail: olivier.voinnet@ibmp-ulp.u-strasbg.fr.
ACKNOWLEDGMENTS
This work was supported by a Ph.D. grant from the French Ministry of Research to G.M., and a post-doctoral fellowship from the Centre National de la Recherche Scientifique to E.A.P. We thank members of the Voinnet Laboratory for critical reading of the manuscript and R. Wagner's team for plant care.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.541307.
REFERENCES
- Allen, E., Xie, Z., Gustafson, A.M., Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., Jan, C., Rajagopalan, R., Bartel, D.P. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Pelletier, G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Blevins, T., Rajeswaran, R., Shivaprasad, P.V., Beknazariants, D., Si-Ammour, A., Park, H.S., Vazquez, F., Robertson, D., Meins F., Jr, Hohn, T., et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche, N., Lauressergues, D., Gasciolli, V., Vaucheret, H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, E.J., Prokhnevsky, A.I., Gopinath, K., Dolja, V.V., Carrington, J.C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes & Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., Macino, G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A.J., Mueller, E., Baulcombe, D.C. Potato virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell. 2000a;12:369–379. doi: 10.1105/tpc.12.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A.J., Rudd, S., Angell, S., Baulcombe, D.C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000b;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Deleris, A., Gallego-Bartolome, J., Bao, J., Kasschau, K.D., Carrington, J.C., Voinnet, O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Lecellier, C.H., Parizotto, E.A., Himber, C., Voinnet, O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunoyer, P., Himber, C., Voinnet, O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Himber, C., Voinnet, O. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 2006;38:258–263. doi: 10.1038/ng1722. [DOI] [PubMed] [Google Scholar]
- Ekwall, K. The RITS complex—A direct link between small RNA and heterochromatin. Mol. Cell. 2004;13:304–305. doi: 10.1016/s1097-2765(04)00057-7. [DOI] [PubMed] [Google Scholar]
- Fusaro, A.F., Matthew, L., Smith, N.A., Curtin, S.J., Dedic-Hagan, J., Ellacott, G.A., Watson, J.M., Wang, M.B., Brosnan, C., Carroll, B.J., et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani, S., Lawerson, T., Woodward, D., Headon, R., Sablowski, R. A link between mRNA turnover and RNA interference in Arabidopsis . Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Lakatos, L., Szittya, G., Silhavy, D., Burgyan, J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos, L., Csorba, T., Pantaleo, V., Chapman, E.J., Carrington, J.C., Liu, Y.P., Dolja, V.V., Calvino, L.F., Lopez-Moya, J.J., Burgyan, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Ding, S.W. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., Carrington, J.C. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Parks, G., Endres, M.W., Baulcombe, D., Bowman, L.H., Pruss, G.J., Vance, V.B. The amplicon-plus system for high-level expression of transgenes in plants. Nat. Biotechnol. 2002;20:622–625. doi: 10.1038/nbt0602-622. [DOI] [PubMed] [Google Scholar]
- Molnar, A., Csorba, T., Lakatos, L., Varallyay, E., Lacomme, C., Burgyan, J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.-B., Jouette, D., Lacombe, A.-M., Nikic, S., Picault, N., et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Muangsan, N., Beclin, C., Vaucheret, H., Robertson, D. Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J. 2004;38:1004–1014. doi: 10.1111/j.1365-313X.2004.02103.x. [DOI] [PubMed] [Google Scholar]
- Pak, J., Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Pantaleo, V., Szittya, G., Burgyan, J. Molecular bases of viral RNA targeting by viral siRNA programmed RISC. J. Virol. 2007;81:3797–3806. doi: 10.1128/JVI.02383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizotto, E.A., Dunoyer, P., Rahm, N., Himber, C., Voinnet, O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes & Dev. 2004;18:2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis . Genes & Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F., Ye, X., Hou, G., Sato, S., Clemente, T.E., Morris, T.J. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana . J. Virol. 2005;79:15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach, F., Vaistij, F.E., Jones, L., Baulcombe, D.C. An RNA-dependent RNA polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen, T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H.A., Fire, A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Steiner, F.A., Thijssen, K.L., Plasterk, R.H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Smardon, A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N., Maine, E.M. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans . Curr. Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Vaistij, F.E., Jones, L., Baulcombe, D.C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., Crete, P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., Baulcombe, D.C. Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Lederer, C., Baulcombe, D.C. A viral movement protein prevents systemic spread of the gene silencing signal in Nicotiana benthamiana . Cell. 2000;103:157–167. doi: 10.1016/s0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M., Krczal, G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., Poethig, R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis . Genes & Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui, M., Irvine, D.V., Martienssen, R.A. Non-coding RNA and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]