Abstract
RNA interference (RNAi) is gene silencing induced by double-stranded RNA of 21–23 nucleotides in length, termed small interfering RNA, or siRNA. RNAi-based techniques have been widely applied to elucidate gene function, identify drug targets, and used in trials as a promising adjunct to silence disease-causing genes. However, emerging evidence suggests unexpected changes in expression of untargeted genes as a consequence of an off-target effect by RNAi in mammalian cells. To date, our understanding of such effects on stem cells is limited. We transfected human fetal femur-derived mesenchymal stem cells using commercially available nonspecific siRNA controls and examined adipocyte differentiation in the cells using morphology, histochemistry, and quantitative real-time PCR to examine the expression of key genes for adipogenic or osteogenic differentiation. We report here the induction of adipocyte differentiation in human mesenchymal stem cells using nonspecific siRNAs raising concerns as to the specificity of RNAi in stem cells and, critically, a need to understand and delineate the rules governing the specificity of RNAi.
Keywords: small interfering RNA, RNA interference, human mesenchymal stem cells, adipocyte differentiation
INTRODUCTION
RNA interference (RNAi) is a potentially powerful method to induce gene silencing using double-stranded RNA (Fire et al. 1998). RNAi is triggered by double-stranded RNA of 21–23 nucleotides (nt) in length, termed small interfering RNA, or siRNA (Zamore et al. 2000). This approach has been widely used to investigate gene function, identify drug targets, and in trials as a promising adjunct to silence disease-causing genes and with the potential therein for the development of specific therapeutics for diseases such as HIV infection (Brummelkamp et al. 2002; Novina et al. 2002).
Stem cells have concomitant potential for self-renewal and capacity for differentiation into specific cell lineages. Mesenchymal stem cells can give rise to cells of the adipogenic, reticular, osteoblastic, myoblastic, and fibroblastic lineages. Due to the developmental plasticity of mesenchymal stem cells there is tremendous interest in their application to replace damaged tissues (Oreffo et al. 2005; Mirmalek-Sani et al. 2006). These progenitor populations offer compelling strategies to replace or restore the function of traumatized, diseased, or lost tissue. siRNAs and RNAi-based techniques have been used to study gene function or to modulate stem cell differentiation in human or animal stem cells (Yang et al. 2001; Hoelters et al. 2005; Hong et al. 2005; Xu et al. 2006).
However, emerging evidence suggests chemically synthetic siRNAs or alternative RNAi-based approaches can lead to unexpected changes in expression of untargeted genes in mammalian cells (Jackson et al. 2003; Couzin 2004; Fedorov et al. 2006). This phenomenon, referred to as the off-target effect by RNAi (Kulkarni et al. 2006), can be caused by either the presence of a cross-hybridizing region in nontargeted mRNAs to the siRNA trigger (Jackson et al. 2003), or translational silencing of unrelated transcripts by siRNA acting as microRNA (Birmingham et al. 2006) or inducing common, nonspecific interferon response (Marques and Williams 2005). Although RNAi-induced gene expression has been observed in mammalian cells, to date, the specificity of action of RNAi in human mesenchymal stem cells remains unknown. We report here the induction of adipocyte differentiation (adipogenesis) in human fetal femur-derived mesenchymal stem cells (fetal MSCs) using commercially available nonspecific siRNA controls with limited sequence homology with known genes in the human, mouse, or rat genomes. These nonspecific siRNAs have been used as negative siRNA controls in a number of studies (Chen and Currie 2006; Rubin et al. 2006; Tu et al. 2006; Ueyama et al. 2006). The current results raise concerns as to the specificity of RNAi in mesenchymal stem cells.
RESULTS
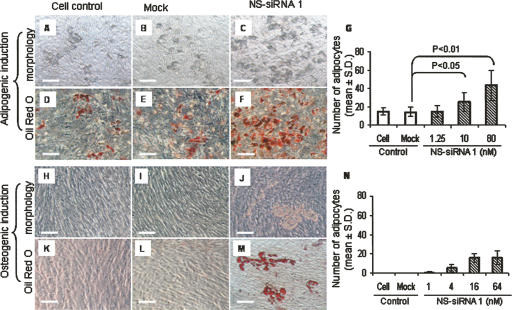
We observed increased adipocyte formation in human preadipocytes transfected with a commercially available siRNA controls (Silencer Negative control #1 siRNA, Ambion) in contrast to cells subjected to mock transfection (Xu et al. 2006). Referred to as nonspecific siRNA control 1, transfection with the control siRNA resulted in significantly increased adipocyte formation in fetal MSCs in contrast to cell or mock transfection controls (Fig. 1). Enhancement of adipogenesis was observed in cells under adipogenic (Fig. 1C,F) or osteogenic conditions (Fig. 1J,M). The enhancement of adipogenesis was dose-dependent under adipogenic (Fig. 1G) or osteogenic conditions (Fig. 1N). Adipocyte formation was not observed in fetal MSCs under basal medium culture as shown in our previous report (Xu et al. 2006). Modulation of fetal MSC osteogenic activity by the nonspecific siRNA control was not observed using alkaline phosphatase staining (date not shown). Similarly, a second siRNA control from Ambion (Silencer Negative control #2 siRNA) showed enhancement of adipogenesis in fetal MSCs, although to a less extent (data not shown).
FIGURE 1.
Adipocyte differentiation induced using nonspecific siRNA control in fetal human femur-derived mesenchymal stem cells (fetal MSCs) under adipogenic (A–G) or osteogenic (H–N) conditions. Cell controls: Cells were grown in adipogenic (A,D) or osteogenic (H,K) conditions. Mock transfection controls: Cells were transfected using Lipofectamine 2000 without siRNA followed by adipogenic (B,E) or osteogenic induction (I,L). NS-siRNA 1: Cells were transfected with the nonspecific siRNA control 1 followed by adipogenic (C,F) or osteogenic induction (J,M). Morphology (A–C,H–J) and Oil Red O staining (D–F,K–M) images were taken from cells following 9 d of adipogenic or osteogenic inductions. Images C, F, J, and M were taken from cells transfected with the nonspecific siRNA at 16 nM. Scale bars = 50 μm. (G,N) Quantification of adipocytes. Adipocytes were identified by accumulation of lipid droplets and staining for lipid with Oil Red O under phase microscopy (10× magnification) in cells following 7 d of adipogenic (G) or osteogenic (N) inductions. Adipocytes were counted from nine randomly selected fields in three wells (three fields a well) for each of treatment group, and data are presented as mean ± standard deviation (SD). The data were taken from a representative study of three experiments using different preparations of fetal MSCs.
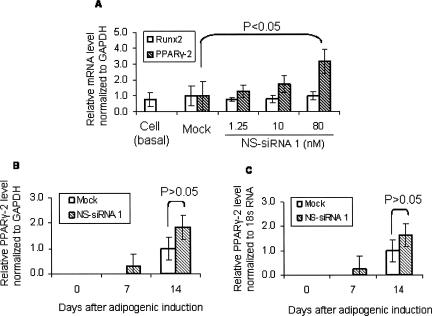
Peroxisome proliferator-activated receptor gamma type 2 (PPARγ-2) is an essential transcription factor for adipogenesis (Saladin et al. 1999) while Runt-related transcription factor 2 (Runx2) is pivotal in osteoblast and chondrocyte differentiation. Suppression of a 14–3–3 binding protein, TAZ (transcriptional coactivator with PDZ-binding motif), using specific siRNA can activate PPARγ-dependent gene transcription and repress Runx2-dependent gene transcription, leading, as a consequence, to the differentiation into adipocytes (Hong et al. 2005). Given the acknowledged roles of PPARγ-2 and Runx2 in the differentiation into adipogenic and osteogenic lineages, we examined expression of the two genes using quantitative real-time PCR (qRT-PCR) in fetal MSCs transfected with the nonspecific siRNA control 1. Glyceraldehydes-3-phosphate dehydrogenase (GAPDH) and 18S ribosomal RNA (18s RNA) were used as endogenous control genes. Runx2, but not PPARγ-2, was detected in fetal MSCs grown in basal cell culture medium (Fig. 2A, Cell [basal]). PPARγ-2 was detected in fetal MSCs under adipogenic induction conditions. Compared to mock transfection controls, transfection with the nonspecific siRNA control 1 promoted expression of PPARγ-2, but not Runx2, in a dose-dependent manner (Fig. 2A), as observed by morphology and histochemistry (Fig. 1). Furthermore, in parallel experiments, we examined the nonspecific siRNA control 1 as well as a specific siRNA against PPARγ (Silencer Validated siRNA ID 5636, Ambion) (Xu et al. 2006) in fetal MSCs and observed enhanced adipogenesis using the nonspecific siRNA while the specific siRNA inhibited adipogenesis (data not shown).
FIGURE 2.
Examining PPARγ-2 and Runx2 mRNA using reverse transcription and qRT-PCR in fetal MSCs. (A) Dose-dependent enhancement of nonspecific siRNA on PPARγ-2, but not Runx2 expression. RNA samples were extracted from cells grown in basal cell culture medium (basal) or cells transfected with the nonspecific siRNA control 1(NS-siRNA 1) at indicated concentrations or without siRNA (Mock), followed by adipogenic induction for 14 d. (B,C) PPARγ-2 expression in cells with mock or NS-siRNA 1 transfection and adipogenic induction for 0, 7, and 14 d, respectively. Each treatment was established in duplicate and each sample was examined in duplicate. PPARγ-2 or Runx2 mRNA levels in cells with mock transfection at day 14 of adipogenic induction (B,C) were taken as 1.0. Relative PPARγ-2 or Runx2 mRNA levels for each treatment are presented as mean ± SD.
DISCUSSION
In the current study we have demonstrated adipocyte differentiation can be induced by transfection with nonspecific siRNA controls used at relatively low concentrations (16 nM) in fetal MSCs under osteogenic induction conditions. It is recognized that siRNAs used at high concentrations (e.g., at 100 nM or higher) can induce nonspecific changes in a large number of common genes in human cell lines (Jackson et al. 2003; Semizarov et al. 2003), and specific siRNA was used by Hong and colleagues at a final concentration of 133 nM with Lipofectamine 2000 as carrier to knock down endogenous target genes in human mesenchymal stem cells (Hong et al. 2005). However, in the current studies, using relatively low concentrations of siRNA, the observed effects of nonspecific siRNAs on adipogenesis appear not to be due to high sensitivity of mesenchymal stem cells to siRNA/lipid mixture, but rather by the nonspecific components.
Global gene expression profiling using microarrays indicate gene expression change in a large number of common genes with limited sequence similarity to siRNA triggers (Jackson et al. 2003) although Brummelkamp and colleagues have shown a siRNA trigger can distinguish one nucleotide mutant between targeted genes (Brummelkamp et al. 2002). In addition, Lin and colleagues reported that a 7-nt motif complementary between a siRNA and an unintended gene can result in degradation of the unintended gene (Lin et al. 2005), supporting a view that partial sequence identity may elicit nonspecific effects in a significant number of currently published siRNA sequences (Snove and Holen 2004). It is possible that siRNAs may have a role as microRNAs in the 3′-UTR of unintended transcriptions (Birmingham et al. 2006; Jackson et al. 2006b). A number of studies indicate RNAi vectors can trigger an interferon response in mammalian cells (Bridge et al. 2003; Pebernard and Iggo 2004). Thus, some motifs, such as the 4-nt “UGGC,” in the RISC-entering strand of siRNAs, have been suggested to be relevant to the toxicity of siRNAs (Fedorov et al. 2006). Similarly, a 7-nt motif “AGGCAGT” is found in the 3′-untranslated region of untargeted genes in which expression is suppressed by the off-target effect of RNAi (Lin et al. 2005). In the current study commercial sensitivity precludes sequence assessment for the degree of nucleotide contiguity between the nonspecific siRNA controls used and human genes, if any.
RNAi has been widely used to study gene function and trialed as a promising adjunct to silence disease-causing genes with the potential therein for the development of specific therapeutics for a number of diseases. Our data suggest a need for caution and, critically, a need to understand and delineate the rules governing the specificity of RNAi. This will be particularly relevant where RNAi-based techniques are used in therapeutic regimes or in stem cells, in which cell differentiation is tightly controlled by a network of biological pathways and subject to exquisite regulation by transcription factors. Nevertheless, off-target search algorithms for siRNA are under development (Snove and Holen 2004; Naito et al. 2005), and chemically modifying sequences, such as 2′-O-methyl modification, to specific positions within siRNAs offer new approaches to reduce what are clearly evident off-target risks of RNAi (Fedorov et al. 2006; Jackson et al. 2006a). RNAi is a powerful and compelling new strategy to elucidate gene function, but such strategies require an awareness of the potential off-target activity of RNAi and a need for new paradigms to harness the full potential of this molecular approach.
MATERIALS AND METHODS
Fetal MSCs
Fetal MSCs were isolated from human fetal femurs, according to guidelines issued by the Polkinghome Report with ethical approval from the Southampton & South West Hampshire Local Research Ethics Committee. Cells were grown in Minimum Essential Medium, Alpha-Modification (α-MEM) containing 10% fetal calf serum (FCS) and 100 units/mL penicillin and 100 μg/mL streptomycin (basal cell culture medium) at 37°C, 5% CO2 in a humidified atmosphere. Cell cultures, at 80% confluence, were detached with trypsin-EDTA and plated into six-well plates for siRNA transfection/adipogenic or osteogenic induction at passage 2 only. Fetal MSCs were induced to differentiate into adipogenic, osteogenic cells under specific culture conditions.
siRNA transfection
Cells plated in six-well plates for 24 h previously became about 50% confluent and were transfected with nonspecific or specific siRNAs at stated concentrations after replacing basal cell culture medium with 2 mL of opti-MEM I reduced serum Medium (Invitrogen Life Technologies) per well. Lipofectamine 2000 (Invitrogen Life Technologies) was used as a carrier at a final concentration of 0.17% (v/v) in a total transfection volume, 2.4 mL/well. Eight hours following addition of siRNA/lipid mixture into cell culture, 2.4 mL of 20% CFS basic cell culture medium/well were added into the cell culture to give a final concentration of 10% CFS in transfection medium. Treatments were established in triplicate. Cells were cultured for an additional 2 d for adipogenic or osteogenic induction.
Adipogenic or osteogenic induction
Adipogenic and osteogenic induction were performed as previously detailed with minor modifications (Mirmalek-Sani et al. 2006). Adipogenic culture: Cells were grown for 3 d in basal cell culture medium containing additional 2 g/L D-glucose and 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone, 1% ITS solution (equal to 10 μg/mL insulin, Sigma-Aldrich), and 100 μM indomethacin, followed by 1 d in basal cell culture medium containing 1% ITS solution. Both cycles were repeated and cell cultures were stopped at day 9 or 14, respectively, for histological staining or total RNA extraction. Osteogenic culture: Cells were maintained in basal cell culture medium containing 100 μM ascorbic acid 2-phosphate and 10 nM dexamethasone and medium changed every 2 d. Osteogenic cultures were stopped after 7 d.
Identification of adipocytes and histological analysis
Adipocytes were identified by accumulation of lipid droplets observed under light microscopy using a Zeiss microscope and following staining for lipid with Oil Red O histochemistry (Mirmalek-Sani et al. 2006). In brief, cells were fixed in Baker's formal calcium, rinsed in 60% isopropanol, and stained using double-filtered Oil Red O solution. Quantification of adipocytes was performed by counting adipocytes (at least five lipid droplets observed in a cell) from nine randomly selected fields (three fields/well) for each treatment or control group. Mean adipocyte numbers were calculated for each group. Osteogenic activity was determined using alkaline phosphatase histochemical staining, demonstrated with Naphthol AS-MX Phosphate and Fast Violet B Salts (Mirmalek-Sani et al. 2006).
Total RNA extraction
Total RNA was extracted using TRI reagent (Sigma) and purified using DNA-free RNA Kit (ZYMO Research) according to the manufacturer's instructions. RNA was dissolved in nuclease-free water and measured using RNA 6000 Nano Assay (Lab-on-a-chip technology, Agilent Technologies). RNA samples were stored at −80°C for reverse transcription and qRT-PCR analysis.
Reverse transcription and qRT-PCR
cDNA synthesis was performed in a total volume of 20 μL containing 125 ng of denatured total RNA, 0.5 mM dNTPs, 25 μg/mL random primers, 1 unit/μL RNasin ribonuclease inhibitor, 10 units/μL M-MLV reverse transcriptase, and 1× M-MLV buffer (Promega). The reaction was performed by incubating at 42°C for 60 min followed by inactivating reverse transcriptase at 95°C for 5 min. GAPDH and 18s RNA were used as endogenous control genes to analyze qRT-PCR data of PPARγ-2 and Runx2. For PPARγ-2, the forward primer was 5′-ACTCTGGGAGATTCTCCTATT-3′ and the reverse primer was 5′-CTCCATAGTGAAATCCAGAAG-3′, used at 500 nM; for Runx2, the forward primer was 5′-GCCTTCAAGGTGGTAGCCC-3′ and the reverse primer was 5′-CGTTACCCGCCATGACAGTA-3′, used at 500 nM; for 18s RNA, the forward primer was 5′-GCCCTGTAATTGGAATGAGTC-3′ and the reverse primer was 5′-TCGCTCTGGTCCGTCTTG-3′, used at 100 nM; and for GAPDH, the forward primer was 5′-CCAGGTGGTCTCCTCTGACTTC-3′ and the reverse primer was 5′-TCATACCAGGAAATGAGCTTGACA-3′ (the primers of GAPDH were designed by Dr. K. Hashimoto, University of Southampton), used at 300 nM. PCR was performed using a 96-Well Optical Reaction Plate and a 7500 Real Time PCR system (Applied Biosystems). For each PCR run, 1 μL of 1:4 diluted cDNA product was mixed with 1× Power SYBR Green PCR Master Mix and PCR primers in total volume of 50 μL. The thermal cycling conditions were comprised of an initial stage of 50°C for 2 min and 95°C for 10 min, 40 cycles of melting at 95°C for 15 sec, and annealing at 60°C for 1 min, and a dissociation stage of 95°C for 15 sec, 60°C for 1 min, and 95°C for 15 sec. Threshold cycle (CT) values for specific and control genes were determined automatically using the 7500 system software. qRT-PCR data were analyzed using the 2-ΔΔCT Method (Livak and Schmittgen 2001). The relative level of a specific mRNA (PPARγ-2 or Runx2) was calculated by subtracting CT values of the control gene (GAPDH or 18s RNA) from the CT values of specific gene (PPARγ-2 or Runx2). PPARγ-2 or Runx2 mRNA levels in mock transfection controls at stated time points were taken as 1.0 in individual experiments.
Statistical analysis
Data collected from adipocyte analysis and qRT-PCR were analyzed using the Student's t-test. P < 0.05 was considered as statistically significant.
ACKNOWLEDGMENTS
We thank Professors Neil Hanley and David Wilson for provision of fetal samples. We thank the BBSRC for Ph.D. studentship funding to S.-H.M.-S. and research support for Y.X.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.527207.
REFERENCES
- Birmingham, A., Anderson, E.M., Reynolds, A., Ilsley-Tyree, D., Leake, D., Fedorov, Y., Baskerville, S., Maksimova, E., Robinson, K., Karpilow, J., et al. 3′-UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Bridge, A.J., Pebernard, S., Ducraux, A., Nicoulaz, A.L., Iggo, R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., Agami, R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Currie, R.W. Small interfering RNA knocks down heat shock factor-1 (HSF-1) and exacerbates pro-inflammatory activation of NF-κB and AP-1 in vascular smooth muscle cells. Cardiovasc. Res. 2006;69:66–75. doi: 10.1016/j.cardiores.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Couzin, J. Molecular biology. RNAi shows cracks in its armor. Science. 2004;306:1124–1125. doi: 10.1126/science.306.5699.1124. [DOI] [PubMed] [Google Scholar]
- Fedorov, Y., Anderson, E.M., Birmingham, A., Reynolds, A., Karpilow, J., Robinson, K., Leake, D., Marshall, W.S., Khvorova, A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hoelters, J., Ciccarella, M., Drechsel, M., Geissler, C., Gulkan, H., Bocker, W., Schieker, M., Jochum, M., Neth, P. Nonviral genetic modification mediates effective transgene expression and functional RNA interference in human mesenchymal stem cells. J. Gene Med. 2005;7:718–728. doi: 10.1002/jgm.731. [DOI] [PubMed] [Google Scholar]
- Hong, J.H., Hwang, E.S., McManus, M.T., Amsterdam, A., Tian, Y., Kalmukova, R., Mueller, E., Benjamin, T., Spiegelman, B.M., Sharp, P.A., et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Burchard, J., Leake, D., Reynolds, A., Schelter, J., Guo, J., Johnson, J.M., Lim, L., Karpilow, J., Nichols, K., et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006a;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A.L., Burchard, J., Schelter, J., Chau, B.N., Cleary, M., Lim, L., Linsley, P.S. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006b;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, M.M., Booker, M., Silver, S.J., Friedman, A., Hong, P., Perrimon, N., Mathey-Prevot, B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- Lin, X., Ruan, X., Anderson, M.G., McDowell, J.A., Kroeger, P.E., Fesik, S.W., Shen, Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marques, J.T., Williams, B.R. Activation of the mammalian immune system by siRNAs. Nat. Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- Mirmalek-Sani, S.H., Tare, R.S., Morgan, S.M., Roach, H.I., Wilson, D.I., Hanley, N.A., Oreffo, R.O. Characterization and multipotentiality of human fetal femur-derived cells: Implications for skeletal tissue regeneration. Stem Cells. 2006;24:1042–1053. doi: 10.1634/stemcells.2005-0368. [DOI] [PubMed] [Google Scholar]
- Naito, Y., Yamada, T., Matsumiya, T., Ui-Tei, K., Saigo, K., Morishita, S. dsCheck: Highly sensitive off-target search software for double-stranded RNA-mediated RNA interference. Nucleic Acids Res. 2005;33:W589–W591. doi: 10.1093/nar/gki419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novina, C.D., Murray, M.F., Dykxhoorn, D.M., Beresford, P.J., Riess, J., Lee, S.K., Collman, R.G., Lieberman, J., Shankar, P., Sharp, P.A. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Oreffo, R.O.C., Cooper, C., Mason, C., Clements, M. Mesenchymal stem cells: Lineage, plasticity, and skeletal therapeutic potential. Stem Cell Rev. 2005;1:169–178. doi: 10.1385/SCR:1:2:169. [DOI] [PubMed] [Google Scholar]
- Pebernard, S., Iggo, R.D. Determinants of interferon-stimulated gene induction by RNAi vectors. Differentiation. 2004;72:103–111. doi: 10.1111/j.1432-0436.2004.07202001.x. [DOI] [PubMed] [Google Scholar]
- Rubin, J., Murphy, T.C., Rahnert, J., Song, H., Nanes, M.S., Greenfield, E.M., Jo, H., Fan, X. Mechanical inhibition of RANKL expression is regulated by H-Ras-GTPase. J. Biol. Chem. 2006;281:1412–1418. doi: 10.1074/jbc.M508639200. [DOI] [PubMed] [Google Scholar]
- Saladin, R., Fajas, L., Dana, S., Halvorsen, Y.D., Auwerx, J., Briggs, M. Differential regulation of peroxisome proliferator activated receptor γ1 (PPARγ1) and PPARγ2 messenger RNA expression in the early stages of adipogenesis. Cell Growth Differ. 1999;10:43–48. [PubMed] [Google Scholar]
- Semizarov, D., Frost, L., Sarthy, A., Kroeger, P., Halbert, D.N., Fesik, S.W. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snove O., Jr, Holen, T. Many commonly used siRNAs risk off-target activity. Biochem. Biophys. Res. Commun. 2004;319:256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
- Tu, Z., Prajapati, S., Park, K.J., Kelly, N.J., Yamamoto, Y., Gaynor, R.B. IKKα regulates estrogen-induced cell cycle progression by modulating E2F1 expression. J. Biol. Chem. 2006;281:6699–6706. doi: 10.1074/jbc.M512439200. [DOI] [PubMed] [Google Scholar]
- Ueyama, T., Geiszt, M., Leto, T.L. Involvement of Rac1 in activation of multicomponent Nox1- and Nox3-based NADPH oxidases. Mol. Cell. Biol. 2006;26:2160–2174. doi: 10.1128/MCB.26.6.2160-2174.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Mirmalek-Sani, S.H., Yang, X., Zhang, J., Oreffo, R.O. The use of small interfering RNAs to inhibit adipocyte differentiation in human preadipocytes and fetal-femur-derived mesenchymal cells. Exp. Cell Res. 2006;312:1856–1864. doi: 10.1016/j.yexcr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Yang, S., Tutton, S., Pierce, E., Yoon, K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., Bartel, D.P. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]