Abstract
Alternative splicing is widely believed to have a major impact on almost all biological processes since it increases proteome complexity and thereby controls protein function. Recently, gene targeting in mice has been used to create in vivo models to study the regulation and consequences of alternative splicing. The evidence accumulated so far argues for a nonredundant, highly specific role of individual splicing factors in mammalian development, and furthermore, demonstrates the importance of distinct protein isoforms in vivo. In this review, we will compare phenotypes of mouse models for alternative splicing to crystallize common themes and to put them into perspective with the available in vitro data.
Keywords: alternative splicing, mouse models
INTRODUCTION TO ALTERNATIVE SPLICING
The recent sequencing of several genomes has enabled an estimation of the number of protein-coding genes in different organisms. The difference in this number is much smaller than expected when comparing higher vertebrates to flies or worms. Therefore, it has been proposed that other mechanisms must exist that significantly contribute to proteome diversity in higher organisms (Maniatis and Tasic 2002; Blencowe 2006). One of the mechanisms that has gained much attention due to its ability to both increase proteome diversity and regulate protein function is alternative splicing. This process is controlled by sequence elements within exons or introns that can be occupied by regulatory proteins that either enhance or inhibit the assembly of the spliceosome. Alternative splicing can generate different isoforms of one single protein with different enzymatic activity, substrate specificity, subcellular localization, or an altered ability to interact with other proteins or DNA. This underlines that alternative splicing not only contributes to proteome diversity, but also has a regulatory function in many physiological processes such as apoptosis or cell-cycle progression, and is critical for the immune response, organ development, and in malignant transformation (Lynch 2004; Venables 2004; Schwerk and Schulze-Osthoff 2005; Srebrow and Kornblihtt 2006) and others, as will be discussed here. As a number of recent reviews cover the splicing process and its regulation in detail (e.g., Black 2003; Matlin et al. 2005), these topics will not be the focus of the present article.
A substantial amount (around 15%) of point mutations in the human genome that are related to diseases is believed to alter the functions of proteins by affecting the process of alternative splicing (Krawczak et al. 1992; Cartegni et al. 2002; Faustino and Cooper 2003). Although this clearly demonstrates the importance of understanding alternative splicing and its impact on an organism as a whole, most research on the regulation of alternative splicing has been carried out using in vitro assays or cell culture-based systems. Only more recently, gene targeting in mice has advanced our knowledge about the biological function of splicing factors, alternative splicing, and the importance of specific protein isoforms generated by alternative splicing in vivo. In this review, we discuss how this new approach using mouse models has influenced the research on alternative splicing and its perception as a fundamental biological process.
SEVERAL WAYS TO INVESTIGATE ALTERNATIVE SPLICING IN VIVO
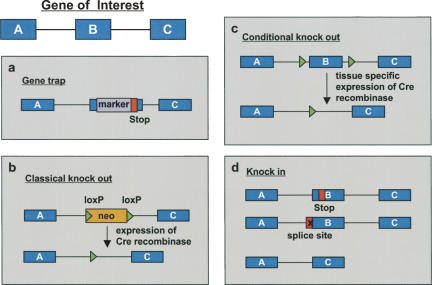
Different approaches have been taken to analyze the impact of alternative splicing in mice, which can be put into two groups: either the activity of a splicing factor is manipulated by its deletion, overexpression, or expression of a dominant-negative mutant, or a certain splice-site choice is manipulated by permanently altering the splice site or by administration of oligonucleotides that redirect the splicing process (Fig. 1; Table 1). The first possibility, the deletion or overexpression of the gene of interest, has been performed for the splicing factors Nova-1, SRp20, ASF/SF2, SC35, hnRNP C, hnRNP U, CUG-BP1, a dominant-negative CELF protein, Brunol1, and U2AF26 (the term splicing factor is used here in a broad sense for RNA-binding proteins even if their main function appears to be regulation of RNA stability or translation, and therefore includes members of the hnRNP family) (Table 2). The analysis of most knockout mouse models is impeded by the finding that they either arrest their development at midgestation or die perinatally. A more thorough analysis therefore requires a conditional gene-targeting strategy and a cell type or tissue-specific deletion of the gene, which has been performed so far only for ASF/SF2 and SC35. The deletion of a splicing factor allows one to identify target genes whose splicing pattern is changed in the mutant mouse, but it also enables one to estimate the contribution of this splicing factor to the development of the whole animal or the formation of specific tissues. In addition, such a model provides the possibility of phenotypically defining cellular or molecular defects provoked by the lack of this factor and to recognize a potential link to a human disease. However, it may be a challenging task to make a connection between a missing splicing factor, an altered splicing pattern of a variety of target genes, and the observed phenotype on a molecular level.
FIGURE 1.
Strategies that have been used to create splicing factor or isoform-specific knockout mice. Three exons (A–C) of the gene of interest are schematically shown as filled boxes (blue). (a) Large-scale random retroviral insertion of marker genes in the genome of murine ES cells, and subsequent sequencing of the integration site has been used to create libraries of targeted ES cells that are commercially available. Using this approach, stop codons, loxP sites, and other alterations can be introduced along with the marker gene into the gene of interest. (b) In a classical knockout approach, the whole gene, or a part thereof, is replaced by a selectable marker, e.g., a neomycin-resistance gene. To avoid potential effects of the neo-cassette, a version flanked by loxP sites can be used that can be removed by expression of the site-specific Cre recombinase. If only parts of the gene of interest are removed, as shown here, the expression of a truncated protein should be taken into consideration. (c) In a conditional knockout approach, the gene of interest is replaced by a version that carries flanking loxP sites (floxed) that can be removed by tissue-specific expression of Cre recombinase. Introduction of the floxed allele also requires the presence of a (removable) selectable marker (data not shown). (d) Knockin mutants are used to replace parts of the gene of interest with an altered version. For example, a stop codon can be introduced to abrogate expression of all isoforms carrying exon B, but allowing expression of A-C isoforms. A similar technique, also resulting in a depletion of isoforms containing exon B, is the destruction of the splice sites of the respective exon. Finally, one exon can be replaced by another exon or an unrelated coding region, e.g., to obtain the expression of an isoform in a tissue, where it is usually not expressed. This technique requires careful consideration of the targeting construct, as exonic regulatory sequences of the deleted exon might be important for tissue-specific splicing of the gene.
TABLE 1.
Comparison of two kinds of mouse models and their use in alternative splicing research
TABLE 2.
Mouse mutants with targeted deletions or transgenic overexpression of splicing factors
An alternative way to investigate the impact of alternative splicing in a mouse model is the selective deletion of a single isoform of a particular protein by either deleting the whole coding exon from the genome, introducing a premature Stop codon, inactivating the splice sites responsible for generating the specific isoform, or simply overexpressing a certain isoform. Either of these techniques has been used to generate mice deficient in specific isoforms of Fibronectin, Integrin, Agrin, VEGF, FGFR2, Dopamine receptor 2, Na-K-2Cl cotransporter, a Ca2+-ATPase, Plectin, Caldesmon, CD45, Stat3, WT1, and Pax6 (Table 3). The obvious advantage of this approach compared with the full knockout of a splicing factor gene is that a direct link between a specific protein isoform and an observed phenotype can be made.
TABLE 3.
Mouse mutants with Isoform specific deletions
Administration of antisense oligonucleotides has also been used to influence the splicing process, i.e., to prevent inclusion of a particular exon in the mature mRNA. In human diseases, where a certain exon is known to carry a nonsense mutation, the oligonucleotide-mediated exclusion of this exon might be a promising therapeutic approach that has been successfully used in mouse models for muscle dystrophy (Gebski et al. 2003; Williams et al. 2006).
In an ideal experiment, both kinds of mouse models can be combined to first identify in vivo targets of a splicing factor and then use an isoform-specific knockout (or overexpression) of one of the targets to demonstrate that an observed phenotype is due to the altered splicing of this specific target. One example for this elegant experimental approach is the recently demonstrated connection between the splicing factor ASF/SF2, the regulation of CaMKIIδ alternative splicing, and a severe defect in the function of adult cardiac muscles (Xu et al. 2005; see below for details).
In addition to the reverse genetic approaches described above, forward genetic screens in mice have identified splicing factors or proteins associated with RNA metabolism to regulate strain-specific susceptibility to certain diseases (e.g., Buchner et al. 2003; Floyd et al. 2003). This technique represents a complementary method to the knockout approaches described in this review, as it uses a given phenotype as a starting point, which is then linked to a certain genomic locus, and finally to one specific gene.
MICE DEFICIENT IN SPLICING FACTORS OR RELATED PROTEINS
Complete and tissue-specific knockout of SR proteins
Members of the SR protein family are essential for constitutive splicing and fulfill regulatory functions in alternative splicing. The common feature of the SR protein family is an N-terminal RNA-binding domain and a C-terminal RS domain consisting largely of arginine-serine repeats. The RS domain of different family members appears to be functionally interchangeable and synthetic RS repeats are sufficient to promote splicing when tethered to a pre-mRNA substrate (Philipps et al. 2003). In vitro experiments showing that most individual SR proteins are able to complement SR protein-deficient S100 extract further suggested a certain degree of redundancy among SR proteins (Tacke and Manley 1999; Hoffman and Lis 2000). However, the identification of distinct DNA sequences in alternatively spliced exons (splicing enhancers) that are responsive to specific SR proteins challenged the notion of redundancy among SR proteins in vivo (Schaal and Maniatis 1999). Moreover, the lethality caused by loss of an SR protein in Drosophila (Ring and Lis 1994) or in the chicken DT40 cell line (Wang et al. 1996) also argues against a large redundancy of SR proteins. To investigate a potential functional overlap in vivo, several mouse models were generated that lack the SR proteins SRp20, SC35, or ASF/SF2. The first model to be published was an SRp20-deficient mouse, which was generated using the loxP–Cre strategy (Jumaa et al. 1999). Unfortunately, a thorough analysis of the mutant animals was hindered by an early embryonic lethality (day 3.5), and a tissue-specific knockout was not performed. Embryos appeared normal until day 2.5 and failed to develop further, which could point either to the initial presence of maternally supplied protein, which is consumed during the first cell divisions, or to a role of SRp20 in blastocyst formation, which takes place between days 2.5 and 3.5. While strongly arguing for an essential, nonredundant role of SRp20 in the early embryonic development, this phenotype also exemplifies the difficulties that can be encountered when a complete knockout rather than a tissue-specific approach is chosen.
Similarly, deletion of two other SR proteins in the germ line of mice, ASF/SF2 or SC35, led to embryonic lethality before day 7.5, which would have impeded further analysis (Wang et al. 2001; Xu et al. 2005). However, these two models were generated using conditional gene-targeting approaches, enabling analysis of animals with tissue-specific deletion of the respective genes. Technical aspects of the generation of conditional knockout mice have been covered by recent reviews (Branda and Dymecki 2004; Xu and Fu 2005), and will therefore not be discussed in detail here. In brief, the gene of interest (or parts of it) is flanked by recognition sites of a site-specific recombinase, e.g., loxP sites, using homologous recombination in ES cells that are then used to generate mice. These mice carrying the floxed allele are further crossed to mice that express the respective recombinase, e.g., Cre recombinase, under the control of a tissue-specific promoter, in order to allow deletion of the targeted allele only in this defined cell type. A large variety of tissue-specific Cre-expressing mice is available, and this technique was used for the deletion of SC35 or ASF/SF2 in heart tissue and SC35 in T cells (Wang et al. 2001; Ding et al. 2004; Xu et al. 2005). Both tissues are interesting targets, since they are well characterized and are functionally dependent on alternative splicing events, some of which have been investigated using cell culture or in vitro systems (e.g., Lynch and Weiss 2000; Ladd et al 2005a). Furthermore, adult cardiomyocytes are mainly nondividing cells representing a valuable model for defects not associated with cell growth.
Using tissue-specific deletion, a striking phenotype was found in the heart of ASF/SF2 knockout mice. Although the mice were born at a Mendelian ratio and started to develop normally, they died at around 6–8 wk after birth due to heart failure. A dilated cardiomyopathy was revealed, in which the heart becomes enlarged and weakened and fails to maintain proper blood circulation. In addition, these mice had a disturbed Ca2+ metabolism starting at about 4 wk of age, which worsened throughout the remaining lifespan, ultimately causing failure of the contractile apparatus. The delayed onset of the disease and the absence of increased apoptosis in ASF/SF2-deficient cardiomyocytes suggested that ASF/SF2 is not essential for cell viability, but might be critical in the transition from the juvenile to an adult heart. Indeed, a molecular analysis demonstrated a failure to induce a switch in the expression from the Ca2+/calmodulin-dependent kinase IIδ (CaMKIIdδ) neuronal isoform A (excluding exon 14, including exons 15 and 16) to isoform B (including exon 14, excluding exons 15 and 16), which normally accompanies adult heart formation. Intriguingly, the transgenic overexpression of the CaMKIIδ isoform A in the heart mimicked the phenotype of the ASF/SF2 deficiency, demonstrating that one particular splicing defect in the ASF/SF2-deficient mice is sufficient to cause heart failure (Xu et al. 2005). However, it remains to be shown whether the effect of ASF/SF2 on CaMKIIδ is direct or mediated by yet another protein.
A more thorough analysis to detect altered alternative splicing in ASF/SF2-deficient cardiomyocytes showed that the splicing pattern of most examined genes was unchanged when compared with wild type. A splicing defect in three genes was observed, but it was evident 20 d postnatal at the earliest, whereas their splicing pattern in wild-type and mutant mice was indistinguishable at earlier time points after birth. It is also worth noting that complete loss of ASF/SF2 led to embryonic lethality, whereas embryonic heart development was not impaired by tissue-specific knockout, which points to a cell type-dependent role of ASF/SF2. These data open up the interesting possibility of ASF/SF2 being only strictly required during developmentally programmed changes in the alternative splicing pattern of very few target genes in distinct cell types, whereas it is negligible for other alternative splicing events and constitutive splicing. This conclusion is further supported by analysis of ASF/SF2-deficient ventricles through expression microarrays, which showed only a few genes to be down-regulated, implicating that nonsense-mediated decay (NMD) is not triggered in the mutant hearts. As a substantial percentage, up to one-third of alternative splicing events has been predicted to result in the inclusion of premature Stop codons causing NMD (Lewis et al. 2003); the absence of a larger number of down-regulated genes suggests that the lack of ASF/SF2 does not affect the global pattern of alternative splicing. This idea is in agreement with earlier studies showing that loss of ASF/SF2 or SRp55 in cell culture or Drosophila only interferes with gene expression or splicing of a very limited amount of targets (Lemaire et al. 2002; Kim et al. 2003).
Another SR protein that was also specifically deleted in the heart is SC35. As for the ASF/SF2 knockout, cardiomyocytes showed rates of proliferation and apoptosis indistinguishable from wild type, suggesting that SC35 is not strictly required for cell survival and proliferation. In contrast to the early postnatal lethality caused by heart-specific deletion of ASF/SF2, mice lacking SC35 expression in the heart showed normal vitality. However, with increasing age, these mice displayed signs of dilated cardiomyopathy, as evidenced by functional and morphological defects. Since no splicing defects in the CaMKIIδ gene could be detected in SC35-deficient cardiomyocytes, the cause of the disease in SC35 knockout mice must be different from the one in ASF/SF2-deficient cells, which further supports the idea that SR proteins fulfill specialized, nonredundant functions. As described for the ASF/SF2-deficient mice, microarray analysis of SC35 knockout hearts did show only a small number of genes to be down-regulated, suggesting that the global splicing pattern is not changed.
The same floxed SC35 mouse that was used to delete SC35 from heart tissue was also used for the ablation of SC35 from the T cell compartment (Wang et al. 2001). As the maturation and activation of T cells is accompanied by well-defined changes in the expression of cell-surface molecules, each step can be easily followed by flow cytometry, which makes T cells an attractive model system. For example, the early T cell development in the thymus is characterized by the transition from double-negative (DN, CD4−CD8−) to double-positive (DP, CD4+CD8+) cells, which then develop into single positive cells (either CD4+ or CD8+) and leave the thymus to home to peripheral lymphoid organs. The deletion of SC35 in T cell precursors provoked a severe loss of thymic cellularity and furthermore blocked the DN to DP transition of the remaining T cells. Naïve peripheral T cells with homozygous SC35 deletion were not detectable, which might be either due to an absolute requirement of SC35 for T cell development or, alternatively, could point to a role of SC35 in peripheral T cell viability or homing. As for the SC35 deleted heart, apoptosis was not increased in SC35-deficient thymocytes, which might support the first possibility. Indeed, the splicing pattern of the transmembrane tyrosine phosphates CD45, which is known to be critically involved in T cell activation and maturation (Hermiston et al. 2003), was shown to be altered in SC35-deficient T cells, but so far it remains to be shown whether this is due to a direct effect of SC35. The in vivo analysis of different CD45 isoforms will be discussed in the section dealing with isoform-specific knockout mice below.
Complete knockout of hnRNP proteins
The hnRNP family of RNA-binding proteins comprises over 20 members that have been implicated in diverse aspects of RNA metabolism such as regulation of RNA stability, nuclear export, transcriptional control, and regulation of constitutive and alternative splicing (Krecic and Swanson 1999). Given the large structural and functional variety of hnRNP proteins, it appears plausible to assume that each family member fulfills a specialized, nonredundant function. Using gene-trap ES cell lines that carry disrupted hnRNP C or hnRNP U alleles, it was shown that these genes are indeed essential for embryonic development, as the lack of either of the two proteins proved to cause early embryonic lethality. In both cases, embryos developed normally through the egg cylinder stage (day 6.5) but failed to initiate gastrulation (Williamson et al. 2000; Roshon and Ruley 2005). Interestingly, cell lines derived from early hnRNP C-deficient embryos were viable and, to some extent, able to differentiate and displayed no obvious defect in the expression or splicing of marker genes (Williamson et al. 2000). The precise mechanism leading to lethality of hnRNP C-deficient embryos remains to be elucidated, but appears not to be based on a strictly cell-autonomous defect. This underlines the value of mouse models to fully appreciate the impact of RNA-binding proteins on mammalian development.
Complete Nova-1 knockout
Alternative splicing has been shown to occur in a particularly high frequency in the nervous system (Yeo et al. 2004), where many of the genes encoding neurotransmitter receptors and ion channels produce several mRNA isoforms. A number of neuron-specific splicing factors are known, among them the Nova family of RNA-binding proteins, which, together with the differential expression of general splicing factors, are required to induce a neuron-specific splicing pattern. Nova proteins bind RNA in a sequence-specific manner to regulate alternative splicing of diverse targets and are exclusively expressed in the central nervous system (Ule et al. 2005). The essential role of Nova-1 in postnatal neuron survival was demonstrated by generation of Nova-1-deficient mice, which die about 1 wk after birth. As these mice appeared normal at birth, Nova-1 does not seem to be required for prenatal neuronal development, but Nova-1-deficient neurons underwent massive apoptosis by day 5 after birth, finally leading to death (Jensen et al. 2000). Analysis of Nova-1-deficient brains revealed a disturbed splicing pattern of the inhibitory receptors GlyRα2 (Glycine receptor α2) and GABARA (γ amino butyric acid receptor), whereas five other mRNAs that are known to be alternatively spliced in neurons showed an unaltered splicing pattern in the mutant mice. Expression of both GlyRα2 and GABAA have been shown to undergo programmed changes in early postnatal stages (Singer et al. 1998; Jensen et al. 2000), and it appears plausible to assume that Nova-1 is required to regulate these changes, whereas it is not essential for their correct expression during embryogenesis, which parallels the situation described for the ASF/SF2 knockout in the heart. A further parallel is the limited effect of the Nova-1 deficiency with respect to the global splicing pattern. Of seven investigated genes, only two showed alterations in splicing, which again points to a specific effect in regulating alternative splicing and against an essential role of individual splicing factors in controlling the global splicing pattern. Whereas it remains to be shown whether the defects in GlyRα2 and/or GABARA alternative splicing are indeed the cause for the observed lethal phenotype, two lines of evidence suggest that the defect in GlyRα2 alternative splicing is directly linked to the absence of Nova-1. First, the defect was observed only in parts of the brain where Nova-1 is actually expressed, whereas it was not evident in other parts of the brain that do not show Nova-1 expression. Furthermore, UV cross-link and minigene assays confirmed the ability of Nova-1 to bind to and to regulate alternative splicing of the GlyRα2 pre-mRNA in a manner consistent with the splicing pattern provoked by Nova-1 deficiency. These experiments demonstrate well how the combined results of knockout mice and in vitro RNA biochemistry can work together to uncover regulatory mechanisms of alternative splicing.
Knockout of Muscleblind-like 1 (Mbnl1)
The Mbnl family of RNA-binding proteins consists of several members, the most abundant of which, Mbnl1, encodes diverse isoforms generated by alternative splicing (Pascual et al. 2006). Isoforms containing exon 3 of the mbnl1 gene have been shown to bind a (CUG)n expansion in the 3′-UTR of myotonic dystrophy protein kinase (DMPK), causing Myotonic Dystrophy type 1 (DM1), a disease associated with muscle degeneration and nervous system dysfunction (Miller et al. 2000). The mutant DMPK transcripts accumulate in distinct nuclear RNA foci and recruit Mbnl proteins into this subnuclear domain (Mankodi et al. 2001). Transgenic mice carrying a CUG repeat in an unrelated gene also developed DM, directly linking the disease phenotype with the CUG expansion (Mankodi et al. 2000). Adeno-associated virus-mediated overexpression of Mbnl1 in skeletal muscle rescued the disease phenotype in this mouse model, providing a direct link of CUG expansion and Mbnl1 expression in development of DM (Kanadia et al. 2006). In order to investigate whether CUG binding and recruitment of Mbnl1 protein into RNA foci was associated with the onset of DM, a mouse model was generated that lacks exon 3 of the Mbnl1 protein to eliminate all isoforms capable of binding the CUG repeat, thus inactivating the protein. The lack of CUG-binding Mbnl1 isoforms should mimic the disease state, if the recruitment of Mbnl1 into RNA foci leads to inactivation of the protein and is responsible for the onset of DM1. Mice lacking exon 3 of Mbnl1 indeed developed a DM-like disease at around 6 wk of age as assessed by morphological and functional assays (Kanadia et al. 2003). A previously identified protein whose alternative splicing pattern is disturbed by aberrant expression of CUG-containing RNAs in mice and in myotonia patients is the skeletal muscle chloride channel ClC-1 (Mankodi et al. 2002). A similarly disturbed ClC-1 splicing pattern was observed in mice lacking Mbnl1 exon 3, suggesting that exon 3 including Mbnl1 is required for the regulation of ClC-1 alternative splicing. Other genes known to be mis-spliced in myotonic conditions also showed splicing defects in the mutant mice, whereas splicing of unrelated genes was not impaired, further supporting the notion that Mbnl1 proteins participate in the regulation of alternative splicing of defined target genes (Kanadia et al. 2003). In vitro and cell culture-based assays were used to confirm a direct involvement of Mbnl proteins in some of these splicing events (Ho et al. 2004). Interestingly, splicing defects that were observed in mice deficient in Mbnl1 exon 3 were also evident in transgenic mice carrying a CUG repeat, but not in mice deficient in Mbnl2 (Lin et al. 2006). In this work, a group of genes was identified that display a concerted splicing switch early after birth, which is prevented by a CUG expansion or the absence of Mbnl1 exon 3 in mice and in human DM patients. The different Mbnl1 model systems exemplify the great contribution of mouse models to understand the molecular mechanisms of human disease.
Knockout and overexpression of CELF family members
A second group of proteins that has been associated with DM1 due to their ability to bind CUG repeats consists of the CUG-BP1 and ETR-3-like factors (CELF). CUG-BP1 expression is increased in individuals with DM1 and the splicing pattern of CUG-BP1 targets is altered in the disease state accordingly (Savkur et al. 2001; Timchenko et al. 2001). In addition to its function as a splicing regulator, CUG-BP1 also regulates the translation of certain target genes (Timchenko et al. 1999). Two transgenic mouse models have been generated to investigate the role of CUG-BP1 in the onset of DM1 in vivo. In the first model, a promoter directing transgene expression mainly to skeletal muscle was used, which led to a CUG-BP1 dose-dependent reduction of muscle tissue, loss of body weight, and embryonic lethality at higher doses of transgene expression (Timchenko et al. 2004). The remaining skeletal muscles in the transgenic mice showed disturbed differentiation and consistent translation of two regulators of myogenesis, p21 and MEF2A (myocyte enhancer factor 2A), was shown to be misregulated. These results suggest a link between the cytoplasmic function of CUG-BP1, translational control of myogenic regulators, and the onset of DM (Timchenko et al. 2004). While this study focused on the regulation of translation by CUG-BP1, a second model was generated that was used to examine the role of CUG-BP1 in controlling alternative splicing. Consistent with the mouse model discussed above, mice with high overexpression of CUG-BP1 in skeletal muscle and heart showed perinatal lethality and disturbed skeletal muscle formation similar to the DM phenotype (Ho et al. 2005). Further analysis revealed splicing defects in at least three target genes that are also observed in the DM state, among them the above-mentioned ClC1. As the splicing defects observed in CUG-BP1 transgenic mice were also seen in mice lacking exon 3 of the Mbnl1 protein, the two proteins appear to have antagonistic roles in these splice-site decisions.
In a further approach to elucidate the function of CELF family members in controlling heart development by regulating alternative splicing, a dominant-negative CELF mutant was expressed under the control of a heart-specific promoter. This CELF mutant localizes exclusively to the nucleus and should therefore only interfere with alternative splicing and not with translational control. Transgenic mice showed disturbed alternative splicing of a specific set of target genes that all contain potential CELF binding sites, whereas the splicing pattern of unrelated RNAs was not changed (Ladd et al. 2005b). The splicing defect became evident at 3 wk of age, which correlated with increased activity of the promoter used for transgene expression. From this time point on, a cardiac enlargement was observed, which gradually increased and led to cardiomyopathy and premature death. Interestingly, cardiac dysfunction, as well as the observed splicing defects, were far more pronounced in female than in male mice, suggesting a gender-specific influence of CELF activity in regulating alternative splicing (Ladd et al. 2005b).
Another member of the CELF family of proteins whose function was investigated in a mouse model is the recently characterized protein Brunol1. It is expressed during late embryogenesis and in the brain and testis of adult mice. Despite its expression in mouse embryos, knockout of Brunol1 had no influence on development, as Brunol1-deficient mice were recovered at a mendelian ratio and were viable and fertile. However, Brunol1-deficient male mice showed a significant reduction in sperm count and motility, which points to a (nonessential) function of Brunol1 in spermatogenesis (Dev et al. 2007).
Taken together, the four mouse models that have been generated to investigate the function of CELF proteins in vivo have demonstrated the essential role of their tightly controlled expression for the development of different tissues, and furthermore, provide a link between CELF proteins and human diseases.
Other knockout mice showing splicing defects: Gfi1 and U2AF26
In addition to splicing factor knockout mice described in the previous sections, other mouse models have been generated that unexpectedly showed defects in splicing. An example is mice deficient in the expression of the transcription factor Gfi1 (growth factor independence-1), which show various defects in the development of the haematopoetic system and sensory tissues (e.g., Karsunky et al. 2002; Hock et al. 2004; Zeng et al. 2004; Fiolka et al. 2006). Gfi1 is a nuclear zinc finger protein that binds DNA in a sequence-specific manner and most likely alters chromatin structure to regulate expression of its target genes (Duan et al. 2005). Several Gfi1 target genes have been identified by expression array and chromatin–immunoprecipitation analysis, but among the known target genes there are no proteins associated with splicing or RNA metabolism. A connection between Gfi1 and alternative splicing was only made recently, when a physical interaction of Gfi1 with a component of the splice machinery, U2AF26, was discovered (Heyd et al. 2006). U2AF26 is one of the homologs of the general splicing factor U2AF35 that has been suggested to be involved in alternative splice-site choice, e.g., in a tissue-specific manner (Shepard et al. 2002; Mollet et al. 2006). Closer analysis revealed a disturbed CD45 isoform expression in T cells of Gfi1-deficient mice, in a manner consistent with the observed reduced response after T cell receptor (TCR) stimulation. Minigene experiments suggested that Gfi1 alters CD45 alternative splicing by negatively regulating U2AF26 activity, and consistent with this hypothesis, similar defects in CD45 alternative splicing and T cell activation as observed in Gfi1-deficient mice were also evident in mice overexpressing U2AF26. Interestingly, even vast overexpression of U2AF26 in T cells had no effect on thymic development, and microarray analysis revealed no major changes in gene expression in thymi of U2AF26 transgenic mice (Heyd et al. 2006; F. Heyd and T. Moroy, unpubl.) indicating that changes in the U2AF26:U2AF35 ratio do not interfere with cell viability as long as both proteins are present.
These results reveal a surprising role of the transcription factor Gfi1 in regulating alternative splicing by direct protein–protein interaction with a component of the spliceosome, and more such surprises can be expected when existing mouse models are re-evaluated using techniques such as splice arrays in order to draw a more complete picture of the regulation of alternative splicing in vivo.
COMPARISON OF KNOCKOUT MICE WITH OTHER ORGANISMS AND IN VITRO DATA
When comparing the phenotypes of the mouse models described so far, some striking similarities become apparent. One common feature is the (embryonic) lethality of most splicing factor knockout models described to date, which contrasts with the viable phenotype of all but one siRNA-mediated knock-downs of splicing-factor homologs in Caenorhabditis elegans, including the direct homologs of ASF/SF2 (lethal), SRp20, and SC35 (Longman et al. 2000). The lethal phenotype in mice clearly demonstrates a nonredundancy among SR proteins, hnRNPs, and tissue-specific factors (Nova-1), and it can therefore be concluded that all splicing factors investigated so far have certain essential functions that cannot be carried out by any other protein.
However, cultured cells established from some of these mice showed almost normal growth and differentiation potential, demonstrating that the respective factor is not per se required for cellular viability. The same conclusion can be drawn from the tissue-specific knockout of ASF/SF2, which allowed normal development of the heart until several weeks after birth. It therefore appears that splicing factors are only essential in certain cell types and in certain developmental stages, whereas they may be dispensable in other tissues. This context dependency might also explain the difference in the viability between knockout mice and C. elegans knock-down mutants: the splicing factors might be essential only during a certain stage of the highly complex vertebrate embryogenesis, whereas loss of one splicing factor can be compensated during the less-complex development of C. elegans. Another explanation for the viable phenotype of splicing-factor knock-down mutants in C. elegans is the technical approach: the siRNA-mediated knock-down might decrease protein levels under detectable amounts, but traces of residual protein could be present and rescue the lethal phenotype.
Core components of the spliceosome or factors essential for snRNP recycling have been deleted or knocked-down in model organisms such as yeast, zebrafish, C. elegans, and Drosophila, which led to lethal phenotypes (Kanaar et al. 1993; Potashkin et al. 1993; Rudner et al. 1996; Zorio and Blumenthal 1999; Trede et al. 2007). Whether this is also the case in mice remains to be shown, as the respective homologs have not yet been analyzed in a gene-targeting approach in the murine system.
These data, together with the microarray analysis mentioned above, demonstrating that a certain splicing factor is not essential to maintain the general pattern of gene expression, suggest that mammalian splicing factors show a certain degree of functional overlap, i.e., they are not required for all cell types under every condition, but apart from that, they all have individual functions that are strictly nonredundant. It is interesting to speculate that SR proteins are redundant for constitutive splicing, but play essential nonredundant functions in the regulation of alternative splicing. This model, in addition to explaining the phenotypes of the mice described above, also fits well with the in vitro data on SR proteins, describing redundancy among the SR domain itself, but different RNA-binding capabilities to regulate distinct targets.
MICE WITH TARGETED DISRUPTION OF A SPECIFIC PROTEIN ISOFORM
Ion transporters, cell surface receptors, and ligands: FGFR2, VEGF, Dopamine Receptor D2, Na-K-2Cl cotransporter, sarco(endo)plasmic reticulum Ca2+ ATPase, and CD45
Fibroblast growth factors (Fgfs) are a large family of intercellular signaling molecules that have been implicated in the development of different species, including mammals. They signal through Fgf receptors (FgfR), a family of four different transmembrane tyrosine kinases whose diversity is further enhanced by alternative splicing. For FgfR2 (as well as FgfR1 and FgfR3), the ligand-binding domain is encoded by two exons, an invariant exon IIIa, and one of two mutually exclusive exons IIIb and IIIc, to generate receptors with different ligand-binding specificities (Orr-Urtreger et al. 1993). The expression of both isoforms is controlled with the IIIb isoform being predominant in epithelia, whereas IIIc isoforms are expressed in the mesenchyme (Orr-Urtreger et al. 1993). Recent evidence suggests that members of the Fox family of RNA-binding proteins are critically involved in regulating alternative splicing of FgfR2, and that this mechanism is conserved among species from zebrafish to mammals (e.g., Baraniak et al. 2006). Deletion of the IIIb exon from the murine genome did not result in the inclusion of exon IIIc instead, but led to a direct fusion of exon IIIa with the next 3′ exon to yield a nonfunctional protein due to inclusion of an in frame Stop codon (De Moerlooze et al. 2000). As both isoforms are expressed in a mutually exclusive manner, the loss of exon IIIb along with the inclusion of a premature Stop codon can be regarded as a tissue-specific knockout of FgfR2 (this problem could have been circumvented by applying a knockin strategy such as was used for the Integrin β1 isoforms, see below). However, unlike the complete loss of FgfR2, which leads to early embryonic lethality (Arman et al. 1998), mice with targeted deletion of exon IIIb developed until birth, but died shortly after due to failure of lung formation, which confirms earlier studies implicating FgfR2 IIIb expression in lung development (Peters et al. 1994; De Moerlooze et al. 2000). A variety of other developmental defects, including the absence of fore- and hindlimbs was also evident in the mutant embryos, whereas other organs such as heart, liver, and kidney were apparently normal (De Moerlooze et al. 2000; Revest et al. 2001). These results are in agreement with an essential role of FgfR2 IIIb in some developmental processes such as limb initiation, but it remains to be established whether this process indeed depends on the correct isoform expression, or whether the observed phenotype rather reflects the complete lack of FgfR2 in certain tissues. In contrast to the formation of a nonfunctional protein caused by the deletion of FgfR2 exon IIIb, the deletion of exon IIIc mainly resulted in inclusion of the IIIb exon. In the heterozygous state, this gain-of-function mutation leads to expression of both isoforms in tissues that usually express only IIIc, rendering these tissues responsive to a further subset of Fgf ligands. Heterozygous IIIc-deficient animals were born at a Mendelian frequency, but showed defects in bone formation as well as abnormalities in lung and kidney development; these animals failed to gain weight after birth and died within 9 d (Hajihosseini et al. 2001). A possible explanation would be that the increased Fgf signaling in these mutant mice provokes immature differentiation of some cell populations, which ultimately leads to failures in organ formation.
A related class of signaling molecules are the vascular endothelial growth factors (Vegf), which are expressed as three isoforms (Vegf120, Vegf164, Vegf188) that differ in the inclusion of two alternatively spliced exons (exons 6 and 7). The importance of Vegf for mammalian development has been underlined by the finding that the heterozygous loss of Vegf already causes embryonic lethality (Carmeliet et al. 1996; Ferrara et al. 1996). Later on, loxP–Cre-mediated knockout or knockin strategies have been employed to exclusively express one of the three isoforms. Interestingly, the viability of the three mutant mice differed considerably: mice only expressing Vegf120 died shortly after birth due to impaired myocardial angiogenesis (Carmeliet et al. 1999), Vegf164-expressing mice appeared normal and exclusive expression of Vegf188 led to reduced viability and fertility (Stalmans et al. 2002). Analysis of angiogenesis in the mutant mice suggested that expression of Vegf120 alone is insufficient to promote angiogenesis, whereas it is normal in mice only expressing Vegf164 and partially disturbed in Vegf188-expressing mice (Carmeliet et al. 1999; Stalmans et al. 2002), which reflects the different degrees of viability in the three mouse models.
Dopamine receptors, especially the D2 subfamily, have been thoroughly investigated due to the involvement of dopamine signaling in neuropsychiatric diseases such as schizophrenia and Parkinson's disease. Dopamine signals through a family of G-protein-coupled receptors that either increase or decrease intracellular cAMP by stimulating or inhibiting adenylyl cyclase, respectively. The D2 subfamily consists of two members, D2L (long) and D2S (short), which differ by an insertion of 29 amino acids in the long isoform, regulating the affinity for different inhibitory G-proteins (Guiramand et al. 1995). Two groups have (simultaneously) followed a knockout approach to define distinct receptor functions by deleting the 29 amino acids insertion specific for the long isoform from the genome (Usiello et al. 2000; Wang et al. 2000). Both groups used conventional knockout strategies with the neo cassette remaining in the targeted allele, which did not affect D2S expression. In both models, deletion of the D2L-specific exon led to an up-regulation of the D2S isoform to wild-type level, whereas other dopamine receptors were not affected. Mutant mice appeared viable and fertile, but showed reduced locomotion in one model (Wang et al. 2000), whereas this defect was not observed in the mouse mutant generated by the second group (Usiello et al. 2000). However, both studies agreed that D2L-deficient mice display altered behavior in response to treatment with different antipsychotic drugs. The difference in the function of the two D2 isoforms in vivo suggested that D2S- and D2L-specific drugs might improve the treatment of neuropathologies in humans (Usiello et al. 2000; Wang et al. 2000; Xu et al. 2002; Lindgren et al. 2003).
Expression of ion transporters is regulated at various levels, including alternative splicing, and the isoform-specific function of some transporters has been investigated in mouse models. The Na-K-2Cl cotransporter (NKCC2) plays a major role in the uptake of ions in the kidney. Four different isoforms are known that differ in their ion-binding capabilities and are differentially expressed in different kidney regions, probably to ensure proper function in areas with varying ion concentrations (Oppermann et al. 2007). Two minor isoforms, A and B, have been targeted in mice using loxP–Cre strategies, introducing premature Stop codons in the respective exons. In contrast to the complete disruption of the entire NKCC2 gene that led to early postnatal death (Takahashi et al. 2000), mice deficient in the A or B isoform showed only mild phenotypes (Oppermann et al. 2006, 2007). Mice deficient in the A isoform showed symptoms consistent with disturbed salt sensing at high Cl concentrations and the B isoform appears to be more active at lower salt concentrations, arguing for specialized functions of the two isoforms. However, the lack of either one of these isoforms seems to be readily compensated by the remaining NKCC2 proteins.
In an attempt to elucidate isoform-specific functions of the sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA2), a knockout mouse model was generated lacking the muscle-specific SERCA2a isoform. In cardiac tissue of mutant mice, a compensatory up-regulation of the SERCA2b isoform was observed, which is not expressed in the heart of wild-type animals. The SERCA2b expression in mutant hearts reached about 50% of total SERCA2 protein in wild type, which is comparable to the amount of SERCA2 protein in mice that carry a heterozygous deletion of the complete gene (Periasamy et al. 1999; Ver Heyen et al. 2001). Mutant mice showed an increased embryonic and neonatal death, which was attributable to cardiac malformation, but the majority of SERCA2a-deficient animals survived this early phenotype. As such an increased mortality was not observed in mice with heterozygous deletion of the SERCA2 gene, the malformation of the embryonic heart appears to be caused by the absence of the SERCA2a isoform rather than the reduced total amount of SERCA2 protein. Mice deficient in SERCA2a that survived to adulthood showed impaired cardiac function and a defect in Ca2+ uptake, which further points to different roles of SERCA2 isoforms in maintaining these processes (Ver Heyen et al. 2001; Antoons et al. 2003).
Another cell-surface receptor that has attracted much attention in recent years due to its ability to regulate immune function is the above-mentioned transmembrane tyrosine phosphate CD45. Several CD45 isoforms are expressed in a tightly controlled manner, and correct isoform expression has been shown to be essential for T cell development and function (Hermiston et al. 2003). Regulation of CD45 alternative splicing has been investigated in great detail using in vitro experiments (e.g., Rothrock et al. 2003, 2005) and a CD45-deficient mouse model has been described (e.g., Byth et al. 1996). In order to investigate the effect of different CD45 isoforms, cDNAs encoding single isoforms were expressed in mice with a CD45 null background. These experiments confirmed that none of the single isoforms was able to fully rescue the CD45 null phenotype with respect to T- or B-cell development and activation, pointing to distinct yet partially overlapping functions of the respective CD45 isoforms in vivo (Kozieradzki et al. 1997; Ogilvy et al. 2003).
Scaffolding and extracellular matrix proteins: Integrin, Fibronectin, Plectin, and Agrin
The Integrin family of adhesion molecules comprises over 20 distinct subunits, some of which are further subjected to alternative splicing. Integrins consist of a noncovalent heterodimeric complex between an α and a β subunit and bind a variety of ligands, among them Fibronectins. They are involved in regulating fundamental processes such as cell adhesion, migration, and differentiation as well as metastasis (Danen and Sonnenberg 2003), and have therefore gained much interest in recent years. Two isoforms, A and B, of the α6 subunit are described, affecting the C-terminal cytoplasmic domain of the protein. These isoforms are formed by inclusion or exclusion of exon A which, when present, introduces a Stop codon to prevent translation of the constitutively present exon B. The expression of Integrin α6 A and B is tightly regulated in a tissue and development-dependent manner, and several lines of evidence pointed to differences in their ability to contribute to development, cell migration, or T cell maturation (Ruiz et al. 1995; Domanico et al. 1997). To address these questions in vivo, a mouse model was constructed lacking the genomic region comprising exon A, leading to the uniform expression of Integrin α6B (Gimond et al. 1998). Normal development and fertility of mutant mice indicated that the α6B isoform is able to fully replace the α6A protein during mouse development. Several morphological and functional tests failed to show differences between wild-type and mutant mice with regard to epidermis and heart development, both of which show highly regulated expression of Integrin α6 isoforms in wild-type mice (Gimond et al. 1998). This work exemplifies a difference between in vitro data, which strongly suggest isoform-specific Integrin α6 functions, and in vivo data, which rather point to a redundant role of the two isoforms. It also underlines the ability of mouse models to complete and refine our view on isoform-specific protein functions.
Another Integrin whose isoform-specific function has been investigated by a knockout mouse model is the β1 Integrin, which has been implicated in a variety of functions ranging from migration of lymphocytes to cardiac muscle development and more general organogenesis. In mice, two isoforms are known, A and D, which differ in their intracellular domain by only 13 amino acids. The less-common D isoform is generated by insertion of a specific exon that introduces a Stop codon in the mRNA in a mechanism similar to the one described for the α6 subunit (van der Flier et al. 1995). Using a loxP–Cre strategy, the exon encoding the D isoform was removed from the genome, resulting in a mouse model that expresses only the Integrin β1A isoform. These mice did not show any severe defects and behaved similar to wild-type littermates with respect to viability and fertility, albeit a mild cardiac dysfunction was noted in male knockout mice (Baudoin et al. 1998). A reduction in Integrin β1D expression as well as cardiac dysfunction were also observed in mice expressing a dominant-negative CELF protein (Ladd et al. 2005b), further providing evidence for a requirement of the β1D isoform for correct heart development. In an effort to also investigate a potential isoform-specific function of the main β1A form, the 3′ end of the cDNA encoding the D isoform was fused to the adjacent 5′ exon and the exon specific for the A isoform was deleted (see Baudoin et al. 1998 for details of the targeting strategy). This knockin mouse mutant was shown to exclusively express the D isoform regardless of tissue or developmental state. Mice homozygous for this mutation showed multiple defects in development and embryonic lethality around day 11.5 (Baudoin et al. 1998). Later experiments suggested compromised nutrition of the embryo caused by malformation of the placenta and decreased muscle formation (both tissues showing exclusive expression of the β1A isoform in wild type) to be the main reasons for the embryonic lethality (Cachaco et al. 2003). Interestingly, some features of the defects observed in development of the neuronal tube resembled those found in Fibronectin deficient embryos (George et al. 1993), which points to an essential interaction between Integrin β1A and Fibronectin in this process.
Fibronectins are essential cell-adhesion proteins in the extracellular matrix, whose expression is regulated by alternative splicing of at least three distinct regions that encode the extra domains A and B (EDA and EDB) and the type III homologies connecting segment (IIICS). As Fibronectins are involved in fundamental processes such as embryogenesis, wound healing and cell migration (George et al. 1993), to name but a few, considerable attention has been paid to the expression of Fibronectin isoforms, and mouse models either lacking EDA or EDB or constitutively expressing EDA have been generated (Fukuda et al. 2002; Muro et al. 2003; Chauhan et al. 2004, 2005). Even though mice either lacking or constitutively expressing the EDA were viable and fertile and showed normal development, they displayed a substantially reduced lifespan when compared with wild-type mice, arguing for a role of correct Fibronectin splicing during aging (Muro et al. 2003). In addition, mice lacking the EDA showed impaired wound healing, reduced atherosclerotic lesions, and reduced motor coordination abilities, which is consistent with a role of EDA including isoforms in tissue remodeling and probably also in the nervous system (Muro et al. 2003; Tan et al. 2004; Chauhan et al. 2005). In contrast, no in vivo defect was found in mice lacking the other alternatively spliced extra domain in Fibronectin, EDB, which was somewhat surprising given the high expression of EDB containing Fibronectin isoforms during embryogenesis (Fukuda et al. 2002).
Different members of the Plectin scaffold protein family are generated by alternative splicing and interact with cytoskeletal filaments, the subplasma membrane skeleton, and focal adhesion contacts to enable cross-talk between these different systems. Plectin isoform 1-deficient mice have been generated and, in contrast to the lethal phenotype of complete Plectin knockout, shown to be fertile and viable (Abrahamsberg et al. 2005). Analysis of different cell populations from the mutant mice revealed impaired migration potential, consistent with a role of Plectin proteins in reorganizing the cytoskeleton. However, as there are at least 11 different Plectin isoforms in mice, isoform-specific functions are only beginning to be understood.
A further extracellular matrix protein whose isoform-specific function has been targeted by the use of mouse models is the heparan sulfate proteoglycan Agrin. In motoneurons, Agrin is released into the synaptic cleft, where it is essential to induce postsynaptic differentiation, i.e., clustering of acetylcholine receptors on the postsynaptic myotubes (Gautam et al. 1996). To distinguish between neuron-specific isoforms (including exons 28 Y+ and 32–33, Z+) and isoforms expressed by muscle tissue (Y+/−, Z−), knockout mice specifically lacking the Y+ or Z+ isoform were generated using the loxP–Cre system. The construct targeting the Z+ isoforms replaced exons 32 and 33 by a loxP-flanked neo cassette, which was excised using Cre recombinase. Mice lacking the Agrin Z+ isoforms were either dead at birth or died perinatally; they failed to move or breathe, most likely due to severe impairment of pre- and postsynaptic differentiation (Burgess et al. 1999). It is interesting to note that a similar targeting strategy, also replacing the exons 32 and 33 with a neo cassette that remains in the genome due to lacking loxP sites, leads to a reduction of total Agrin expression to < 10% of wild-type level, probably due to reduced stability of the primary transcript (Gautam et al. 1996). As effects of a remaining neo cassette on target gene expression are not uncommon, this example demonstrates the great advantage of the loxP–Cre system over conventional knockout strategies. Mice with reduced total Agrin expression displayed the same phenotype as mice lacking only the Z+ isoforms, despite normal levels of Z− Agrins in the latter animals (Gautam et al. 1996), questioning the requirement for Z− Agrin in vivo. In contrast, mice lacking the Y+ isoforms were found to be viable and fertile, and behaved like wild type in all applied assays. While these studies clearly identify Z+ Agrins to be key players in formation of the neuromuscular junction, the question remains unanswered whether Z− isoforms have any specific function, and which role is played by the Y exon. As Z− isoforms are expressed in tissues such as muscle, kidney, and lung, it appears improbable to have no function at all, but such a function has yet to be identified.
Transcription factors: WT1, STAT3β, and Pax6(5a)
The Wilms' Tumor 1 (WT1) gene encodes a zinc finger protein with the potential to both repress and enhance transcription; it acts as tumor suppressor and has been implicated in kidney development. Multiple different isoforms are generated from one gene by means of alternative splicing and alternative translation start sites. Two variants have attracted particular interest, as in vitro results suggest distinct functions (Laity et al. 2000). They differ by only three amino acids (KTS) that are inserted or left out at the end of exon 9 by use of different splice sites. By selectively mutating these splice sites, mice have been generated that express only one of the two isoforms at wild-type level. The mutation that has been used to delete the + KTS isoform has been shown to induce Frasier syndrome in human patients, a disease characterized by glumerular sclerosis and male-to-female sex reversal (Hammes et al. 2001).
Newborn mutant mice of both genotypes showed severe kidney abnormalities and died shortly after birth, demonstrating that both isoforms are required for kidney development. In accordance with the phenotype of Frasier syndrome patients, mice carrying the same mutation leading to exclusive expression of the − KTS isoform developed along the female pathway also in XY animals, whereas female development was not affected. In contrast, mice only expressing the + KTS isoform showed disturbed development of the gonads in both sexes, arguing for a distinct role of both isoforms in sex determination. In addition, + KTS and – KTS isoforms may also have some overlapping functions, as the development of other organs known to be effected by complete WT1 knockout appears normal in the isoform-specific mutant mice (Hammes et al. 2001).
The Pax6 transcriptional activator has been shown to be essential for eye development. It is conserved among species, but in vertebrates a splice variant exists that includes exon 5a, which has been suggested to alter DNA-binding specificity (Epstein et al. 1994). The function of exon 5a was investigated through its loxP–Cre-mediated removal. Heterozygous and homozygous mutant mice showed Aniridia (also present in humans with mutations in the Pax6 gene), a condition marked by misdevelopment of the iris and enlarged pupils (Singh et al. 2002). As the eyes of newborn mutant mice were undistinguishable from wild-type mice, it appears that the expression of Pax6(5a) is not necessary for embryonic eye development, but is essential at later differentiation stages and/or maintenance of the adult eye. This is in agreement with studies showing that during embryonic development Pax6(5a) is only expressed to a small fraction of total Pax6, but is strongly up-regulated after birth (Singh et al. 2002). As expression of Pax6 has also been shown in the nose, the pancreas, and the nervous system, it will be interesting to determine how these tissues are affected by the lack of Pax6(5a).
The Stat family of transcription factors plays a central role in the immune system, as its members mediate signaling through cytokine receptors. Stat3, for example, mediates IL-6 signaling, which plays a role in the acute-phase response, the early phase of an infection. Two isoforms of Stat3 are known, α and β, which arise by alternative splice-acceptor sites in exon 23, and which differ in their C-terminal transactivation domains (Schaefer et al. 1995). The cryptic splice-acceptor site in exon 23 that gives rise to Stat3β expression was mutated in mice to yield animals that only express Stat3α. These animals appeared normal and fertile and showed no defect in the development of immune tissues (Yoo et al. 2002). However, after a challenge with LPS (Lipopolysaccharide), the mutant mice showed impaired recovery and increased mortality, which was likely due to septic shock. Further analysis suggested that Stat3β negatively regulates a subset of LPS-responsive genes to attenuate the response to endotoxic challenge. In accordance with this view, Stat3β expression in wild-type mice was found to be induced after administration of LPS (Yoo et al. 2002), which might also explain why the absence of Stat3β has only little effect in the basal state but becomes important after LPS challenge.
WHAT CAN BE LEARNED FROM ISOFORM-SPECIFIC MOUSE MODELS?
What are the common themes that emerge from the new knowledge gathered from mouse models, and how consistent are they in relation to the wealth of available in vitro data in splicing research? Clearly, the vast majority of the mouse models display distinct phenotypes, strongly supporting the view that alternative splicing does contribute substantially to the regulation of biological processes in vivo. However, it should be kept in mind that the experimental strategies to generate particular mouse models were chosen after in vitro data suggested specific functions of splicing factors or alternatively spliced isoforms. It is interesting to note that most isoform-specific knockout models display a less-severe phenotype than the mutants with a complete knockout. A good example is the three isoform-specific Vegf knockout models, which show distinct phenotypes ranging from early lethality and disturbed angiogenesis to no impairment in viability, suggesting functional overlap between different isoforms of the same protein. This observation could be taken as a good indicator that alternative splicing does not radically change protein function, but rather introduces subtle changes. An alternative explanation would be that different isoforms of one protein fulfill completely different functions, and the loss of one provokes a less-pronounced phenotype than the loss of the whole protein.
Interestingly, the models that displayed redundancy among alternatively spliced isoforms all showed lethal phenotypes when the whole gene was deleted (Integrins, Fibronectins, Tropomyosin, and GABA receptor γ subunit). This could indicate that redundancy among isoforms represents a safety measure: in that case, loss of one exon, or the ability to include it, could be compensated by the respective other isoform without any impairment of biological function. Alternatively, isoforms that appear redundant in laboratory animals may have highly specialized functions during other-than-normal conditions, such as stress response or environmental changes that are likely to be overlooked in standard analysis.
Another interesting aspect to note from isoform-specific knockout mice is the splicing pattern of the remaining gene. In some cases, an up-regulation of the remaining isoform takes place, but in other models, such a compensatory mechanism is not observed. How these compensatory effects are regulated is poorly understood, and it is likely that mouse models will prove useful to learn more about this phenomenon.
MODELS TO INVESTIGATE MECHANISMS OF ALTERNATIVE SPLICING IN VIVO
A further interesting experimental setup that can be used in alternative splicing research are mice that carry reporter genes to monitor alternative splice-site decisions, rather than focusing on phenotypic effects. Such reporter constructs are usually designed in such a way that expression of a GFP reporter is dependent on a particular splice decision, e.g., two GFP exons are separated by another exon, which has to be excluded in order to allow GFP expression. Such strategies have been used in cell-culture experiments to investigate cis-acting RNA elements and trans-acting splicing factors involved in a particular splicing event (e.g., Sheives and Lynch 2002). However, these studies have the limitation of cell-culture experiments, i.e., it is uncertain whether immortalized cells indeed represent a physiologically relevant condition. Therefore, several mouse models were generated that express a splicing-sensitive GFP reporter that can be analyzed under in vivo conditions. An additional advantage over cell-culture experiments is that splicing can be monitored in every relevant tissue at the same time (Crenshaw et al. 1987; Ellis et al. 2004; for a review on technical aspects, see Kemp et al. 2005). In one such model, the tissue-specific silencing of the FgfR2 exon IIIb has been investigated and revealed the requirement of distinct cis-acting elements in different tissues (Bonano et al. 2006). A similar technique was also used to monitor uptake of antisense oligomers into different mouse tissues. To this end, a GFP reporter gene whose expression was dependent on the silencing of an interrupting exon was expressed in transgenic mice, and the ability of antisense oligomers directed against the splice site of the interrupting exon to restore GFP expression was assessed (Sazani et al. 2002).
CONCLUSION AND FUTURE PERSPECTIVES
To date, the current approaches to study alternative splicing and its impact on physiological processes are mainly based on in vitro or cell-culture assays. Therefore, suitable in vivo models are required not only to verify the relevance of the current concepts, but also to further our understanding in splicing research. Research over the past 20 yr has demonstrated that gene targeting in the mouse is a very powerful approach to generate in vivo models. Research in the splicing field has only just begun to adopt this approach, but has already produced surprising results and new insights. In this review we have summarized what has been learned from mouse models with regard to alternative splicing. We can conclude that mouse mutants have been shown to be highly valuable tools to refine, adjust, and verify the current concepts of alternative splicing. It has enabled the generation of new experimental strategies ranging from reporter mouse strains with the possibility to monitor splice-site decisions in vivo to mutants modeling human diseases. Still, many questions remain open, and the majority of the key issues have not yet been addressed. For instance, none of the core components of the spliceosome nor the majority of the tissue-specific splicing factors have been investigated in a gene-targeting approach. In addition, processes such as proliferation and differentiation are accompanied by well-defined changes in transcription, but the implication of alternative splicing in such cellular programs has so far not been systematically analyzed. Mouse models seem to be the first choice to address such questions in vivo, and therefore, many more interesting and probably surprising results are to be expected in the near future. The data discussed here may only be the proverbial tip of an iceberg, as the use of mouse models in splicing research has only just begun.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.554607.
REFERENCES
- Abrahamsberg, C., Fuchs, P., Osmanagic-Myers, S., Fischer, I., Propst, F., Elbe-Burger, A., Wiche, G. Targeted ablation of plectin isoform 1 uncovers role of cytolinker proteins in leukocyte recruitment. Proc. Natl. Acad. Sci. 2005;102:18449–18454. doi: 10.1073/pnas.0505380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoons, G., Ver Heyen, M., Raeymaekers, L., Vangheluwe, P., Wuytack, F., Sipido, K.R. Ca2+ uptake by the sarcoplasmic reticulum in ventricular myocytes of the SERCA2b/b mouse is impaired at higher Ca2+ loads only. Circ. Res. 2003;92:881–887. doi: 10.1161/01.RES.0000069032.81501.98. [DOI] [PubMed] [Google Scholar]
- Arman, E., Haffner-Krausz, R., Chen, Y., Heath, J.K., Lonai, P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl. Acad. Sci. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak, A.P., Chen, J.R., Garcia-Blanco, M.A. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell. Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudoin, C., Goumans, M.J., Mummery, C., Sonnenberg, A. Knockout and knockin of the β1 exon D define distinct roles for integrin splice variants in heart function and embryonic development. Genes & Dev. 1998;12:1202–1216. doi: 10.1101/gad.12.8.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Bonano, V.I., Oltean, S., Brazas, R.M., Garcia-Blanco, M.A. Imaging the alternative silencing of FGFR2 exon IIIb in vivo. RNA. 2006;12:2073–2079. doi: 10.1261/rna.248506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda, C.S., Dymecki, S.M. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Buchner, D.A., Trudeau, M., Meisler, M.H. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science. 2003;301:967–969. doi: 10.1126/science.1086187. [DOI] [PubMed] [Google Scholar]
- Burgess, R.W., Nguyen, Q.T., Son, Y.J., Lichtman, J.W., Sanes, J.R. Alternatively spliced isoforms of nerve- and muscle-derived agrin: Their roles at the neuromuscular junction. Neuron. 1999;23:33–44. doi: 10.1016/s0896-6273(00)80751-5. [DOI] [PubMed] [Google Scholar]
- Byth, K.F., Conroy, L.A., Howlett, S., Smith, A.J., May, J., Alexander, D.R., Holmes, N. CD45-null transgenic mice reveal a positive regulatory role for CD45 in early thymocyte development, in the selection of CD4+CD8+ thymocytes, and B cell maturation. J. Exp. Med. 1996;183:1707–1718. doi: 10.1084/jem.183.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachaco, A.S., Chuva de Sousa Lopes, S.M., Kuikman, I., Bajanca, F., Abe, K., Baudoin, C., Sonnenberg, A., Mummery, C.L., Thorsteinsdottir, S. Knock-in of integrin β 1D affects primary but not secondary myogenesis in mice. Development. 2003;130:1659–1671. doi: 10.1242/dev.00394. [DOI] [PubMed] [Google Scholar]
- Carmeliet, P., Ferreira, V., Breier, G., Pollefeyt, S., Kieckens, L., Gertsenstein, M., Fahrig, M., Vandenhoeck, A., Harpal, K., Eberhardt, C., et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Carmeliet, P., Ng, Y.S., Nuyens, D., Theilmeier, G., Brusselmans, K., Cornelissen, I., Ehler, E., Kakkar, V.V., Stalmans, I., Mattot, V., et al. Impaired myocardial angiogenesis and ischemic cardiomyopathy in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Nat. Med. 1999;5:495–502. doi: 10.1038/8379. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., Krainer, A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Chauhan, A.K., Iaconcig, A., Baralle, F.E., Muro, A.F. Alternative splicing of fibronectin: A mouse model demonstrates the identity of in vitro and in vivo systems and the processing autonomy of regulated exons in adult mice. Gene. 2004;324:55–63. doi: 10.1016/j.gene.2003.09.026. [DOI] [PubMed] [Google Scholar]
- Chauhan, A.K., Moretti, F.A., Iaconcig, A., Baralle, F.E., Muro, A.F. Impaired motor coordination in mice lacking the EDA exon of the fibronectin gene. Behav. Brain Res. 2005;161:31–38. doi: 10.1016/j.bbr.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Crenshaw E.B., 3rd, Russo, A.F., Swanson, L.W., Rosenfeld, M.G. Neuron-specific alternative RNA processing in transgenic mice expressing a metallothionein-calcitonin fusion gene. Cell. 1987;49:389–398. doi: 10.1016/0092-8674(87)90291-1. [DOI] [PubMed] [Google Scholar]
- Danen, E.H., Sonnenberg, A. Integrins in regulation of tissue development and function. J. Pathol. 2003;201:632–641. doi: 10.1002/path.1472. [DOI] [PubMed] [Google Scholar]
- De Moerlooze, L., Spencer-Dene, B., Revest, J., Hajihosseini, M., Rosewell, I., Dickson, C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Dev, A., Nayernia, K., Meins, M., Adham, I., Lacone, F., Engel, W. Mice deficient for RNA-binding protein brunol1 show reduction of spermatogenesis but are fertile. Mol. Reprod. Dev. 2007 doi: 10.1002/mrd.20742. [DOI] [PubMed] [Google Scholar]
- Ding, J.H., Xu, X., Yang, D., Chu, P.H., Dalton, N.D., Ye, Z., Yeakley, J.M., Cheng, H., Xiao, R.P., Ross, J., et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004;23:885–896. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanico, S.Z., Pelletier, A.J., Havran, W.L., Quaranta, V. Integrin α 6A β 1 induces CD81-dependent cell motility without engaging the extracellular matrix migration substrate. Mol. Biol. Cell. 1997;8:2253–2265. doi: 10.1091/mbc.8.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Z., Zarebski, A., Montoya-Durango, D., Grimes, H.L., Horwitz, M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 2005;25:10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, P.D., Smith, C.W., Kemp, P. Regulated tissue-specific alternative splicing of enhanced green fluorescent protein transgenes conferred by α-tropomyosin regulatory elements in transgenic mice. J. Biol. Chem. 2004;279:36660–36669. doi: 10.1074/jbc.M405380200. [DOI] [PubMed] [Google Scholar]
- Epstein, J.A., Glaser, T., Cai, J., Jepeal, L., Walton, D.S., Maas, R.L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes & Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Faustino, N.A., Cooper, T.A. Pre-mRNA splicing and human disease. Genes & Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Ferrara, N., Carver-Moore, K., Chen, H., Dowd, M., Lu, L., O'Shea, K.S., Powell-Braxton, L., Hillan, K.J., Moore, M.W. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- Fiolka, K., Hertzano, R., Vassen, L., Zeng, H., Hermesh, O., Avraham, K.B., Duhrsen, U., Moroy, T. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 2006;7:326–333. doi: 10.1038/sj.embor.7400618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd, J.A., Gold, D.A., Concepcion, D., Poon, T.H., Wang, X., Keithley, E., Chen, D., Ward, E.J., Chinn, S.B., Friedman, R.A., et al. A natural allele of Nxf1 suppresses retrovirus insertional mutations. Nat. Genet. 2003;35:221–228. doi: 10.1038/ng1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, T., Yoshida, N., Kataoka, Y., Manabe, R., Mizuno-Horikawa, Y., Sato, M., Kuriyama, K., Yasui, N., Sekiguchi, K. Mice lacking the EDB segment of fibronectin develop normally but exhibit reduced cell growth and fibronectin matrix assembly in vitro. Cancer Res. 2002;62:5603–5610. [PubMed] [Google Scholar]
- Gautam, M., Noakes, P.G., Moscoso, L., Rupp, F., Scheller, R.H., Merlie, J.P., Sanes, J.R. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–535. doi: 10.1016/s0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- Gebski, B.L., Mann, C.J., Fletcher, S., Wilton, S.D. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum. Mol. Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
- George, E.L., Georges-Labouesse, E.N., Patel-King, R.S., Rayburn, H., Hynes, R.O. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- Gimond, C., Baudoin, C., van der Neut, R., Kramer, D., Calafat, J., Sonnenberg, A. Cre-loxP-mediated inactivation of the α6A integrin splice variant in vivo: Evidence for a specific functional role of α6A in lymphocyte migration but not in heart development. J. Cell Biol. 1998;143:253–266. doi: 10.1083/jcb.143.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiramand, J., Montmayeur, J.P., Ceraline, J., Bhatia, M., Borrelli, E. Alternative splicing of the dopamine D2 receptor directs specificity of coupling to G-proteins. J. Biol. Chem. 1995;270:7354–7358. doi: 10.1074/jbc.270.13.7354. [DOI] [PubMed] [Google Scholar]
- Guo, H., Wang, C.L. Specific disruption of smooth muscle caldesmon expression in mice. Biochem. Biophys. Res. Commun. 2005;330:1132–1137. doi: 10.1016/j.bbrc.2005.03.089. [DOI] [PubMed] [Google Scholar]
- Hajihosseini, M.K., Wilson, S., De Moerlooze, L., Dickson, C. A splicing switch and gain-of-function mutation in FgfR2-IIIc hemizygotes causes Apert/Pfeiffer-syndrome-like phenotypes. Proc. Natl. Acad. Sci. 2001;98:3855–3860. doi: 10.1073/pnas.071586898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes, A., Guo, J.K., Lutsch, G., Leheste, J.R., Landrock, D., Ziegler, U., Gubler, M.C., Schedl, A. Two splice variants of the Wilms' tumor 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106:319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
- Hermiston, M.L., Xu, Z., Weiss, A. CD45: A critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 2003;21:107–137. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- Heyd, F., ten Dam, G., Moroy, T. Auxiliary splice factor U2AF26 and transcription factor Gfi1 cooperate directly in regulating CD45 alternative splicing. Nat. Immunol. 2006;7:859–867. doi: 10.1038/ni1361. [DOI] [PubMed] [Google Scholar]
- Ho, T.H., Charlet, B.N., Poulos, M.G., Singh, G., Swanson, M.S., Cooper, T.A. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, T.H., Bundman, D., Armstrong, D.L., Cooper, T.A. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- Hock, H., Hamblen, M.J., Rooke, H.M., Schindler, J.W., Saleque, S., Fujiwara, Y., Orkin, S.H. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- Hoffman, B.E., Lis, J.T. Pre-mRNA splicing by the essential Drosophila protein B52: Tissue and target specificity. Mol. Cell. Biol. 2000;20:181–186. doi: 10.1128/mcb.20.1.181-186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homanics, G.E., Harrison, N.L., Quinlan, J.J., Krasowski, M.D., Rick, C.E., de Blas, A.L., Mehta, A.K., Kist, F., Mihalek, R.M., Aul, J.J., et al. Normal electrophysiological and behavioral responses to ethanol in mice lacking the long splice variant of the γ2 subunit of the γ-aminobutyrate type A receptor. Neuropharmacology. 1999;38:253–265. doi: 10.1016/s0028-3908(98)00177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, K.B., Dredge, B.K., Stefani, G., Zhong, R., Buckanovich, R.J., Okano, H.J., Yang, Y.Y., Darnell, R.B. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Jumaa, H., Wei, G., Nielsen, P.J. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 1999;9:899–902. doi: 10.1016/s0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- Kanaar, R., Roche, S.E., Beall, E.L., Green, M.R., Rio, D.C. The conserved pre-mRNA splicing factor U2AF from Drosophila: Requirement for viability. Science. 1993;262:569–573. doi: 10.1126/science.7692602. [DOI] [PubMed] [Google Scholar]
- Kanadia, R.N., Johnstone, K.A., Mankodi, A., Lungu, C., Thornton, C.A., Esson, D., Timmers, A.M., Hauswirth, W.W., Swanson, M.S. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia, R.N., Shin, J., Yuan, Y., Beattie, S.G., Wheeler, T.M., Thornton, C.A., Swanson, M.S. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsunky, H., Zeng, H., Schmidt, T., Zevnik, B., Kluge, R., Schmid, K.W., Duhrsen, U., Moroy, T. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 2002;30:295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- Kemp, P.R., Ellis, P.D., Smith, C.W. Visualization of alternative splicing in vivo. Methods. 2005;37:360–367. doi: 10.1016/j.ymeth.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Kim, S., Shi, H., Lee, D.K., Lis, J.T. Specific SR protein-dependent splicing substrates identified through genomic SELEX. Nucleic Acids Res. 2003;31:1955–1961. doi: 10.1093/nar/gkg286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozieradzki, I., Kundig, T., Kishihara, K., Ong, C.J., Chiu, D., Wallace, V.A., Kawai, K., Timms, E., Ionescu, J., Ohashi, P., et al. T cell development in mice expressing splice variants of the protein tyrosine phosphatase CD45. J. Immunol. 1997;158:3130–3139. [PubMed] [Google Scholar]
- Krawczak, M., Reiss, J., Cooper, D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Krecic, A.M., Swanson, M.S. hnRNP complexes: Composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Ladd, A.N., Stenberg, M.G., Swanson, M.S., Cooper, T.A. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 2005a;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- Ladd, A.N., Taffet, G., Hartley, C., Kearney, D.L., Cooper, T.A. Cardiac tissue-specific repression of CELF activity disrupts alternative splicing and causes cardiomyopathy. Mol. Cell. Biol. 2005b;25:6267–6278. doi: 10.1128/MCB.25.14.6267-6278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity, J.H., Chung, J., Dyson, H.J., Wright, P.E. Alternative splicing of Wilms' tumor suppressor protein modulates DNA binding activity through isoform-specific DNA-induced conformational changes. Biochemistry. 2000;39:5341–5348. doi: 10.1021/bi9926678. [DOI] [PubMed] [Google Scholar]
- Lemaire, R., Prasad, J., Kashima, T., Gustafson, J., Manley, J.L., Lafyatis, R. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: A novel function for SR proteins. Genes & Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P., Green, R.E., Brenner, S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X., Miller, J.W., Mankodi, A., Kanadia, R.N., Yuan, Y., Moxley, R.T., Swanson, M.S., Thornton, C.A. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Lindgren, N., Usiello, A., Goiny, M., Haycock, J., Erbs, E., Greengard, P., Hokfelt, T., Borrelli, E., Fisone, G. Distinct roles of dopamine D2L and D2S receptor isoforms in the regulation of protein phosphorylation at presynaptic and postsynaptic sites. Proc. Natl. Acad. Sci. 2003;100:4305–4309. doi: 10.1073/pnas.0730708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman, D., Johnstone, I.L., Caceres, J.F. Functional characterization of SR and SR-related genes in Caenorhabditis elegans . EMBO J. 2000;19:1625–1637. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, K.W. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- Lynch, K.W., Weiss, A. A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol. Cell. Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Tasic, B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Mankodi, A., Logigian, E., Callahan, L., McClain, C., White, R., Henderson, D., Krym, M., Thornton, C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- Mankodi, A., Urbinati, C.R., Yuan, Q.P., Moxley, R.T., Sansone, V., Krym, M., Henderson, D., Schalling, M., Swanson, M.S., Thornton, C.A. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Mankodi, A., Takahashi, M.P., Jiang, H., Beck, C.L., Bowers, W.J., Moxley, R.T., Cannon, S.C., Thornton, C.A. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- Matlin, A.J., Clark, F., Smith, C.W. Understanding alternative splicing: Toward a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Miller, J.W., Urbinati, C.R., Teng-Umnuay, P., Stenberg, M.G., Byrne, B.J., Thornton, C.A., Swanson, M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet, I., Barbosa-Morais, N.L., Andrade, J., Carmo-Fonseca, M. Diversity of human U2AF splicing factors. FEBS J. 2006;273:4807–4816. doi: 10.1111/j.1742-4658.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- Muro, A.F., Chauhan, A.K., Gajovic, S., Iaconcig, A., Porro, F., Stanta, G., Baralle, F.E. Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J. Cell Biol. 2003;162:149–160. doi: 10.1083/jcb.200212079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvy, S., Louis-Dit-Sully, C., Cooper, J., Cassady, R.L., Alexander, D.R., Holmes, N. Either of the CD45RB and CD45RO isoforms are effective in restoring T cell, but not B cell, development and function in CD45-null mice. J. Immunol. 2003;171:1792–1800. doi: 10.4049/jimmunol.171.4.1792. [DOI] [PubMed] [Google Scholar]
- Oppermann, M., Mizel, D., Huang, G., Li, C., Deng, C., Theilig, F., Bachmann, S., Briggs, J., Schnermann, J., Castrop, H. Macula densa control of renin secretion and preglomerular resistance in mice with selective deletion of the B isoform of the Na, K, 2Cl co-transporter. J. Am. Soc. Nephrol. 2006;17:2143–2152. doi: 10.1681/ASN.2006040384. [DOI] [PubMed] [Google Scholar]
- Oppermann, M., Mizel, D., Kim, S.M., Chen, L., Faulhaber-Walter, R., Huang, Y., Li, C., Deng, C., Briggs, J., Schnermann, J., et al. Renal function in mice with targeted disruption of the A isoform of the Na-K-2Cl co-transporter. J. Am. Soc. Nephrol. 2007;18:440–448. doi: 10.1681/ASN.2006091070. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger, A., Bedford, M.T., Burakova, T., Arman, E., Zimmer, Y., Yayon, A., Givol, D., Lonai, P. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2) Dev. Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- Pascual, M., Vicente, M., Monferrer, L., Artero, R. The Muscleblind family of proteins: An emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Periasamy, M., Reed, T.D., Liu, L.H., Ji, Y., Loukianov, E., Paul, R.J., Nieman, M.L., Riddle, T., Duffy, J.J., Doetschman, T., et al. Impaired cardiac performance in heterozygous mice with a null mutation in the sarco(endo)plasmic reticulum Ca2+-ATPase isoform 2 (SERCA2) gene. J. Biol. Chem. 1999;274:2556–2562. doi: 10.1074/jbc.274.4.2556. [DOI] [PubMed] [Google Scholar]
- Peters, K., Werner, S., Liao, X., Wert, S., Whitsett, J., Williams, L. Targeted expression of a dominant negative FGF receptor blocks branching morphogenesis and epithelial differentiation of the mouse lung. EMBO J. 1994;13:3296–3301. doi: 10.1002/j.1460-2075.1994.tb06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps, D., Celotto, A.M., Wang, Q.Q., Tarng, R.S., Graveley, B.R. Arginine/serine repeats are sufficient to constitute a splicing activation domain. Nucleic Acids Res. 2003;31:6502–6508. doi: 10.1093/nar/gkg845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potashkin, J., Naik, K., Wentz-Hunter, K. U2AF homolog required for splicing in vivo. Science. 1993;262:573–575. doi: 10.1126/science.8211184. [DOI] [PubMed] [Google Scholar]
- Revest, J.M., Spencer-Dene, B., Kerr, K., De Moerlooze, L., Rosewell, I., Dickson, C. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev. Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- Ring, H.Z., Lis, J.T. The SR protein B52/SRp55 is essential for Drosophila development. Mol. Cell. Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshon, M.J., Ruley, H.E. Hypomorphic mutation in hnRNP U results in post-implantation lethality. Transgenic Res. 2005;14:179–192. doi: 10.1007/s11248-004-8147-8. [DOI] [PubMed] [Google Scholar]
- Rothrock, C., Cannon, B., Hahm, B., Lynch, K.W. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Rothrock, C.R., House, A.E., Lynch, K.W. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner, D.Z., Kanaar, R., Breger, K.S., Rio, D.C. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc. Natl. Acad. Sci. 1996;93:10333–10337. doi: 10.1073/pnas.93.19.10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, P., Wiles, M.V., Imhof, B.A. Alpha 6 integrins participate in pro-T cell homing to the thymus. Eur. J. Immunol. 1995;25:2034–2041. doi: 10.1002/eji.1830250735. [DOI] [PubMed] [Google Scholar]
- Savkur, R.S., Philips, A.V., Cooper, T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Sazani, P., Gemignani, F., Kang, S.H., Maier, M.A., Manoharan, M., Persmark, M., Bortner, D., Kole, R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- Schaal, T.D., Maniatis, T. Selection and characterization of pre-mRNA splicing enhancers: Identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, T.S., Sanders, L.K., Nathans, D. Cooperative transcriptional activity of Jun and Stat3 β, a short form of Stat3. Proc. Natl. Acad. Sci. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk, C., Schulze-Osthoff, K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Sheives, P., Lynch, K.W. Identification of cells deficient in signaling-induced alternative splicing by use of somatic cell genetics. RNA. 2002;8:1473–1481. [PMC free article] [PubMed] [Google Scholar]
- Shepard, J., Reick, M., Olson, S., Graveley, B.R. Characterization of U2AF6, a splicing factor related to U2AF35 . Mol. Cell. Biol. 2002;22:221–230. doi: 10.1128/MCB.22.1.221-230.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, J.H., Talley, E.M., Bayliss, D.A., Berger, A.J. Development of glycinergic synaptic transmission to rat brain stem motoneurons. J. Neurophysiol. 1998;80:2608–2620. doi: 10.1152/jn.1998.80.5.2608. [DOI] [PubMed] [Google Scholar]
- Singh, S., Mishra, R., Arango, N.A., Deng, J.M., Behringer, R.R., Saunders, G.F. Iris hypoplasia in mice that lack the alternatively spliced Pax6(5a) isoform. Proc. Natl. Acad. Sci. 2002;99:6812–6815. doi: 10.1073/pnas.102691299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srebrow, A., Kornblihtt, A.R. The connection between splicing and cancer. J. Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- Stalmans, I., Ng, Y.S., Rohan, R., Fruttiger, M., Bouche, A., Yuce, A., Fujisawa, H., Hermans, B., Shani, M., Jansen, S., et al. Arteriolar and venular patterning in retinas of mice selectively expressing VEGF isoforms. J. Clin. Invest. 2002;109:327–336. doi: 10.1172/JCI14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke, R., Manley, J.L. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 1999;11:358–362. doi: 10.1016/S0955-0674(99)80050-7. [DOI] [PubMed] [Google Scholar]
- Takahashi, N., Chernavvsky, D.R., Gomez, R.A., Igarashi, P., Gitelman, H.J., Smithies, O. Uncompensated polyuria in a mouse model of Bartter's syndrome. Proc. Natl. Acad. Sci. 2000;97:5434–5439. doi: 10.1073/pnas.090091297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, M.H., Sun, Z., Opitz, S.L., Schmidt, T.E., Peters, J.H., George, E.L. Deletion of the alternatively spliced fibronectin EIIIA domain in mice reduces atherosclerosis. Blood. 2004;104:11–18. doi: 10.1182/blood-2003-09-3363. [DOI] [PubMed] [Google Scholar]
- Timchenko, N.A., Welm, A.L., Lu, X., Timchenko, L.T. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPβ mRNA and regulates translation of C/EBPβ isoforms. Nucleic Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko, N.A., Cai, Z.J., Welm, A.L., Reddy, S., Ashizawa, T., Timchenko, L.T. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- Timchenko, N.A., Patel, R., Iakova, P., Cai, Z.J., Quan, L., Timchenko, L.T. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- Trede, N.S., Medenbach, J., Damianov, A., Hung, L.H., Weber, G.J., Paw, B.H., Zhou, Y., Hersey, C., Zapata, A., Keefe, M., et al. Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc. Natl. Acad. Sci. 2007;104:6608–6613. doi: 10.1073/pnas.0701919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule, J., Ule, A., Spencer, J., Williams, A., Hu, J.S., Cline, M., Wang, H., Clark, T., Fraser, C., Ruggiu, M., et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Usiello, A., Baik, J.H., Rouge-Pont, F., Picetti, R., Dierich, A., LeMeur, M., Piazza, P.V., Borrelli, E. Distinct functions of the two isoforms of dopamine D2 receptors. Nature. 2000;408:199–203. doi: 10.1038/35041572. [DOI] [PubMed] [Google Scholar]
- van der Flier, A., Kuikman, I., Baudoin, C., van der Neut, R., Sonnenberg, A. A novel β 1 integrin isoform produced by alternative splicing: Unique expression in cardiac and skeletal muscle. FEBS Lett. 1995;369:340–344. doi: 10.1016/0014-5793(95)00814-p. [DOI] [PubMed] [Google Scholar]
- Venables, J.P. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Ver Heyen, M., Heymans, S., Antoons, G., Reed, T., Periasamy, M., Awede, B., Lebacq, J., Vangheluwe, P., Dewerchin, M., Collen, D., et al. Replacement of the muscle-specific sarcoplasmic reticulum Ca2+-ATPase isoform SERCA2a by the nonmuscle SERCA2b homologue causes mild concentric hypertrophy and impairs contraction-relaxation of the heart. Circ. Res. 2001;89:838–846. doi: 10.1161/hh2101.098466. [DOI] [PubMed] [Google Scholar]
- Vrhovski, B., Lemckert, F., Gunning, P. Modification of the tropomyosin isoform composition of actin filaments in the brain by deletion of an alternatively spliced exon. Neuropharmacology. 2004;47:684–693. doi: 10.1016/j.neuropharm.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Wang, H.Y., Xu, X., Ding, J.H., Bermingham J.R., Jr, Fu, X.D. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- Wang, J., Takagaki, Y., Manley, J.L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes & Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Xu, R., Sasaoka, T., Tonegawa, S., Kung, M.P., Sankoorikal, E.B. Dopamine D2 long receptor-deficient mice display alterations in striatum-dependent functions. J. Neurosci. 2000;20:8305–8314. doi: 10.1523/JNEUROSCI.20-22-08305.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J.H., Sirsi, S.R., Latta, D.R., Lutz, G.J. Induction of dystrophin expression by exon skipping in mdx mice following intramuscular injection of antisense oligonucleotides complexed with PEG-PEI copolymers. Mol. Ther. 2006;14:88–96. doi: 10.1016/j.ymthe.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Williamson, D.J., Banik-Maiti, S., DeGregori, J., Ruley, H.E. hnRNP C is required for postimplantation mouse development but is dispensable for cell viability. Mol. Cell. Biol. 2000;20:4094–4105. doi: 10.1128/mcb.20.11.4094-4105.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R., Parlow, A.F., Wang, Y. The effects of dopamine and D2 receptor antagonists on pituitary hormone secretion are intact in mice lacking dopamine D2L receptor. Brain Res. 2002;939:95–99. doi: 10.1016/s0006-8993(02)02559-3. [DOI] [PubMed] [Google Scholar]
- Xu, X., Fu, X.D. Conditional knockout mice to study alternative splicing in vivo. Methods. 2005;37:387–392. doi: 10.1016/j.ymeth.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Xu, X., Yang, D., Ding, J.H., Wang, W., Chu, P.H., Dalton, N.D., Wang, H.Y., Bermingham J.R., Jr, Ye, Z., Liu, F., et al. ASF/SF2-regulated CaMKIIδ alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Yeo, G., Holste, D., Kreiman, G., Burge, C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, J.Y., Huso, D.L., Nathans, D., Desiderio, S. Specific ablation of Stat3β distorts the pattern of Stat3-responsive gene expression and impairs recovery from endotoxic shock. Cell. 2002;108:331–344. doi: 10.1016/s0092-8674(02)00636-0. [DOI] [PubMed] [Google Scholar]
- Zeng, H., Yucel, R., Kosan, C., Klein-Hitpass, L., Moroy, T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23:4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio, D.A., Blumenthal, T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans . RNA. 1999;5:487–494. doi: 10.1017/s1355838299982225. [DOI] [PMC free article] [PubMed] [Google Scholar]