INTRODUCTION
Information retrieval for life science research (a broad rubric encompassing many traditional disciplines such as biochemistry, botany, cell biology, and molecular biology [1]) often involves the use of combinations of multiple information resources. Such combinations have been called “workflows” [2, 3] and may include factual databases such as Genbank [4], literature databases such as Entrez-PubMed [5], and analysis tools such as the Basic Local Alignment Search Tool (BLAST) [6]. Information resources can be combined in different ways toward the same goal; varying combinations may produce different results for the same research question. Combinations that produce different results may appear equivalent to a scientifically sophisticated user who lacks knowledge of metadata about the resources that may indicate the possibility of varying results. In addition, a user who pursues only a single combination of resources may not even realize that another combination might produce different results.
This study's objective was to compare the results of three intuitively plausible and seemingly similar workflows for retrieving gene function information, with the goal of illustrating the importance of library science in bioinformatics and the need for a multidisciplinary team approach to authoring, vetting, and using life science workflows.
METHODS
Microarray analysis is a high-throughput experimental technique that engenders significant information retrieval requirements [7]. One use of microarrays is analyzing gene expression: raw data from the microarray are statistically analyzed to determine which genes show significant changes in expression, with one or more lists of genes as the final result. Interpreting the biological meaning of this result often necessitates retrieving information from other sources about the function of the listed genes. Microarray analysis, therefore, is one example of a domain in which information from the biological literature must be integrated with information contained in sequence and other databases.
For some microarray analyses, each gene has a related representative DNA sequence. The identifier of that DNA sequence (its nucleotide sequence accession number, hereafter, “accession number”) may be used to search for information about the function of the associated gene. This study compared three workflows that used accession numbers as starting points and utilized linkages among PubMed and other Entrez databases [8]. Although using accession numbers to search for gene function information has problems [9], the workflows compared here have been selected as simple, intuitively plausible strategies similar to some of those the authors have seen used in practice. Other workflows, using other starting points or information resources, are also possible and potentially useful.
This study used a list of 251 accession numbers representing genes determined to be of interest in a microarray experiment related to muscle recovery after immobilization (NIH grant AG18881) [10–12]. The genes on the list represented an example of real-world microarray results for which researchers might need to retrieve gene function information. The list of accession numbers was used as the test-set against which workflows were executed and their results compared.
Description of the three workflows
The three workflows are depicted in Figure 1 (available online). Each starts with an accession number (e.g., M29293), denoted as “xxxxxxx.”
Workflow 1: PubMed only
The Entrez-PubMed “Secondary Source” or SI field (which identifies secondary data sources and associated accession numbers discussed in MEDLINE articles) [13] was searched using a query of the form genbank/xxxxxxx[si]. The result was a set of PubMed records, represented here as a set of PubMed IDs (PMIDs). For example, the query “genbank/ M29293[si]” retrieved PMID 2532363.
Workflow 2: Nucleotide-PubMed
Entrez-Nucleotide [14] was searched using a query of the form “xxxxxxx” and retrieved nucleotide sequence records that might provide links to other resources. Two types of links, PubMed links and PubMed Central links, were pursued. PubMed links led to Entrez-PubMed and a set of PMIDs. PubMed Central links led to PubMed Central (the Entrez full-text repository) [15] and a set of PubMed Central records. These records had a PubMed links option, which provided a set of PMIDs corresponding to the PubMed Central records. For example, the query “M29293” led, via the PubMed links, to PMID 2532363 and via the PubMed Central links, to PMIDs 15644144 and 2532363.
Workflow 3: Gene-PubMed
Entrez-Gene [16] was searched using a query of the form “xxxxxxx[NACC]” ([NACC] was used to unambiguously declare xxxxxxx an accession number). The result was the record for a gene that might provide links to other resources. As before, only PubMed links and PubMed Central links were pursued. For example, the query “M29293 [NACC]” retrieved an entry for the gene Snrpn. That gene entry included both PubMed links and PubMed Central links. In this example, both the PubMed links and the PubMed Central links led to PMIDs 12477932 and 2532363.
Workflow comparison procedures
Previously, the 251 accession numbers were searched using Java implementations of the 3 workflows, and results were partially reported [17]. Between July 14 and 24, 2006, the search results were manually verified and updated. For each workflow, the PMIDs retrieved by each accession number were recorded. For workflows 2 and 3, whether the PMIDs could be retrieved via the PubMed links or PubMed Central links was also recorded.
Three aspects of the workflows were compared: which and how many accession numbers successfully retrieved one or more PMIDs, which and how many PMIDs were retrieved, and which and how many unique pairings between a particular accession number and a particular PMID (hereafter, “accession number–PMID pairings”) were produced. The overall output of each of the three workflows was compared. In addition, for workflows 2 and 3, the results of following the PubMed links and PubMed Central links paths were compared. Because workflow 1 involved direct search of PubMed, this workflow had no alternative paths to the literature.
Statistical analysis
Agreement between pairs of workflows was assessed using Cohen's kappa [18] (denoted Κ). Statistical calculations were performed using SPSS [19]. The P value for each individual comparison was multiplied by nine to adjust for multiple comparisons [20]; adjusted P values < 0.05 were considered significant. Significant comparisons were interpreted as suggested by Byrt [18].
RESULTS
Tables 1, 2, and 3 present the aggregate study results. Figures 2, 3, and 4 present the results of comparisons 1, 2, and 3, respectively.
Table 1 Workflow results and comparisons
Table 2 Assessment of agreement between workflows
Table 3 Comparison of alternate paths within workflows
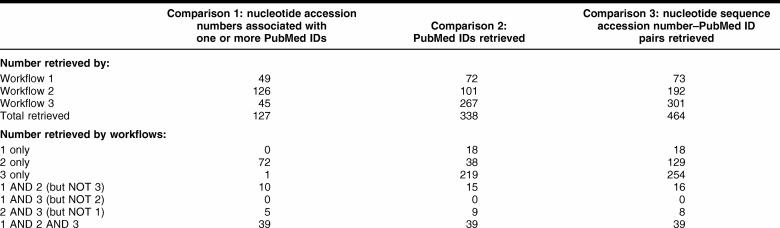
Comparison 1: Which and how many accession numbers were successfully used to retrieve one or more PubMed IDs (PMIDs) using the different workflows?
Overall results
PMIDs were associated with 127 accession numbers: 49 by workflow 1, 126 by workflow 2, and 45 by workflow 3. In terms of overlap, 39 accession numbers were associated with PMIDs by all 3 workflows.
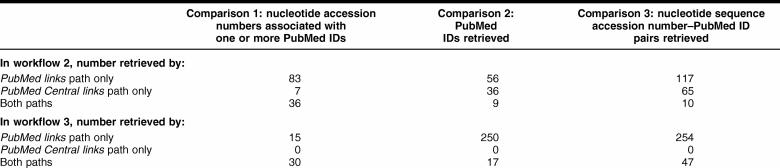
PubMed links and PubMed Central links paths
In workflow 2, 83 accession numbers were associated with PMIDs via PubMed links only, 7 via PubMed Central links only, and 36 via both. In workflow 3, 15 accession numbers were associated with PMIDs via PubMed links only, none via PubMed Central links only, and 30 via both.
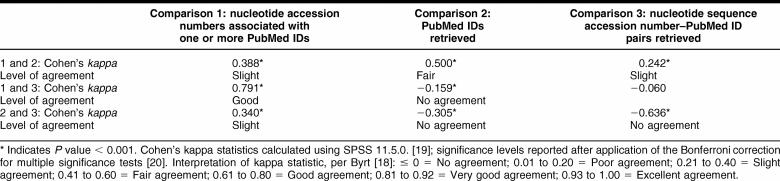
Agreement between workflows
Agreement between workflows was assessed regarding the accession numbers for which they retrieved PMIDs. The agreement between workflows 1 and 2 (Κ = 0.388, P < 0.001) and between 2 and 3 (Κ = 0.340, P < 0.001) was slight. Workflows 2 and 3 showed good agreement (Κ = 0.791, P < 0.001).
Comparison 2: Which and how many PMIDs were retrieved using the different workflows?
Overall results
A total of 338 PMIDs were retrieved: 72 by workflow 1, 101 by workflow 2, and 267 by workflow 3. Thirty-nine PMIDs were retrieved by all 3 workflows.
PubMed links and PubMed Central links paths
Workflow 2 retrieved 56 PMIDs via PubMed links only, 36 via PubMed Central links only, and 9 via both. In workflow 3, 250 PMIDs were retrieved via PubMed links only, none via PubMed Central links only, and 17 via both.
Agreement between the workflows
Agreement between workflows was assessed regarding which PMIDs they retrieved. The agreement between workflows 1 and 2 was fair (Κ = 0.500, P < 0.001). There was no agreement between workflows 1 and 3 (Κ = −0.159, P < 0.001) or between workflows 2 and 3 (Κ = −0.305, P < 0.001).
Comparison 3: Which and how many accession number–PMID pairs were produced using the different workflows?
A workflow results in an accession number–PMID pairing when inputting the accession number to the workflow retrieves the PMID. The purpose of the workflows here was to retrieve literature on the function of the genes associated with each of the accession numbers; therefore, the accession number–PMID pairings were of particular interest.
Overall results
A total of 464 distinct accession number–PMID pairs were retrieved: 73 from workflow 1, 192 from workflow 2, and 301 from workflow 3. Overlap between the 3 workflows was fairly low, including 39 pairs resulting from all 3 workflows.
PubMed links and PubMed Central links paths
In workflow 2, 117 accession number–PMID pairs resulted from the PubMed links only, 65 resulted from the PubMed Central links only, and 10 pairs resulted from both paths. In workflow 3, 254 pairs resulted from the PubMed links only, none resulted from the PubMed Central links only, and 47 resulted from both paths.
Agreement between the workflows
Agreement between workflows was assessed regarding which accession number–PMID pairings they produced. The agreement between workflows 1 and 2 was slight (Κ = 0.242, P < 0.001). There was no agreement between workflows 2 and 3 (Κ = −0.636, P < 0.001), and the comparison between workflows 1 and 3 was not statistically significant.
DISCUSSION
The results show the three workflows are neither strictly equivalent nor even nearly equivalent in the sense of strong agreement or overlapping of results. The significant differences among the workflows might surprise an otherwise scientifically sophisticated user who is not an expert in the use of these information resources.
In this case, the existing Help documentation for the information resources can account for differences in the workflow output. The PubMed Secondary Source or SI field documentation accounts for differences between workflows 1 and 2. According to PubMed's Help information [13], the SI field and the PubMed links to GenBank are generated differently and are themselves not linked. The SI field identifies GenBank accession numbers discussed in MEDLINE articles, while the GenBank reference field (which for a given record includes citations that discuss the associated sequence) is used to create the PubMed links to GenBank. The Entrez Gene documentation accounts for differences between workflow 3 and workflows 1 and 2. The Entrez Gene PubMed Links documentation indicates that some Entrez Gene PubMed links are generated from GeneRIFs, as indicated by the PubMed (GeneRIF) option [21], and that the GeneRIF mechanism is a way to let scientists themselves add to the functional annotation of genes [22].
Although such documentation is available, the biologist using or designing workflows may not know about it. It is no more reasonable to expect biologists to be experts in the metadata of biological information resources than it is to expect librarians to be experts in biology. Thus, because even simple, apparently similar information retrieval workflows may produce different results, a multidisciplinary team approach to authoring, vetting, and using life science workflows is needed. Such teams must include experts in the primary science and experts in the metadata characterizing the information resources.
The importance of librarians as metadata experts in life science research was recognized by the Human Genome Project in 1997 [23]. Unfortunately, almost a decade later, the library remains largely excluded from the mainstream of life science research: very few universities offer bioinformatics end-user support services through the library [24]; demand is generally not great for such services when offered [25]; and molecular biology students in particular do not choose the library as their preferred source of information about bioinformatics databases [26].
The life science information space is growing extremely rapidly, largely facilitated by “the breakdown of the traditional barriers between academic disciplines and the application of technologies across these disciplines” [27]. Similarly, breaking down the barriers between “scientist” and “librarian” and fostering the interdisciplinary and synergistic combination of their respective expertise in the development and use of life science workflows are crucial to achieving full and optimal exploitation of the life science information space.
Supplementary Material
Footnotes
* Based on a presentation at MLA '06, the 106th Annual Meeting of the Medical Library Association, Phoenix, AZ, May 19–24, 2006.
† This research was supported by a Medical Library Association Donald A. B. Lindberg Research fellowship and the National Library of Medicine Biomedical and Health Informatics Research Training grant 2-T15-LM07089-11.
Supplemental Figures 1, 2, 3, and 4 are available with the online version of this journal.
REFERENCES
- EverythingBio. Definition of life science. [Web document]. EverythingBio.com. [cited 25 Jan 2007]. <http://www.everythingbio.com/glos/definition.php?word=life+science>. [Google Scholar]
- Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, and Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006 Jul. 1;34. (Web server issue). W729–W732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, and Li P. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004 Nov 22;20(17). 3045–54. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, and Wheeler DL. GenBank. Nucleic Acids Res. 2005 Jan 1; 33(database issue):D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US National Library of Medicine,. National Center for Biotechnology Information. PubMed overview. [Web document]. Bethesda, MD: The Library. [rev. 30 Jun 2006; cited 14 Aug 2006]. <http://www.ncbi.nlm.nih.gov/entrez/query/static/overview.html>. [Google Scholar]
- Ye J, McGinnis S, and Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006 Jul 1;34(Web server issue). W6–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masys DR. Linking microarray data to the literature. Nat Genet. 2001 May; 28(1):9–10. [DOI] [PubMed] [Google Scholar]
- US National Library of Medicine,. National Center for Biotechnology Information. Databases. [Web document]. Bethesda, MD: The Library. [rev. 17 Jan 2006; cited 1 Dec 2006]. <http://www.ncbi.nlm.nih.gov/Database/>. [Google Scholar]
- Xuan W, Watson SJ, and Meng F. GeneInfoMiner—a Web server for exploring biomedical literature using batch sequence ID. Bioinformatics 2005 21(16):3452–3. [DOI] [PubMed] [Google Scholar]
- Pattison JS, Folk LC, Madsen RW, Childs TE, and Booth FW. Transcriptional profiling identifies extensive downregulation of extracellular matrix gene expression in sarcopenic rat soleus muscle. Physiol Genomics. 2003 Sep 29; 15(1):34–43. [DOI] [PubMed] [Google Scholar]
- Pattison JS, Folk LC, Madsen RW, Childs TE, Spangenburg EE, and Booth FW. Expression profiling identifies dysregulation of myosin heavy chains IIb and IIx during limb immobilization in the soleus muscles of old rats. J Physiol. 2003 Dec 1; 553(pt 2):357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison JS, Folk LC, Madsen RW, and Booth FW. Selected contribution: identification of differentially expressed genes between young and old rat soleus muscle during recovery from immobilization-induced atrophy. J Appl Physiol. 2003 Nov; 95(5):2171–9. [DOI] [PubMed] [Google Scholar]
- US National Library of Medicine,. National Center for Biotechnology Information. PubMed help. [Web document]. Bethesda, MD: The Library. [rev. 8 Aug 2006; cited 14 Aug 2006]. <http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=helppubmed.chapter.pubmedhelp>. [Google Scholar]
- US National Library of Medicine,. National Center for Biotechnology Information. Entrez nucleotide. [Web document]. Bethesda, MD: The Library. [rev. 17 Jan 2006; cited 14 Aug 2006]. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide>. [Google Scholar]
- US National Library of Medicine,. National Center for Biotechnology Information. PubMed Central overview. [Web document]. Bethesda, MD: The Library. [rev. Jan 7 2005; cited Aug 17 2006]. <http://www.pubmedcentral.nih.gov/about/intro.html>. [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, and Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005 Jan 1; 33(database issue):D54–D58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick T, Folk LC, and Craven CK. Asymmetries in retrieval of gene function information. Presented at: MLA '06, 106th Annual Meeting of the Medical Library Association, “Transformations A-Z”; Phoenix, AZ; May 19–24, 2006. [Google Scholar]
- Byrt T. How good is that agreement? Epidemiology. 1996 Sep; 7(5):561. [DOI] [PubMed] [Google Scholar]
- SPSS. SPSS for Windows, release 11.5.0. SPSS, 2002. [Google Scholar]
- Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ 1995 Jan;310(6973);170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US National Library of Medicine,. National Center for Biotechnology Information. Entrez Gene help: integrated access to genes of genomes in the reference sequence collection: finding data related to Entrez gene in other Entrez databases. [Web document]. Bethesda, MD: The Library. [rev. 13 Nov 2006; cited 30 Nov 2006]. <http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=helpgene.section.EntrezGene.Finding_Data_Related>. [Google Scholar]
- US National Library of Medicine,. National Center for Biotechnology Information. GeneRIF—Gene Reference Into Function. [Web document]. Bethesda, MD: The Library. [rev. 30 Nov 2006; cited 30 Nov 2006]. <http://www.ncbi.nlm.nih.gov/projects/GeneRIF/GeneRIFhelp.html>. [Google Scholar]
- Human genome project [report no.]. JSR-97–315. [Web document]. The Mitre Corporation. [rev. 1997; cited 29 Jan 2007]. <http://www.ornl.gov/sci/techresources/Human_Genome/publicat/miscpubs/Jason/>. [Google Scholar]
- Messersmith DJ, Benson DA, and Geer RC. A Web-based assessment of bioinformatics end-user support services at US universities. J Med Libr Assoc. 2006 Jul; 94(3):299–305.E156– E187. [PMC free article] [PubMed] [Google Scholar]
- Geer RC. Broad issues to consider for library involvement in bioinformatics. J Med Libr Assoc. 2006 Jul; 94(3):286–98.E152–E155. [PMC free article] [PubMed] [Google Scholar]
- Brown C.. Where do molecular biology graduate students find information? Sci Technol Libr. 2005;25(3):89–104. [Google Scholar]
- Welsh E, Jirotka M, and Gavaghan D. Post-genomic science: cross-disciplinary and large-scale collaborative research and its organizational and technological challenges for the scientific research process. Philos Trans Roy Soc Lond A. 2006 Jun 15; 364(1843):1533–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.