Abstract
The Drosophila splicing factor RBP1 participates together with TRA and TRA-2 in the regulation of alternative splicing of doublesex (dsx) pre-mRNA. It does so by recognizing RBP1 RNA target sequences in the dsx pre-mRNA. RBP1 belongs to the Ser–Arg-rich (SR) protein family of splicing factors, which have in common a N-terminal RNA recognition motif-type RNA binding domain, a Gly-rich region, and a C-terminal SR domain. Using a tissue culture transfection assay, we demonstrate that the Gly residues within the Gly-rich domain, the ribonucleoprotein motifs within the RNA recognition motif RNA binding domain, and the SR domain are required for regulation of dsx splicing by RBP1 in vivo. Furthermore, using a two-hybrid system, we show protein–protein interactions between RBP1 and itself and between RBP1 and TRA-2. The SR domain and the Gly residues within the Gly-rich domain of RBP1 were found to be involved in these protein–protein interactions. Our results suggest that RBP1 and TRA-2 function in regulation of dsx splicing by forming a complex.
Keywords: alternative pre-mRNA splicing, Ser–Arg-rich protein, protein–protein interaction, sex determination
Regulation of alternative pre-mRNA splicing involves both trans-acting protein factors and cis-acting sequence elements. Among the few trans-acting factors identified so far are the Ser–Arg-rich (SR) proteins, a family of splicing factors, several of which are conserved from Drosophila to humans. The SR protein family contains at least six evolutionary conserved members (1), which are known as SRp20 [RBP1 in Drosophila (2), X16 in mice (3)], SRp30a [ASF/SF2 in humans (4, 5)], SRp30b [SC35 in humans (6, 7)], SRp40 (8, 9), SRp55 [B52 in Drosophila (10, 11)], and SRp75 (12). Several additional SR proteins have been described recently (9, 13). SR proteins have been shown to regulate various splicing decisions both in vitro and in vivo. These include splicing of Drosophila doublesex (dsx) pre-mRNA (14, 15), simian virus 40 early pre-mRNA (16), adenovirus E1A pre-mRNA (2), troponin T pre-mRNA (17), and thalassemic human β-globin pre-mRNA (18, 19). In addition to involvement in alternative splicing, SR proteins can also function as general splicing factors in vitro (4, 20).
A common feature of the SR proteins is their domain structure. The SR proteins, including RBP1, share a N-terminal RNA recognition motif (RRM)-type RNA binding domain, a Gly-rich region, and a C-terminal SR domain (1). In vitro, the RRM domain, which is also found in numerous other RNA binding proteins, is required for binding of ASF/SF2 to RNA (35, 39). SR domains, which are highly phosphorylated, have been reported to be involved in protein–protein interactions (23) and in the intranuclear localization of Drosophila SU(WA) and TRA proteins to nuclear speckles, regions of the nucleus enriched in splicing factors (24). The function of the Gly-rich region is unknown.
The identification of the functional domains in SR proteins necessary for the regulation of alternative splicing can help to understand which molecular interactions are required in splicing regulation. Both binding of SR proteins to their target pre-mRNAs and protein–protein interactions of SR proteins with both regulatory and general splicing factors are expected to be important in determining splice site choices. These ideas are supported by (i) the finding that RNA target sequences selected by the SR proteins RBP1, ASF/SF2, and SC35 in vitro are present in the proximity of regulated splice sites and are required for splicing regulation by these SR proteins (15, 25) and (ii) by evidence that SR proteins can form homooligomeric and heterooligomeric complexes and can interact with other splicing factors like TRA, TRA-2, and U1-70K (21, 23). Consequently, both the RRM domains and SR domains of SR proteins could be essential for regulation of alternative splicing. To date, the domains of SR proteins that are required for regulation of alternative splicing in vivo are unknown.
To address the question of functional domains in splicing regulators in vivo, we have employed regulation of alternative splicing of the Drosophila dsx pre-mRNA by the Drosophila SR protein RBP1 as an experimental system. The dsx gene plays an important role in Drosophila sex determination by expressing male-specific and female-specific DSX proteins that regulate sexual differentiation (22). The dsx expression pattern is regulated at the level of sex-specific alternative splicing, leading to the incorporation of a female-specific exon into the dsx mRNA in females. In males, the female exon is spliced out and male-specific exons are incorporated instead. Regulation of these splicing decisions occurs at the female-specific 3′ splice site, which is selectively activated in females (26–28). RBP1 has been shown to participate in the activation of the female-specific 3′ splice site in vivo by binding to RBP1 RNA target sequences located in the proximity of the female-specific 3′ splice site. In Drosophila tissue culture cells, overexpression of RBP1 activates female-specific dsx splicing, and mutations within the RBP1 target sequences inhibit activation by RBP1 (15). Activation of the female-specific 3′ splice site also involves the factors TRA and TRA-2 in Drosophila (26–28). dsx repeat elements recognized by TRA and TRA-2 are found close to or overlap with RBP1 RNA target sequences within the dsx repeat region (15), and in the presence of TRA and TRA-2 the RBP1 protein can be cross-linked to dsx repeat elements (29). These observations raise the question whether RBP1, TRA, and TRA-2 function by forming a complex.
The RBP1 tissue culture system (15) is well suited to study structural elements of an SR protein essential for the regulation of alternative splicing in vivo. Here, we present an in vivo analysis of the regions of RBP1 required for the regulation of dsx alternative splicing. Testing of RBP1 variants in the transfection assay allowed us to identified elements essential for splicing regulation within the RRM domain, the Gly-rich region, and the SR domain of RBP1. We show that, on the molecular level, the SR domain and elements within the Gly-rich region affect protein–protein interactions of RBP1 with TRA-2 and with RBP1 in the two-hybrid system.
MATERIALS AND METHODS
Plasmid Constructs.
All mutant RBP1 transfection constructs are based on derivatives of the rbp1-a cDNA (2). ΔSR was generated by PCR using the primers 5′-ATCATATGCCGCGATATAGGGA-3′ and 5′-GCGGATCCTTACCTGTAGCGTCCGGAACC-3′. ΔRRM-Gly was generated by PCR using the primers 5′-ATCATATGTCACGTTCGCCACGTC-3′ and 5′-GCGGATCCTTAATGACGATCCCGAGAATC-3′. ΔPvu was generated by deleting PvuI fragments between nucleotides 968 and 2310 of the rbp1 sequence. Δ115-135, Δ115-124, Δ125-135, Δ107-114, Sub 109,111-115, +1, +2, +3, +3 SRSR, and +1 R-K were generated by inserting synthetic fragments between the BspEI and EcoRV sites of the rbp1 cDNA. The following primer pairs were used to generate synthetic fragments; Δ115-135, 5′-GTTCCGGACGCTACAGGTCACGTTCGCCACGTCGCTC-3′ and 5′-ACGATATCCTTTTGGAAACATTATCGGGAGCGACGTGGCG-3′; Δ115-124, 5′-GTTCCGGACGCTACAGGTCACGTTCGCCACGTCGCTC-3′ and 5′-ACGATATCCTTTTGGAAACATTAATGACGATCCCGAGAATCCGAGCGACTTCGACGTCGGGAGCGACGTGGCG-3′; Δ125-135, 5′-GTTCCGGACGCTACAGGTCACGTTCGCCACGTCGCTC-3′ and 5′-ACGATATCCTTTTGGAAACATTAATCGCGCGAGAAGCTGCGGCTGCGGGGCGATCGGGAGCGACGTGGCG-3′; Δ107-114, 5′-GTTCCGGACGCTACAGGTCGCCCCGCAGCCGCAGCTTCTG-3′ and 5′-ACGATATCCTTTTGGAAACATTAATGACGATCCCGAGAATCCGAGCGACTTCGACGATCGCGCGAGAAGCTGCGGC-3′; Sub 109,111-115, 5′-GTTCCGGACGCTACAGGTCACGTCGTCCATCGTCCCGCTCGCGACCCCGCAGCCGCAGCTTCTCG-3′ and 5′-ACGATATCCTTTTGGAAACATTAATGACGATCCCGAGAATCCGAGCGACTTCGACGATCGCGCGAGAAGCTGCGGC-3′; +1, 5′-GTTCCGGACGCTACAGGTCATAATGTTTCCAAAAGG-3′ and 5′-ACGATATCCTTTTGGAAACATTA-3′; +2, 5′-GTTCCGGACGCTACAGGTCACGTTAATGTTTCCAAAAGG-3′ and 5′-ACGATATCCTTTTGGAAACATTA-3′; +3, 5′-GTTCCGGACGCTACAGGTCACGTTCGTAATGTTTCCAAAAGG-3′ and 5′-ACGATATCCTTTTGGAAACATTA-3′; and +3 SRSR, 5′-GTTCCGGACGCTACTCAAGGTCGCGTTAATGTTTCCAAAAGG-3′ and 5′-ACGATATCCTTTTGGAAACATTA-3′. Constructs Gly S-T, Gly R-K, Gly G-A and Sub ribonucleoprotein (RNP) were generated by site-directed mutagenesis using the U.S.E. mutagenesis system (Pharmacia). The following mutagenic primers were used: Gly S-T, 5′-ACCTGTAGCGTCCGGTACCAGTGCGACCAGTAGTGCCGCCTTCTCCGCGCCGGCGATCGCGCGTGCGACCCGAAGACAT-3′; Gly R-K, 5′-ACGTGGCGAACGTGACTTGTACTTTCCGGAACCAGACTTACCACTACTGCCGCCTTCTCCCTTCTTCTTATCCTTCGACTTACCCGAAGACATCTC-3′; Gly G-A, 5′-GTGACCTGTAGCGTGCGGAAGCAGAGCGAGCACTACTGGCGGCTTCTGCGCGCCGGCGATCGCGCGAGCGAGCCGAAGACATCTCTA-3′; and Sub RNP, 5′-GTCACGGCGATCCTCGTCTTCGACATCGGCGTCACCTGGTGGATTGC-3′ and 5′-CCAGGTTTTCCCACGTCCACCTTGCAGGCC-3′. Construct +1 R-K was made by inserting the synthetic BspEI–EcoRV fragment of construct +1 into construct Gly R-K. All cDNA mutations were confirmed by sequencing and built into pAct RBP1 transfection constructs as described (15). For the two-hybrid experiments, constructs were made using the expression vectors pEG 202 and pJG4-5 (30) and are referred to as bait and prey, respectively. The rbp1, tra (31), and tra-2 (32, 33) bait constructs were generated by inserting cDNA fragments generated by PCR into the EcoRI site of pEG 202. PCR was performed using Pfu polymerase (Stratagene) and PCR products were sequenced. The following PCR primer pairs were used: RBP1, 5′-GGGAATTCCCGCGATATAGGGAGT-3′ and M13-20 primer (5′-GTAAAACGACGGCCAGT-3′); tra, 5′-GGGAATTCATGAAAATGGATGCCGACAG-3′ and 5′-TAGGATCCGAATGGAGTATGTTCAATATGG-3′; tra-2, 5′-GGGAATTCATGGATAGGGAGCCACTC-3′ and 5′-TAGAATTCTTAATAGCGCGATGAAGTTCG-3′. The bicoid bait construct pRFHM-1 is as described (30). The RBP1 prey constructs were generated by inserting cDNA fragments generated by PCR into the EcoRI site of pJG4-5. RBP1 PCR fragments were generated using the primer pair described for the RBP1 bait construct. All other plasmids are as described (15, 28).
Transfection and RNase Protection Experiments.
Transfections into SL2 cells and RNase protection experiments were done as described previously (15, 28). pAct expression constructs (100 ng) were transfected. RNase protections were done using the dsx wild-type (wt) riboprobe (28). Quantitation of RNase protection products was done by densitometry.
Two-Hybrid Experiments.
Two-hybrid experiments were done using a LexA/B42-system described in detail elsewhere (30). We used yeast strain EGY48 and the lacZ reporter construct pSH18-34. All bait constructs were assayed for lack of background activation of both the Leu and the lacZ reporter. Expression of bait fusion proteins was confirmed by the repression assay as described (30).
RESULTS
To define essential functional domains of RBP1 we began by testing the ability of RBP1 expression constructs carrying deletions and substitutions in the RRM domain, the Gly-rich region, and the SR domain to activate the female-specific dsx splicing pattern in tissue culture. For this purpose, RBP1 expression constructs were cotransfected together with a dsx minigene construct into Drosophila Schneider (SL2) cells as described recently (15). Overexpression of several RBP1 protein variants was confirmed by Western analysis (data not shown). dsx splicing products were analyzed by RNase protection assay (28) using a dsx riboprobe that detects the spliced female-specific exon, spliced common exon, spliced male-specific exon, and unspliced dsx pre-mRNA (see Figs. 2 and 3).
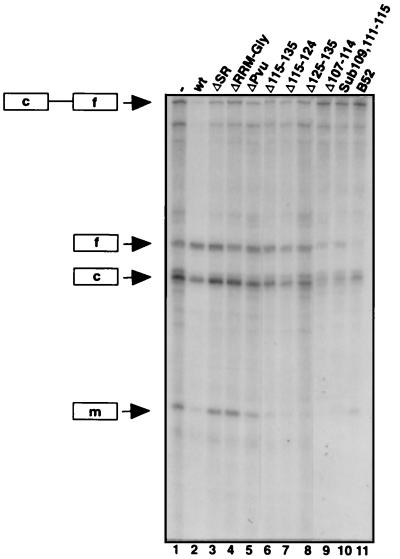
Figure 2.
Defining functional domains of RBP1 in splicing regulation by deletion analysis. The autoradiogram shows RNase protection assays probing dsx splicing in tissue culture in the presence of various RBP1 expression constructs. RNase protection products corresponding to (top to bottom) unspliced dsx pre-mRNA (c–f), spliced female-specific dsx exon (f), spliced common dsx exon (c), and spliced male-specific dsx exon (m) are indicated to the left of the autoradiogram. The RBP1 expression constructs (Fig. 1) transfected in each experiment are indicated above lanes 2–11. Lane 1 shows a control experiment lacking cotransfected RBP1 (—).
Figure 3.
(A) Defining a minimal functional SR domain. The RNase protection assay shows dsx splicing in the absence of cotransfected RBP1 (lane 1, —) and in the presence of wt RBP1 (lane 2) and ΔSR RBP1 (lane 3). Extended ΔSR constructs are tested in lanes 4–8. Constructs used are indicated above each lane and are described in more detail in Fig. 1. (B) Identifying RBP1 amino acid residues essential for dsx regulation in vivo. The RNase protection assay compares the regulatory activity of RBP1 constructs carrying amino acid substitutions in the Gly-rich region (lanes 3, 4, and 6) and in the RRM domain (lane 5) to the activity of a wt RBP1 construct (lane 2). RBP1 constructs (Fig. 1) are indicated above each lane. Lane 1 shows a control experiment lacking cotransfected RBP1.
The SR Domain Is Essential for RBP1 Function.
It was shown previously that, while translation of spliced RBP1 mRNA produces the full-length RBP1 protein, translation of unspliced RBP1 pre-mRNA gives rise to a shortened RBP1 protein lacking the C-terminal SR domain (2). To determine the significance of the SR domain and the shortened RBP1 expression variant for splicing regulation, we tested a deletion mutant ΔSR (Fig. 1) lacking the SR domain in the transfection assay. To assess the importance of the other two domains, a deletion mutant lacking the RRM domain and the Gly-rich region, ΔRRM-Gly (Fig. 1) was tested. As shown recently (15), transfection of a wt RBP1 construct results in an increase of female-specific splicing product and a decrease of male-specific splicing product (Fig. 2, lane 2) compared with the control experiment lacking a cotransfected RBP1 construct (Fig. 2, lane 1). In contrast, both deletion constructs ΔSR (Fig. 2, lane 3) and Δ RRM-Gly (Fig. 2, lane 4) are inactive in switching dsx splicing. A B52 (Drosophila SRp55) control construct did not activate female-specific dsx splicing (Fig. 2, lane 11), as shown previously (15). In vitro RNA binding experiments using wt RBP1 protein and ΔSR RBP1 protein expressed in Escherichia coli showed that deletion of the SR domain does not inhibit binding of RBP1 to RBP1 RNA target sequences (data not shown). These results suggest that the SR domain or parts of it are necessary, but not sufficient, for splicing regulation.
Figure 1.
Schematic drawings of RBP1 protein variants and summary of their activity in dsx splicing regulation. RBP1 domains are indicated by boxes and amino acid sequences are shown for the RNP motifs (RNP2 and RNP1) within the RRM, for the Gly-rich region (Gly), and for the SR domain. Amino acid positions within the SR domain are designated above each construct and amino acid substitutions are represented in italics. Construct designations are given to the left and activity of the constructs in switching dsx splicing from the male to the female pattern is indicated to the right of each construct. The dsx splicing assays were quantified by densitometry, and the intensity ratio of female to male RNase protection products (f/m) and standard errors are given at the right margin. For the negative control experiments in lanes 1 and 11 of Fig. 2, f/m ratios of 1.5 ± 0.2 and 0.75, respectively, were obtained.
A Minimal Functional RBP1 SR Domain.
Since no essential sequence elements within SR domains have been described to date, we investigated the effects of deleting various segments of the SR domain of RBP1 on dsx splicing regulation in vivo. For this purpose, the RBP1 SR domain extending from amino acid residues 107 to 135 was arbitrarily subdivided into three segments, segment 1 from residues 107–114, segment 2 from residues 115–124, and segment 3 from residues 125–135 (Fig. 1). Fig. 2 (lanes 6–9) shows that deletions of any single segment or deletion of segments 2 and 3 combined do not abolish activation of female-specific dsx splicing by RBP1. We noticed that several residues within segment 1 are especially conserved between RBP1 and its presumed mammalian homolog, SRp20 (X16) (13). However, no inhibition of RBP1 function was observed upon substitution of the conserved amino acids as in the construct Sub 109,111-115 (Fig. 2, lane 10). Taken together, the above experiments suggest that the SR domain of RBP1 is required for its activation of dsx female splicing and that there must be functional redundancy within the SR domain, since this set of contiguous deletions spanning the SR domain had no effect in this assay.
To try to define a minimal functional SR domain, we proceeded by adding back single amino acid residues to the nonfunctional construct ΔSR. As shown in Fig. 3A (lane 4), addition of a single serine residue to the ΔSR construct as in construct +1 (Fig. 1) restores the regulatory activity of RBP1 close to the activity of wt RBP1 protein, as determined by the ratio of female to male RNase protection products (Fig. 1). It should be noted that the Gly-rich region immediately adjacent to the SR domain also contains serine and arginine residues. To determine whether arginine residues within the Gly-rich region contribute to the activity of construct +1, we substituted six of the eight arginine residues within the Gly-rich region with lysines to create construct +1 R-K. Construct +1 R-K was found to be inactive (Fig. 3A, lane 8) in switching dsx splicing, suggesting that arginine residues within the Gly-rich region are part of the minimal functional SR domain. The larger constructs +2 and +3 (Fig. 1) show a regulatory activity (Fig. 3A, lanes 5 and 6) very similar to construct +1, as determined by the ratio of female to male protection products (Fig. 1). These experiments indicate a minimal functional SR domain in RBP1 with regard to regulation of dsx splicing. To determine whether the amino acid sequence at the C terminus of these minimal functional constructs is essential, construct +3 SRSR (Fig. 1) was generated. We found that construct +3 SRSR does activate female-specific dsx splicing (Fig. 3A, lane 7), suggesting that the exact sequence of serine and arginine residues is not crucial for function.
Testing the Requirement of the RRM Domain.
The RRM domain in other proteins has been shown to be an RNA binding domain. Since regulation of dsx splicing by RBP1 involves RBP1 RNA target sequences located in the proximity of the regulated female-specific 3′ splice site (15), we expected the RRM domain to be essential for RBP1 function. To test the requirement of the RRM domain, we introduced amino acid substitutions into the essential and conserved sequence motifs RNP2 and RNP1 within the RRM domain of the RBP1 expression construct. Since aromatic residues within the RNP motifs are known to crosslink to RNA (34), phenylalanines and a tyrosine in RNP2 and RNP1 were substituted with aspartic acid residues (Fig. 1). Analogous substitutions were shown previously to inhibit crosslinking of ASF/SF2 to RNA (35). Overexpression of the RRM mutant protein was confirmed by Western blot analysis (not shown). As shown in Fig. 3B (lane 5) the RRM mutant is nonfunctional in the dsx splicing assay, suggesting an essential role of the RRM domain in dsx regulation by RBP1.
Determining Essential Elements Within the Gly-Rich Region.
To determine whether elements within the Gly-rich region are required for dsx regulation, we generated the construct ΔPvu lacking parts of the Gly-rich region and the SR domain (Fig. 1). As shown in Fig. 2 lane 5, construct ΔPvu shows a reduced regulatory activity compared with the wt construct (Fig. 2, lane 2). Since a deletion of any segment within the SR domain did not abolish RBP1 function, this result would suggest that the Gly-rich region may contain essential functional elements.
The Gly-rich region, 32 aa long, is largely comprised of three amino acid residues: glycines (7 residues), serines (9 residues), and arginines (10 residues). To determine whether any one of these amino acid constituents are required for dsx regulation, we generated substitution mutants. To change the overall character of the protein as little as possible, glycines were substituted with alanines, serines with threonines, and arginines with lysines (Fig. 1). We found that the glycine substitution mutant is nonfunctional (Fig. 3B, lane 4) whereas the serine (Fig. 3B, lane 6) and arginine (Fig. 3B, lane 3) substitutions do not affect dsx activation by RBP1. We confirmed overexpression of the glycine mutant protein in the transfected cells by Western analysis. The glycine mutant protein was found to be overexpressed to the same level as other RBP1 protein variants (data not shown). Thus, our analysis using substitution mutants confirms an essential role of the Gly-rich region and indicates that the glycine residues within this domain are crucial for RBP1 function.
A Role of RBP1 Residues Essential for dsx Regulation in Interactions of RBP1 with TRA-2 and with Itself.
To determine whether the essential RBP1 domains identified in the tissue culture splicing assays are involved in protein–protein interactions between RBP1 and TRA/TRA-2 in vivo, we examined possible interactions between RBP1, TRA, and TRA-2 using a yeast two-hybrid system. To test for interactions, we transformed constructs expressing various RBP1 proteins each fused to a B42 transcription activation domain (referred to as “prey”) together with either wt rbp1–, tra–, tra-2–, or bicoid–LexA fusion constructs (referred to as “bait”) into yeast cells. The two-hybrid fusion proteins used in our study include independent nuclear localization signals encoded by the expression vectors to ensure entry of the proteins into the nucleus. Transformants were grown on galactose/5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) plates to assay for activation of a lacZ reporter gene due to interacting bait–prey fusion proteins. Assays were done in parallel on glucose/X-Gal plates (data not shown) to confirm dependence of the signals observed on expression of the galactose inducible prey fusion proteins.
As shown in Fig. 4, wt RBP1 can interact with wt RBP1 and with TRA-2 as indicated by the blue staining, supporting involvement of RBP1 in dsx splicing regulation in a complex with TRA-2. No interactions with TRA and BICOID were observed, confirming specificity of interaction.
Figure 4.
Mutations in RBP1 residues essential for dsx splicing regulation affect interaction of RBP1 with RBP1 and with TRA-2 in vivo. The photograph shows interactions in the yeast two-hybrid system between bait and prey fusion proteins as detected by activation of a lacZ reporter construct in colonies grown on galactose/X-Gal/CM–Ura, –His, and –Trp plates. Interaction assays were done in triplicate. RBP1, tra, tra-2, and bicoid bait constructs as indicated to the left were tested for interaction with wt RBP1, Gly G-A RBP1, Sub RNP RBP1, ΔPvu RBP1, ΔSR RBP1, and +1 RBP1 prey constructs as indicated on top. RBP1 variants are as described in Fig. 1. Some colonies transformed with both the wt RBP1 bait and prey constructs remained white (data not shown), probably because they fail to express the wt RBP1 bait protein.
Next we tested the ability of several mutant RBP1 prey constructs to interact with wt RBP1, TRA, TRA-2, and the BICOID control in the two-hybrid assay (Fig. 4). The various RBP1 prey proteins were found to be expressed at similar levels in the yeast cells as determined by Western analysis (data not shown). The RRM RNA binding mutation (Sub RNP) had little effect on the protein–protein interaction as compared with the wt RBP1 prey construct, resulting in a slightly weaker interaction signal. However, deletion of the RBP1 SR domain (ΔSR) results in a loss of interaction with TRA-2 and RBP1 consistent with a recent study reporting an involvement of SR domains in protein–protein interactions (23). Surprisingly, when testing the ΔPvu construct, which lacks a large part of the Gly-rich domain, we observed a stronger interaction signal by a factor of 2 with RBP1 and with TRA-2 as compared with the wt construct. Consistent with this observation, the RBP1 Gly → Ala mutant prey construct (Gly G-A) also showed a stronger signal by a factor of 1.6 compared with the wt RBP1 prey construct both in combination with the wt RBP1 and the TRA-2 bait constructs. The signal intensities were quantitated by scanning. No interaction was detected when testing the +1 prey construct. However, since overexpression of the +1 construct switches dsx splicing in the tissue culture assay, the +1 protein possibly participates in weaker interactions. In summary, our analysis suggests that the SR domain and the glycine residues within the Gly-rich domain participate in interaction of RBP1 with TRA-2 and with itself.
DISCUSSION
Here we present an analysis of the functional domains of an alternative splicing factor in vivo. We demonstrate functional requirements for the RRM domain, the Gly-rich region, and the SR domain of RBP1 in dsx splicing regulation and we have identified essential amino acid residues. Moreover, we present in this study evidence for an involvement of the SR domain and of the Gly residues within the Gly-rich domain in protein–protein interactions between RBP1 and TRA-2 and between RBP1 and itself.
We have demonstrated a requirement for the RNP motifs within the RRM type RNA binding domain for activation of female-specific dsx splicing by RBP1. This finding is consistent with earlier evidence showing that mutations within RBP1 RNA target sequences in the proximity of the female-specific 3′ splice site cause inhibition of activation of female-specific dsx splicing by RBP1 and inhibition of binding of RBP1 to dsx pre-mRNA (15). Taken together, these data strongly suggest that RBP1 participates in regulation of dsx splicing by binding to RBP1 RNA target sequences. In our analysis we have introduced mutations into all aromatic residues in both RNP motifs. Recently, in an in vitro study of SF2/ASF, more limited substitutions of only two aromatic residues within the RNP1 motif clearly reduced crosslinking of SF2/ASF to RNA, but only slightly reduced the splice site switching activity of SF2/ASF. However, a deletion mutant of SF2/ASF lacking the complete RRM domain lost its ability to switch the use of splice sites in vitro (35). Thus, both conserved RNP motifs in RBP1 appear to be essential for function, which is consistent with studies showing that Tyr-13 in the RNP2 motif of the U1-A protein is involved in RNA binding (36) and that aromatic residues in both RNP motifs of U1-A are in contact with RNA (37).
We also show that a RBP1 protein lacking the SR domain is inactive in regulation of dsx splicing. Since the ΔSR protein binds to RBP1 RNA target sequences efficiently in vitro (data not shown), the SR domain of RBP1 does not appear to be involved in RNA–protein interactions. SR domains have been implicated in protein–protein interactions between splicing factors (23) and also in localization of proteins to nuclear speckles (24). In the two-hybrid system, deletion of the RBP1 SR domain abolishes protein–protein interactions with TRA-2, which is known to be involved in dsx regulation in vivo (26–28) and in vitro (38), suggesting that the inhibitory effect observed in tissue culture reflects blockage of splicing regulation. A striking result of our investigation of SR residues is the apparent absence of any requirement for a defined sequence segment involving SR residues, suggesting a sequence independent functional redundancy. This functional redundancy is best demonstrated by our finding that arginines within the Gly-rich region are only essential for RBP1 function when they are part of a shortened minimal SR domain, whereas, in a full-length RBP1 protein, their presence is not crucial. It is interesting to notice that the shortened RBP1 protein variant generated by translation of unspliced RBP1 pre-mRNA in Drosophila, because it lacks an SR domain (2), is probably functionally different from full-length RBP1 protein in vivo. Similarly, expression variants of ASF/SF2 lacking SR residues were also found to be inactive in splicing regulation in vitro (39), indicating a basic functional similarity between Drosophila and human SR proteins in vitro and in vivo.
In this paper we report evidence for an essential role of the Gly-rich region shared by all known SR proteins and also found in other splicing factors like TRA-2 and U2AF. We show that a deletion extending into the Gly-rich region inhibits the regulatory function of RBP1. The glycine residues are this region’s unique feature, and substitutions of the Glys by alanines were found to abolish regulation of dsx splicing by RBP1. Our study points also to an interesting structural and functional difference between RBP1 and ASF/SF2. Whereas in RBP1 the Gly-rich region is located next to the SR domain (2), in ASF/SF2 it is found in the N-terminal half of the protein between the RRM domain and an additional RRM-like domain not present in RBP1 (4, 5). In vitro, deletion of the Gly-rich region in ASF/SF2 did not affect the ability of ASF/SF2 to switch splicing of simian virus 40 RNA (39). Thus, the Gly-rich regions of RBP1 and ASF/SF2 might also have different functions, or the function of the Gly-rich region is not detected in vitro. On the molecular level, we show that in RBP1, the Gly residues affect interactions of RBP1 with RBP1 and with TRA-2 in vivo in the two-hybrid assay. Both the deletion mutant ΔPvu and the Gly → Ala substitution mutant cause an increase in the interaction signal, suggesting that a specific contribution of the glycine residues and not a change in the overall protein structure caused by the mutations is being detected. Although the glycine mutant proteins were found to be expressed at levels similar to other RBP1 protein variants in the yeast two-hybrid system and in transfected Drosophila cells, a subtle effect of the glycine mutations on the expression or stability of RBP1 cannot be excluded. The glycine residues could be involved in direct interactions between RBP1 and other splicing factors, or they could be required to position the SR domain in a steric orientation, which is optimal for protein–protein interactions. It is conceivable that Gly-rich regions function as flexible hinges between protein domains. However, it should be noticed that for example U2AF35 (40) and hnRNP A1 (41) exhibit extensive glycine stretches at their C termini. More significantly, the SR proteins RBP1, SC35, hSRp20, and 9G8 contain Gly-rich regions overlapping with SR domains known to participate in protein–protein interactions, and in our study we have shown that arginine residues within the Gly-rich region are essential for RBP1 function if they are part of a minimal SR domain. These observations support a possible involvement of the glycine residues in protein–protein interactions. Based on the stronger interaction signal obtained with the RBP1 glycine mutants, it is tempting to suggest that in RBP1 the glycine residues may modulate the binding affinity between RBP1 and interacting proteins.
Our discovery of protein–protein interactions between RBP1 and itself and between RBP1 and TRA-2 provides new insights into the mechanism of dsx splicing regulation. In a previous study we observed that RBP1 RNA target sequences occur pairwise in the polypyrimidine tract of the female-specific 3′ splice site and next to the repeat motifs recognized by TRA/TRA-2 within the dsx repeat region (15). Dimerization of RBP1 as detected in the two-hybrid system is consistent with earlier indications for cooperative binding of RBP1 to multiple RBP1 RNA target sequences and with evidence for oligomerization of RBP1 in gel-shift experiments (15). In particular, the detection of a RBP1-RBP1 interaction in vivo supports the model that the pair of RBP1 RNA target sequences within the polypyrimidine tract of the female-specific 3′ splice site is recognized by a RBP1 dimer. Furthermore, interaction of RBP1 with TRA-2 suggests that RBP1 and TRA-2 may also recognize pairwise combinations of RBP1 RNA target sequences and dsx repeat motifs in a cooperative manner. Cooperative binding of RBP1 and TRA/TRA-2 to the repeat region was also suggested recently (29). At present, we can conclude that in Drosophila, among the factors regulating dsx splicing, RBP1, TRA-2, and TRA are in close contact with the dsx repeat region and, since TRA-2 and TRA have been shown to interact (42), they are likely to form a complex. These findings suggest that a TRA/TRA-2-dependent complex assembled on the dsx repeat region in a HeLa in vitro system (14) may have a functional counterpart in Drosophila. However, at least some of the proteins associating with the dsx repeat region in Drosophila are likely to be significantly different from those identified in the heterologous system (29). Interaction between RBP1 and TRA-2 may also imply that RBP1 and TRA-2 provide direct contact between the dsx repeat region recognized by RBP1, TRA and TRA-2 and the regulated female-specific 3′ splice site recognized by RBP1. However, contact between these regulatory elements may as well involve additional factors yet to be identified.
Acknowledgments
We thank Roger Brent’s lab for two-hybrid system ingredients and recipes, William Mattox for the tra-2 cDNA, Michael McKeown for the tra cDNA, Guennet Bohm for the preparation of media and laboratory supplies, and Lisa Ryner for comments on this manuscript. This work was supported by a grant from the National Institutes of Health to B.S.B.. V.H. was supported in part by a fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
Abbreviations: SR, Ser–Arg-rich; RRM, RNA recognition motif; RNP, ribonucleoprotein; wt, wild type.
References
- 1.Zahler A M, William L S, Stolk J A, Roth M B. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 2.Kim Y-J, Zuo P, Manley J L, Baker B S. Genes Dev. 1992;6:2569–2579. doi: 10.1101/gad.6.12b.2569. [DOI] [PubMed] [Google Scholar]
- 3.Ayane M, Preuss U, Köhler G, Nielsen P J. Nucleic Acids Res. 1991;19:1273–1278. doi: 10.1093/nar/19.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge H, Zuo P, Manley J L. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 5.Krainer A R, Mayeda A, Kozak D, Binns G. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 6.Fu X-D, Maniatis T. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 7.Vellard M, Sureau A, Soret J, Martinerie C, Perbal B. Proc Natl Acad Sci USA. 1992;89:2511–2515. doi: 10.1073/pnas.89.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond R H, Du K, Lee V M, Mohn K L, Haber B A, Tewari D S, Taub R. J Biol Chem. 1993;268:15185–15192. [PubMed] [Google Scholar]
- 9.Screaton G R, Cáceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell JI, Krainer A R. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champlin D T, Frasch M, Saumweber H, Lis J T. Genes Dev. 1991;5:1611–1621. doi: 10.1101/gad.5.9.1611. [DOI] [PubMed] [Google Scholar]
- 11.Roth M B, Zahler A M, Stolk J A. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahler A M, Neugebauer K M, Stolk J A, Roth M B. Mol Cell Biol. 1993;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavaloc Y, Popielarz M, Fuchs J-P, Gattoni R, Stévenin J. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian M, Maniatis T. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 15.Heinrichs V, Baker B S. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge H, Manley J L. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 17.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayeda A, Zahler A M, Krainer A R, Roth M B. Proc Natl Acad Sci USA. 1992;89:1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cáceres J F, Stamm S, Helfman D M, Krainer A R. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 20.Krainer A R, Conway G C, Kozak D. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 21.Wu J Y, Maniatis T. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 22.Baker B S. Nature (London) 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- 23.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Nature (London) 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Bingham P M. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 25.Tacke R, Manley J L. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedley M L, Maniatis T. Cell. 1991;65:579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- 27.Hoshijima K, Inoue K, Higuchi I, Sakamoto H, Shimura Y. Science. 1991;252:833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- 28.Ryner L C, Baker B S. Genes Dev. 1991;5:2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- 29.Lynch K W, Maniatis T. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 30.Golemis E A, Gyuris J, Brent R. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. New York: Wiley; 1994. pp. 13.14.1–13.14.14. [Google Scholar]
- 31.Boggs R T, Gregor P, Idriss S, Belote J M, McKeown M. Cell. 1987;50:739–747. doi: 10.1016/0092-8674(87)90332-1. [DOI] [PubMed] [Google Scholar]
- 32.Amrein H, Gorman M, Nöthiger R. Cell. 1988;55:1025–1035. doi: 10.1016/0092-8674(88)90247-4. [DOI] [PubMed] [Google Scholar]
- 33.Goralski T J, Edstrom J E, Baker B S. Cell. 1989;56:1011–1018. doi: 10.1016/0092-8674(89)90634-x. [DOI] [PubMed] [Google Scholar]
- 34.Merrill B M, Stone K L, Cobianchi F, Wilson S H, Williams K R. J Biol Chem. 1988;263:3307–3313. [PubMed] [Google Scholar]
- 35.Cáceres J F, Krainer A R. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jessen T-H, Oubridge C, Teo C H, Pritchard C, Nagai K. EMBO J. 1991;10:3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oubridge C, Ito N, Evans P R, Teo C-H, Nagai K. Nature (London) 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 38.Tian M, Maniatis T. Science. 1992;256:237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- 39.Zuo P, Manley J L. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, Zamore P D, Carmo-Fonseca M, Lamond A I, Green M R. Proc Natl Acad Sci USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cobianchi F, SenGupta D N, Zmudzka B Z, Wilson S H. J Biol Chem. 1986;261:3536–3543. [PubMed] [Google Scholar]
- 42.Inoue K, Hoshijima K, Higuchi I, Sakamoto H, Shimura Y. Proc Natl Acad Sci USA. 1992;89:8092–8096. doi: 10.1073/pnas.89.17.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]